

| Basic Information | |
|---|---|
| Species | Arabidopsis thaliana |
| Cazyme ID | AT1G10760.1 |
| Family | CBM45 |
| Protein Properties | Length: 1400 Molecular Weight: 156582 Isoelectric Point: 5.6937 |
| Chromosome | Chromosome/Scaffold: 1 Start: 3581034 End: 3590928 |
| Description | alpha-glucan water dikinase, chloroplast precursor, putative, expressed |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM45 | 394 | 472 | 8e-21 |
| LHWALSQKGGEWLDPPSDILPPNSLPVRGAVDTKLTITSTDLPSPVQTFELEIEGDSYKGMPFVLNAGERWIKNNDSDF | |||
| CBM45 | 120 | 195 | 8.4e-24 |
| LHWGAILDNKENWVLPSRSPDRTQNFKNSALRTPFVKSGGNSHLKLEIDDPAIHAIEFLIFDESRNKWYKNNGQNF | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1400 Download |
| MSNSVVHNLL NRGLIRPLNF EHQNKLNSSV YQTSTANPAL GKIGRSKLYG KGLKQAGRSL 60 VTETGGRPLS FVPRAVLAMD PQAAEKFSLD GNIDLLVEVT STTVREVNIQ IAYTSDTLFL 120 HWGAILDNKE NWVLPSRSPD RTQNFKNSAL RTPFVKSGGN SHLKLEIDDP AIHAIEFLIF 180 DESRNKWYKN NGQNFHINLP TERNVKQNVS VPEDLVQIQA YLRWERKGKQ MYNPEKEKEE 240 YEAARTELRE EMMRGASVED LRAKLLKKDN SNESPKSNGT SSSGREEKKK VSKQPERKKN 300 YNTDKIQRKG RDLTKLIYKH VADFVEPESK SSSEPRSLTT LEIYAKAKEE QETTPVFSKK 360 TFKLEGSAIL VFVTKLSGKT KIHVATDFKE PVTLHWALSQ KGGEWLDPPS DILPPNSLPV 420 RGAVDTKLTI TSTDLPSPVQ TFELEIEGDS YKGMPFVLNA GERWIKNNDS DFYVDFAKEE 480 KHVQKDYGDG KGTAKHLLDK IADLESEAQK SFMHRFNIAA DLVDEAKSAG QLGFAGILVW 540 MRFMATRQLV WNKNYNVKPR EISKAQDRLT DLLQDVYASY PEYRELLRMI MSTVGRGGEG 600 DVGQRIRDEI LVIQRKNDCK GGIMEEWHQK LHNNTSPDDV VICQALMDYI KSDFDLSVYW 660 KTLNDNGITK ERLLSYDRAI HSEPNFRGEQ KDGLLRDLGH YMRTLKAVHS GADLESAIQN 720 CMGYQDDGEG FMVGVQINPV SGLPSGYPDL LRFVLEHVEE KNVEPLLEGL LEARQELRPL 780 LLKSHDRLKD LLFLDLALDS TVRTAIERGY EQLNDAGPEK IMYFISLVLE NLALSSDDNE 840 DLIYCLKGWQ FALDMCKSKK DHWALYAKSV LDRSRLALAS KAERYLEILQ PSAEYLGSCL 900 GVDQSAVSIF TEEIIRAGSA AALSSLVNRL DPVLRKTANL GSWQVISPVE VVGYVIVVDE 960 LLTVQNKTYD RPTIIVANRV RGEEEIPDGA VAVLTPDMPD VLSHVSVRAR NGKICFATCF 1020 DSGILSDLQG KDGKLLSLQP TSADVVYKEV NDSELSSPSS DNLEDAPPSI SLVKKQFAGR 1080 YAISSEEFTS DLVGAKSRNI GYLKGKVPSW VGIPTSVALP FGVFEKVISE KANQAVNDKL 1140 LVLKKTLDEG DQGALKEIRQ TLLGLVAPPE LVEELKSTMK SSDMPWPGDE GEQRWEQAWA 1200 AIKKVWASKW NERAYFSTRK VKLDHDYLCM AVLVQEVINA DYAFVIHTTN PSSGDSSEIY 1260 AEVVKGLGET LVGAYPGRSL SFICKKNNLD SPLVLGYPSK PIGLFIRRSI IFRSDSNGED 1320 LEGYAGAGLY DSVPMDEEDQ VVLDYTTDPL ITDLSFQKKV LSDIARAGDA IEKLYGTAQD 1380 IEGVIRDGKL YVVQTRPQV* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
