You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004899_03766
You are here: Home > Sequence: MGYG000004899_03766
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900555635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900555635 | |||||||||||
| CAZyme ID | MGYG000004899_03766 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18158; End: 19672 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 149 | 453 | 1.3e-95 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.61e-52 | 140 | 456 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.59e-23 | 126 | 454 | 47 | 361 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd14948 | BACON | 7.36e-14 | 29 | 107 | 5 | 83 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
| pfam13004 | BACON | 1.84e-07 | 53 | 107 | 4 | 61 | Putative binding domain, N-terminal. The BACON (Bacteroidetes-Associated Carbohydrate-binding Often N-terminal) domain is an all-beta domain found in diverse architectures, principally in combination with carbohydrate-active enzymes and proteases. These architectures suggest a carbohydrate-binding function which is also supported by the nature of BACON's few conserved amino-acids. The phyletic distribution of BACON and other data tentatively suggest that it may frequently function to bind mucin. Further work with the characterized structure of a member of glycoside hydrolase family 5 enzyme, Structure 3ZMR, has found no evidence for carbohydrate-binding for this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT35080.1 | 2.51e-279 | 1 | 504 | 12 | 516 |
| AVM57519.1 | 4.53e-277 | 1 | 504 | 58 | 562 |
| ADD61911.1 | 2.43e-269 | 24 | 504 | 3 | 484 |
| QRX62664.1 | 1.73e-250 | 9 | 504 | 21 | 512 |
| 1814455A | 1.03e-243 | 1 | 457 | 11 | 467 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQY_A | 6.20e-155 | 124 | 489 | 25 | 384 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 4YHE_A | 5.53e-131 | 115 | 504 | 1 | 385 | NativeBacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a],4YHE_B Native Bacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a] |
| 4YHG_A | 4.45e-130 | 115 | 504 | 1 | 385 | NativeBacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a],4YHG_B Native Bacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a] |
| 2JEP_A | 1.80e-83 | 123 | 486 | 35 | 393 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 3NDY_A | 3.93e-65 | 120 | 486 | 9 | 340 | Thestructure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_B The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_C The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_D The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDZ_A The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_B The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_C The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_D The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 1.82e-77 | 120 | 486 | 37 | 398 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P28623 | 8.15e-63 | 120 | 486 | 40 | 371 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| Q12647 | 3.67e-59 | 119 | 486 | 21 | 362 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P28621 | 1.26e-58 | 120 | 460 | 39 | 348 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P23660 | 1.77e-53 | 120 | 469 | 24 | 347 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
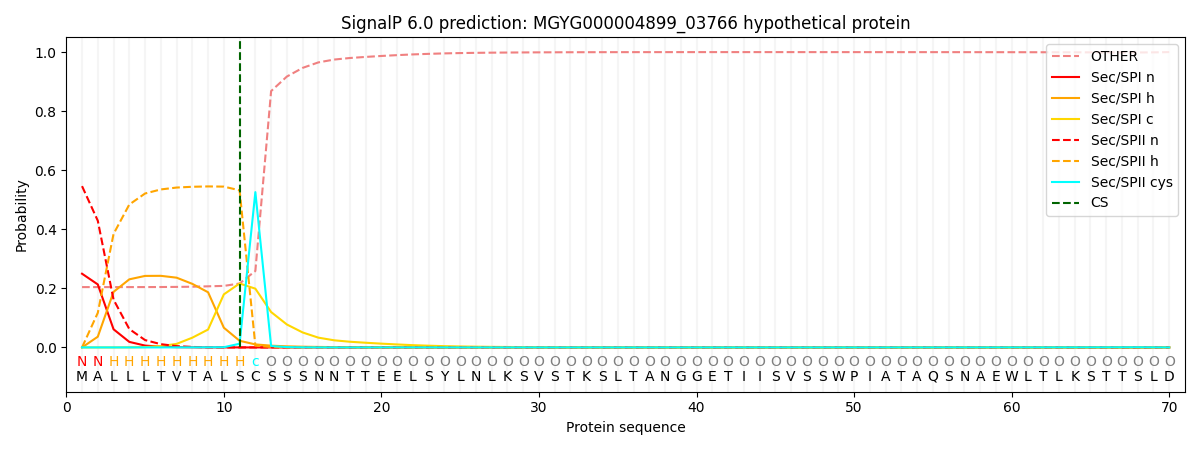
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.209404 | 0.241290 | 0.548791 | 0.000191 | 0.000149 | 0.000176 |
