

| Basic Information | |
|---|---|
| Species | Ricinus communis |
| Cazyme ID | 30026.m001490 |
| Family | CBM57 |
| Protein Properties | Length: 2046 Molecular Weight: 227813 Isoelectric Point: 6.9345 |
| Chromosome | Chromosome/Scaffold: 30026 Start: 399945 End: 421385 |
| Description | receptor-like kinase in flowers 1 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM57 | 1454 | 1620 | 1.4e-23 |
| VYINCGGSNEMVNRNTYEGDGEDSGGAARFYQHRNNWGFSSTGDFMGDSDDRNLASNIIISSSSNLSGLYRTARLSPLSLTYYRYCLVNGSYNITLYFAE ILFTNEKHYNSLGRRIFDIYIQDILVEKDFNIKTNASGVLTPTKITYAVNVSKSILEIRFYWAGKGT | |||
| CBM57 | 397 | 568 | 2e-26 |
| LYINCGGENMEVNGNTYEGDGDVGGGASTFYPSNNGWGFSSTGDFMDDDDFLNEAYIAESPSSLISNGLYRTARIAPLSLTYYRQCLIDGNYTVSLHFAE IQFTNDSTYNSLGRRLFNIYIQNDMVEQNFNIADEANGVATLIKKMYNATVTNNILEIRLSWAGKGTTRIPD | |||
| Full Sequence |
|---|
| Protein Sequence Length: 2046 Download |
| MFFKHAFVVL VLALSYLGTQ RFAAAVLPQD EVDALNLITR KMGASGWNFN ADACQDNVIP 60 IQPTDPERNI SCNCNFPNNT CHIVSLKIKR FSLPGELPPE LVQLSFLEHI DFAYNYLNGS 120 IPREWASIPL KFISVLANRL SGNIPTHLEN FTNLTSLDLE LNQFSGNVPR ELGKLVNLRI 180 LKLSSNKLSG NLPVELAELR NLTDFRINDN NFTGSIPDSI QNWRQLGRLE MQGSGLEGPI 240 PSSVSILEKL TDLRISDINV TNQAFPDLIN ITGLSRLILR NCKISGNIPS YIWTMSRLRV 300 LDLSFNNLHG ELPNAITTET NRLLYIFLNG NFLSGVIPFF SSGLNIDLSY NNFTRQDQPA 360 CRQTDIRLNL FRSSSMGNDI GGACAKSFDC DRHWHSLYIN CGGENMEVNG NTYEGDGDVG 420 GGASTFYPSN NGWGFSSTGD FMDDDDFLNE AYIAESPSSL ISNGLYRTAR IAPLSLTYYR 480 QCLIDGNYTV SLHFAEIQFT NDSTYNSLGR RLFNIYIQND MVEQNFNIAD EANGVATLIK 540 KMYNATVTNN ILEIRLSWAG KGTTRIPDSG VYGPLISAIS IDPHFKPPSG GGNTKIAPII 600 VGVLGSCLII LALGLIVWKR YFRAKNGRQK DFEGLEIQTV SFTLKQIKTA TNNFAPANKI 660 GEGGFGPVYK GLLADNTVIA VKQLSSKSNQ GNREFLNEIG VISCMQHPNL VKLHGCCIEG 720 NQLLLVYEYM ENNSLAHTLL GPEDRCLKLD WQTRQRICVG IAKGLAYLHE ESTLKIVHRD 780 IKATNVLLDK HLNPKISDFG LAKLDSEEKT HISTRVAGTI GYMAPEYALW GYLTYKADIY 840 SFGIVALEIV SGKHNMSRGP ESNFGCLLDW ACHLQQGGKL MELVDEKLGS EFKKVEAERM 900 IKVALLCTNG SASLRPIMSE VVSMLEGTKT IPDVIPEESS YNEDLRFKAI REHHKEIRSQ 960 SLRSQNHTST SSGWSRLDSS SATTHDLYEI KSELYANAKA KGDQDKQMDS QVSTSFHMTA 1020 SLIRMMLYLP KYFLISLLAL SCFGRQRIAA VTLPEDEGLF LSFFLSFFGL GFINNHMGSK 1080 VLVLCVCIVE ALNQITQKMG ASGWSFSGDD ACGSEVDQSI GGRRNITCNC QFLNNTCHIL 1140 LLEIKNFSLP GVLPPQLIQL PNLESIDFAY NYLTGSIPQE WTSMQLKFIS VLVNRLSGTI 1200 PTYLEDFTSL TYLNLEANQF SGLVPQELGK LVNLNSLILC SNNLSGNLPM QLAELKNLTD 1260 FRISDNNFNG SIPDFIGSWR QLQRLELQAS GLRGPIPSSI SLLENLTDLR ISDIKGATQA 1320 FPNLSSMTNL KRLVLRNCNI SGEIPPYIWG MNNLLTLDLS YNNLRGKPPN SIDNKHLLFL 1380 FLSHNLLNGD IPLFRKETDV DLSYNNFTRQ SPANPACQQT RGMNLNLYRS SSMRNDLGGA 1440 CMEDFICNEY WHSVYINCGG SNEMVNRNTY EGDGEDSGGA ARFYQHRNNW GFSSTGDFMG 1500 DSDDRNLASN IIISSSSNLS GLYRTARLSP LSLTYYRYCL VNGSYNITLY FAEILFTNEK 1560 HYNSLGRRIF DIYIQDILVE KDFNIKTNAS GVLTPTKITY AVNVSKSILE IRFYWAGKGT 1620 TRIPRIGMYG PLISAISAVN LNFRPRTGGR KTKVAPIVIG VIVSCLIFST LGVIWWRHHS 1680 KVKNKRHKDL EGLEIQIASF TLKQIKDATD NFDSSNKIGE GGFGPVYKGS LADGTGIAVK 1740 QLSSKSSQGN REFLNEIGMI SCLQHPNLVK LHGCCIEEDQ LLLVYEYMEN NSLARALFGA 1800 ADKQLKLDWQ TRHKICVGVA RGLAFLHEES SLRIVHRDIK GTNILLDKNL NPKISDFGLA 1860 KLDEKDKTHI STRIAGTIGY IAPEYALWGY LTYKADVYSF GIVALEIVSG RNNMNRGPES 1920 KFTCLLDWAC QLQKCGNLME LVDEKLGSEF NKAEAERMIK VALLCTNDTP SVRPTMSEVV 1980 GMLEGTRFVP DVIANESNNT EDLRFKIIRD HLKSMTSDSS GISEDHDSKS IVCRRGSSCV 2040 CTSTDL |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
