

| Basic Information | |
|---|---|
| Species | Ricinus communis |
| Cazyme ID | 30190.m011310 |
| Family | GT35 |
| Protein Properties | Length: 949 Molecular Weight: 107415 Isoelectric Point: 6.4058 |
| Chromosome | Chromosome/Scaffold: 30190 Start: 109896 End: 118780 |
| Description | alpha-glucan phosphorylase 2 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT35 | 264 | 444 | 0 |
| ALSQLGFEFEVLQEQEGDAALGNGGLARLSACQMDSLATLDYPAWGYGLRYQYGLFRQVILDGYQHEQPDYWLNFGNPWEIERVHVTYPVKFYGTVEEEG FNGGKRQVWVPKETVEAVAYDNPIPGYGTRNTITLRLWSAKPSDQNDMESFNTGDYINAVVNRQRAETISSVLYPDDRSYQ | |||
| GT35 | 461 | 943 | 0 |
| LIVEVMRVLVDEEHISWNRSWDIVCKIFSFTTHTVSPAGLEKIPVDLLESLLPRHLQIIYDINFNFIEELKKRIGLDYDRLSRMSIVEEGAVKSIRMANL SIVCSHSVNGVSKVHAELLKTRVFKDFYELWPQKFDYKTNGVTQRRWIVVSNPSLCVLISKWLGTEAWIRDVDLLAGLQDYVTNADLHQEWNMVRKVNKT RLAEYIEAMSGIKVSVDAMFDVQIKRIHEYKRQLLNILGIIHRYDCIKNMEKNDRRNVVPRVCIIGGKAAPGYEMAKKIIKLCHAVAEKINNDPDVGDLM KLIFIPDYNVSVAELVIPGADLSQHISTSGHEASGTSSMKFLMNGCLLLATADGSTVEIIEEIGKDNVFLFGAKIHEVPVLREKGSALKVPLQFARVVRM VRNGYFGFEDYFKSLCDSVENGNDFYLLGCDFESYLEAQAAADKAYVDQEKWTRMSILSTAGSGRFSSDRTIEEYADRSWGIE | |||
| Full Sequence |
|---|
| Protein Sequence Length: 949 Download |
| MYPITLPLSH SKLSFPSIKP FSTPSNNLST ATSYVSFRSF ASTSESLIAS TTTTAVAVTT 60 IQTANLSDPD ATANATAFVI HARNRIGLLQ VITRVFKLLG LRIEKATVEL EGDHFAKTFY 120 VTDSHGNRIE DAESLDKIKK ALIDAIDGGD EANEVKKVGC TQRGVAVRRR LGSSSEGKAK 180 VDRMFGLMDR FLENDPSSLQ KDILDHALAH SVRDRLIERW HDTQIYFKRK DPKRIYFLSL 240 EYLMGRSLSN SVINLGVRDQ YADALSQLGF EFEVLQEQEG DAALGNGGLA RLSACQMDSL 300 ATLDYPAWGY GLRYQYGLFR QVILDGYQHE QPDYWLNFGN PWEIERVHVT YPVKFYGTVE 360 EEGFNGGKRQ VWVPKETVEA VAYDNPIPGY GTRNTITLRL WSAKPSDQND MESFNTGDYI 420 NAVVNRQRAE TISSVLYPDD RSYQAWLFLF YWSVTAYALN LIVEVMRVLV DEEHISWNRS 480 WDIVCKIFSF TTHTVSPAGL EKIPVDLLES LLPRHLQIIY DINFNFIEEL KKRIGLDYDR 540 LSRMSIVEEG AVKSIRMANL SIVCSHSVNG VSKVHAELLK TRVFKDFYEL WPQKFDYKTN 600 GVTQRRWIVV SNPSLCVLIS KWLGTEAWIR DVDLLAGLQD YVTNADLHQE WNMVRKVNKT 660 RLAEYIEAMS GIKVSVDAMF DVQIKRIHEY KRQLLNILGI IHRYDCIKNM EKNDRRNVVP 720 RVCIIGGKAA PGYEMAKKII KLCHAVAEKI NNDPDVGDLM KLIFIPDYNV SVAELVIPGA 780 DLSQHISTSG HEASGTSSMK FLMNGCLLLA TADGSTVEII EEIGKDNVFL FGAKIHEVPV 840 LREKGSALKV PLQFARVVRM VRNGYFGFED YFKSLCDSVE NGNDFYLLGC DFESYLEAQA 900 AADKAYVDQE KWTRMSILST AGSGRFSSDR TIEEYADRSW GIEPCRCPF |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd04300 | GT1_Glycogen_Phosphorylase | 0 | 206 | 942 | 781 | + This is a family of oligosaccharide phosphorylases. It includes yeast and mammalian glycogen phosphorylases, plant starch/glucan phosphorylase, as well as the maltodextrin phosphorylases of bacteria. The members of this family catalyze the breakdown of oligosaccharides into glucose-1-phosphate units. They are important allosteric enzymes in carbohydrate metabolism. The allosteric control mechanisms of yeast and mammalian members of this family are different from that of bacterial members. The members of this family belong to the GT-B structural superfamily of glycoslytransferases, which have characteristic N- and C-terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. | ||
