You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004759_01449
You are here: Home > Sequence: MGYG000004759_01449
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; | |||||||||||
| CAZyme ID | MGYG000004759_01449 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2652; End: 4517 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 139 | 595 | 1.2e-51 | 0.6371191135734072 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1554 | ATH1 | 7.64e-07 | 170 | 527 | 177 | 548 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXE33659.1 | 6.41e-74 | 140 | 587 | 332 | 788 |
| QHW32096.1 | 6.33e-73 | 112 | 587 | 434 | 917 |
| AWS40658.1 | 3.24e-64 | 140 | 587 | 305 | 762 |
| BCB76148.1 | 2.55e-63 | 114 | 587 | 232 | 715 |
| QKW17790.1 | 2.90e-53 | 8 | 587 | 23 | 608 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EAB_A | 1.57e-06 | 310 | 463 | 381 | 546 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 2EAD_A | 1.57e-06 | 310 | 463 | 381 | 546 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2EAE_A | 1.57e-06 | 310 | 463 | 380 | 545 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 5.41e-07 | 299 | 600 | 388 | 688 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
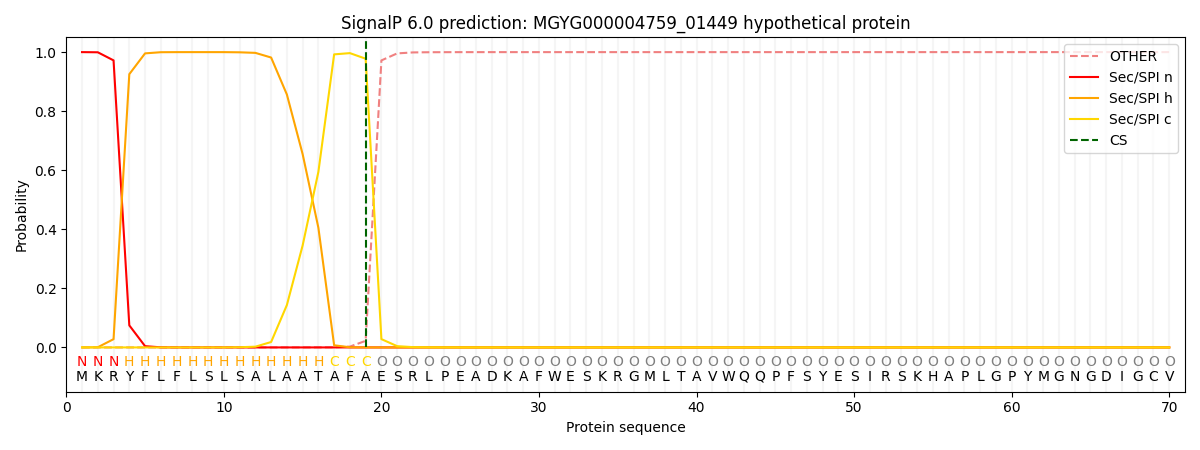
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000409 | 0.998748 | 0.000168 | 0.000240 | 0.000208 | 0.000179 |