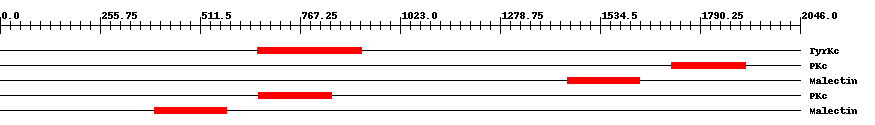 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| smart00219 | TyrKc | 3.0e-46 | 659 | 925 | 273 | + Tyrosine kinase, catalytic domain. Phosphotransferases. Tyrosine-specific kinase subfamily. | ||
| cd00180 | PKc | 7.0e-47 | 1718 | 1907 | 194 | + Catalytic domain of Protein Kinases. Protein Kinases (PKs), catalytic (c) domain. PKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine or tyrosine residues on protein substrates. The PK family is part of a larger superfamily that includes the catalytic domains of RIO kinases, aminoglycoside phosphotransferase, choline kinase, phosphoinositide 3-kinase (PI3K), and actin-fragmin kinase. PKs make up a large family of serine/threonine kinases, protein tyrosine kinases (PTKs), and dual-specificity PKs that phosphorylate both serine/threonine and tyrosine residues of target proteins. Majority of protein phosphorylation, about 95%, occurs on serine residues while only 1% occurs on tyrosine residues. Protein phosphorylation is a mechanism by which a wide variety of cellular proteins, such as enzymes and membrane channels, are reversibly regulated in response to certain stimuli. PKs often function as components of signal transduction pathways in which one kinase activates a second kinase, which in turn, may act on other kinases; this sequential action transmits a signal from the cell surface to target proteins, which results in cellular responses. The PK family is one of the largest known protein families with more than 100 homologous yeast enzymes and 550 human proteins. A fraction of PK family members are pseudokinases that lack crucial residues for catalytic activity. The mutiplicity of kinases allows for specific regulation according to substrate, tissue distribution, and cellular localization. PKs regulate many cellular processes including proliferation, division, differentiation, motility, survival, metabolism, cell-cycle progression, cytoskeletal rearrangement, immunity, and neuronal functions. Many kinases are implicated in the development of various human diseases including different types of cancer. | ||
| pfam11721 | Malectin | 6.0e-48 | 1452 | 1636 | 189 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| cd00180 | PKc | 5.0e-50 | 660 | 849 | 194 | + Catalytic domain of Protein Kinases. Protein Kinases (PKs), catalytic (c) domain. PKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine or tyrosine residues on protein substrates. The PK family is part of a larger superfamily that includes the catalytic domains of RIO kinases, aminoglycoside phosphotransferase, choline kinase, phosphoinositide 3-kinase (PI3K), and actin-fragmin kinase. PKs make up a large family of serine/threonine kinases, protein tyrosine kinases (PTKs), and dual-specificity PKs that phosphorylate both serine/threonine and tyrosine residues of target proteins. Majority of protein phosphorylation, about 95%, occurs on serine residues while only 1% occurs on tyrosine residues. Protein phosphorylation is a mechanism by which a wide variety of cellular proteins, such as enzymes and membrane channels, are reversibly regulated in response to certain stimuli. PKs often function as components of signal transduction pathways in which one kinase activates a second kinase, which in turn, may act on other kinases; this sequential action transmits a signal from the cell surface to target proteins, which results in cellular responses. The PK family is one of the largest known protein families with more than 100 homologous yeast enzymes and 550 human proteins. A fraction of PK family members are pseudokinases that lack crucial residues for catalytic activity. The mutiplicity of kinases allows for specific regulation according to substrate, tissue distribution, and cellular localization. PKs regulate many cellular processes including proliferation, division, differentiation, motility, survival, metabolism, cell-cycle progression, cytoskeletal rearrangement, immunity, and neuronal functions. Many kinases are implicated in the development of various human diseases including different types of cancer. | ||
| pfam11721 | Malectin | 4.0e-54 | 395 | 579 | 188 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004672 | protein kinase activity |
| GO:0005515 | protein binding |
| GO:0005524 | ATP binding |
| GO:0006468 | protein phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_002305716.1 | 0 | 32 | 1015 | 1 | 1008 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002305716.1 | 0 | 1089 | 2045 | 1 | 977 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002317385.1 | 0 | 1 | 1021 | 1 | 1019 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002317385.1 | 0 | 1025 | 2046 | 1 | 985 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002522276.1 | 0 | 1 | 2046 | 1 | 2046 | kinase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3ulz_A | 0 | 1696 | 1985 | 16 | 308 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 3ulz_A | 0 | 638 | 927 | 16 | 308 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 3uim_A | 0 | 1696 | 1985 | 16 | 308 | A Chain A, Structural Basis For The Impact Of Phosphorylation On Plant Receptor- Like Kinase Bak1 Activation |
| PDB | 3uim_A | 0 | 638 | 927 | 16 | 308 | A Chain A, Structural Basis For The Impact Of Phosphorylation On Plant Receptor- Like Kinase Bak1 Activation |
| PDB | 3tl8_H | 0 | 1696 | 1985 | 24 | 316 | B Chain B, The Avrptob-Bak1 Complex Reveals Two Structurally Similar Kinaseinteracting Domains In A Single Type Iii Effector |