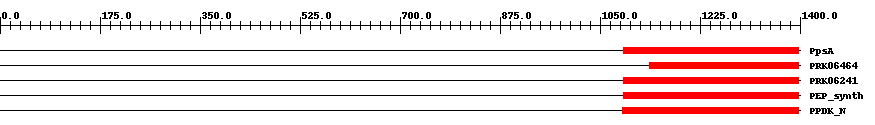 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG0574 | PpsA | 4.0e-11 | 1091 | 1397 | 339 | + Phosphoenolpyruvate synthase/pyruvate phosphate dikinase [Carbohydrate transport and metabolism] | ||
| PRK06464 | PRK06464 | 1.0e-15 | 1136 | 1397 | 311 | + phosphoenolpyruvate synthase; Validated | ||
| PRK06241 | PRK06241 | 7.0e-17 | 1091 | 1397 | 334 | + phosphoenolpyruvate synthase; Validated | ||
| TIGR01418 | PEP_synth | 1.0e-21 | 1091 | 1398 | 342 | + phosphoenolpyruvate synthase. Also called pyruvate,water dikinase and PEP synthase. The member from Methanococcus jannaschii contains a large intein. This enzyme generates phosphoenolpyruvate (PEP) from pyruvate, hydrolyzing ATP to AMP and releasing inorganic phosphate in the process. The enzyme shows extensive homology to other enzymes that use PEP as substrate or product. This enzyme may provide PEP for gluconeogenesis, for PTS-type carbohydrate transport systems, or for other processes [Energy metabolism, Glycolysis/gluconeogenesis]. | ||
| pfam01326 | PPDK_N | 3.0e-35 | 1089 | 1398 | 335 | + Pyruvate phosphate dikinase, PEP/pyruvate binding domain. This enzyme catalyzes the reversible conversion of ATP to AMP, pyrophosphate and phosphoenolpyruvate (PEP). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005515 | protein binding |
| GO:0005524 | ATP binding |
| GO:0005739 | mitochondrion |
| GO:0005983 | starch catabolic process |
| GO:0007623 | circadian rhythm |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAD31337.1 | 0 | 1 | 1399 | 1 | 1358 | AC007354_10 Strong similarity to gb |
| GenBank | AAF17665.1 | 0 | 46 | 1399 | 160 | 1540 | AC009398_14 F20B24.19 [Arabidopsis thaliana] |
| RefSeq | NP_563877.1 | 0 | 1 | 1399 | 1 | 1399 | SEX1 (STARCH EXCESS 1); alpha-glucan, water dikinase [Arabidopsis thaliana] |
| RefSeq | XP_002270485.1 | 0 | 1 | 1399 | 1 | 1470 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002527902.1 | 0 | 1 | 1399 | 1 | 1469 | alpha-glucan water dikinase, chloroplast precursor, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2ols_A | 0.0003 | 1093 | 1398 | 20 | 336 | A Chain A, The Crystal Structure Of The Phosphoenolpyruvate Synthase From Neisseria Meningitidis |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| HO777523 | 898 | 505 | 1400 | 0 |
| HO794398 | 752 | 651 | 1400 | 0 |
| HO794398 | 142 | 510 | 651 | 0 |
| EX118207 | 304 | 985 | 1287 | 0 |
| HO644248 | 444 | 960 | 1400 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |