You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004586_02109
You are here: Home > Sequence: MGYG000004586_02109
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
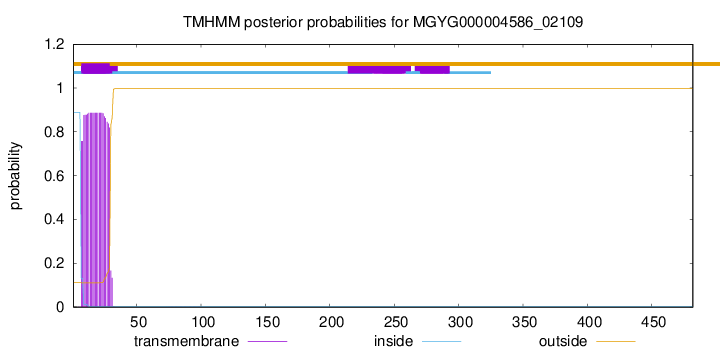
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; | |||||||||||
| CAZyme ID | MGYG000004586_02109 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 979; End: 2427 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 4.43e-22 | 45 | 406 | 10 | 317 | Glycosyl hydrolases family 8. |
| cd14256 | Dockerin_I | 1.57e-14 | 423 | 479 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| COG3405 | BcsZ | 2.96e-13 | 82 | 405 | 52 | 341 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam00404 | Dockerin_1 | 2.02e-09 | 424 | 479 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| PRK11097 | PRK11097 | 7.36e-09 | 45 | 285 | 30 | 236 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL17008.1 | 1.95e-166 | 1 | 456 | 1 | 448 |
| BCK01002.1 | 8.15e-131 | 36 | 415 | 116 | 500 |
| QNU66099.1 | 4.63e-119 | 37 | 415 | 520 | 905 |
| BBI36306.1 | 3.70e-111 | 29 | 415 | 68 | 439 |
| AZS18334.1 | 4.01e-109 | 29 | 421 | 1028 | 1419 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XD0_A | 1.41e-90 | 29 | 412 | 52 | 406 | ApoStructure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4],5XD0_B Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4] |
| 1V5C_A | 1.59e-62 | 38 | 415 | 30 | 384 | Thecrystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 [Bacillus sp. (in: Bacteria)],1V5D_A The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)],1V5D_B The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)] |
| 7CJU_A | 2.07e-62 | 38 | 415 | 36 | 390 | Crystalstructure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7CJU_B Crystal structure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7XGQ_A Chain A, chitosanase [Bacillus sp. K17-2],7XGQ_B Chain B, chitosanase [Bacillus sp. K17-2] |
| 1CEM_A | 7.72e-34 | 83 | 415 | 60 | 359 | ChainA, CELLULASE CELA (1,4-BETA-D-GLUCAN-GLUCANOHYDROLASE) [Acetivibrio thermocellus],1IS9_A Chain A, endoglucanase A [Acetivibrio thermocellus] |
| 1KWF_A | 5.27e-33 | 83 | 415 | 60 | 359 | ChainA, Endoglucanase A [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19254 | 2.75e-90 | 29 | 412 | 52 | 406 | Beta-glucanase OS=Niallia circulans OX=1397 GN=bgc PE=3 SV=1 |
| P29019 | 8.92e-66 | 29 | 436 | 77 | 461 | Endoglucanase OS=Bacillus sp. (strain KSM-330) OX=72575 PE=1 SV=1 |
| A3DC29 | 6.21e-39 | 83 | 449 | 92 | 441 | Endoglucanase A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celA PE=1 SV=1 |
| P37701 | 1.48e-32 | 31 | 449 | 55 | 430 | Endoglucanase 2 OS=Ruminiclostridium josui OX=1499 GN=celB PE=3 SV=1 |
| P37699 | 6.34e-31 | 39 | 449 | 63 | 430 | Endoglucanase C OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCC PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
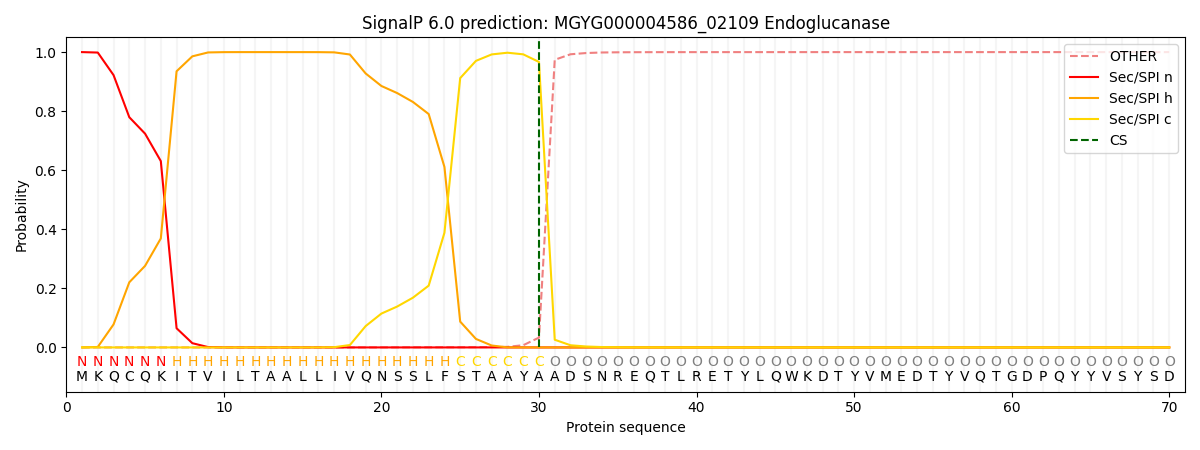
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000360 | 0.998794 | 0.000233 | 0.000226 | 0.000187 | 0.000166 |