| TIGR02093 | P_ylase | 0 | 207 | 942 | 776 | + glycogen/starch/alpha-glucan phosphorylases. This family consists of phosphorylases. Members use phosphate to break alpha 1,4 linkages between pairs of glucose residues at the end of long glucose polymers, releasing alpha-D-glucose 1-phosphate. The nomenclature convention is to preface the name according to the natural substrate, as in glycogen phosphorylase, starch phosphorylase, maltodextrin phosphorylase, etc. Name differences among these substrates reflect differences in patterns of branching with alpha 1,6 linkages. Members include allosterically regulated and unregulated forms. A related family, TIGR02094, contains examples known to act well on particularly small alpha 1,4 glucans, as may be found after import from exogenous sources [Energy metabolism, Biosynthesis and degradation of polysaccharides]. | ||
| pfam00343 | Phosphorylase | 0 | 264 | 944 | 720 | + Carbohydrate phosphorylase. The members of this family catalyze the formation of glucose 1-phosphate from one of the following polyglucoses; glycogen, starch, glucan or maltodextrin. | ||
| PRK14986 | PRK14986 | 0 | 195 | 946 | 794 | + glycogen phosphorylase; Provisional | ||
| COG0058 | GlgP | 0 | 207 | 944 | 774 | + Glucan phosphorylase [Carbohydrate transport and metabolism] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004645 | phosphorylase activity |
| GO:0005975 | carbohydrate metabolic process |
| GO:0008152 | metabolic process |
| GO:0016597 | amino acid binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI30609.1 | 0 | 188 | 948 | 1 | 813 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_001757919.1 | 0 | 188 | 945 | 1 | 810 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_002273615.1 | 0 | 184 | 948 | 1 | 817 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002305114.1 | 0 | 184 | 949 | 1 | 818 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002509431.1 | 0 | 1 | 949 | 1 | 949 | glycogen phosphorylase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1ygp_B | 0 | 180 | 945 | 37 | 878 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 1ygp_A | 0 | 180 | 945 | 37 | 878 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 4el5_A | 0 | 207 | 946 | 43 | 820 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 4el0_A | 0 | 207 | 946 | 43 | 820 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 4eky_A | 0 | 207 | 946 | 43 | 820 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
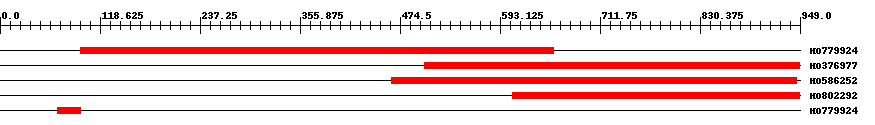 | ||||
| Hit | Length | Start | End | EValue |
| HO779924 | 618 | 96 | 656 | 0 |
| HO376977 | 447 | 504 | 949 | 0 |
| HO586252 | 484 | 464 | 945 | 0 |
| HO802292 | 347 | 608 | 949 | 0 |
| HO779924 | 28 | 68 | 95 | 0.98 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |