

| Basic Information | |
|---|---|
| Species | Arabidopsis thaliana |
| Cazyme ID | AT4G19970.1 |
| Family | GT77 |
| Protein Properties | Length: 716 Molecular Weight: 82076.1 Isoelectric Point: 7.9243 |
| Chromosome | Chromosome/Scaffold: 4 Start: 10818242 End: 10825381 |
| Description | regulatory protein, putative, expressed |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT77 | 149 | 346 | 0 |
| LKHVIAVCLDIKAYDQCLKVHPHCYLINATDSDQLSGPNRFMTPGYLKLIWRRMDLLRQVIGLGYNFIFTDADILWLRDPFPRFFPDADFQITCDDYNGR PSDKKNHVNSGFTYVKANNKTSKFYKYWIRSSRKFPGKHDQDVFNFIKNDLHVEKLGIKMRFFDTVYFGGFCQPSRDINVVNTMHANCCIGLDNKVNN | |||
| GT77 | 489 | 686 | 0 |
| LQHVVVVCLDSKAFARCSQLHPNCYYLKTTGTDFSGEKLFATPDYLKMMWRRIELLTQVLEMGYNFIFTDADIMWLRDPFPRLYPDGDFQMACDRFFGDP HDSDNWVNGGFTYVKSNHRSIEFYKFWYNSRLDYPKMHDQDVFNQIKHKALVSEIGIQMRFFDTVYFGGFCQTSRDINLVCTMHANCCVGLAKKLHDL | |||
| Full Sequence |
|---|
| Protein Sequence Length: 716 Download |
| MKSTNDESPS LGYDFAGVSI KDGKSKPPAP VICYDGLFGG RDVIRIVLLV TTVTLSCLLF 60 YKSANNPLNM VFSPWKTDCY ASKLTNESSS KTEPKKEPVS ELERVLMNAA MEDNTVIITA 120 LNQAWAEPNS TFDVFRESFK VGIETERLLK HVIAVCLDIK AYDQCLKVHP HCYLINATDS 180 DQLSGPNRFM TPGYLKLIWR RMDLLRQVIG LGYNFIFTDA DILWLRDPFP RFFPDADFQI 240 TCDDYNGRPS DKKNHVNSGF TYVKANNKTS KFYKYWIRSS RKFPGKHDQD VFNFIKNDLH 300 VEKLGIKMRF FDTVYFGGFC QPSRDINVVN TMHANCCIGL DNKVNNLKAA LEDWKRYVSL 360 NTTVSETKWN IPPSFLDYGS AIGQKEVKKI LVLVLGLAAC LLLYKTAYPL HQELDVNNLS 420 SRPLLDHTSS SSPLTRSKSI SFREVLENAS TENRTVIVTT LNQAWAEPNS LFDLFLESFR 480 IGQGTKKLLQ HVVVVCLDSK AFARCSQLHP NCYYLKTTGT DFSGEKLFAT PDYLKMMWRR 540 IELLTQVLEM GYNFIFTDAD IMWLRDPFPR LYPDGDFQMA CDRFFGDPHD SDNWVNGGFT 600 YVKSNHRSIE FYKFWYNSRL DYPKMHDQDV FNQIKHKALV SEIGIQMRFF DTVYFGGFCQ 660 TSRDINLVCT MHANCCVGLA KKLHDLNLVL DDWRNYLSLS EPVKNTTWSV PMKCT* 720 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam03407 | Nucleotid_trans | 9.0e-62 | 148 | 347 | 208 | + Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. | ||
| pfam03407 | Nucleotid_trans | 7.0e-77 | 487 | 686 | 209 | + Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003674 | molecular_function |
| GO:0005575 | cellular_component |
| GO:0008150 | biological_process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAM61239.1 | 0 | 33 | 377 | 18 | 366 | regulatory protein [Arabidopsis thaliana] |
| GenBank | AAM61239.1 | 0 | 376 | 714 | 19 | 364 | regulatory protein [Arabidopsis thaliana] |
| RefSeq | NP_193730.1 | 0 | 1 | 715 | 1 | 715 | unknown protein [Arabidopsis thaliana] |
| RefSeq | NP_199295.2 | 0 | 37 | 377 | 24 | 366 | unknown protein [Arabidopsis thaliana] |
| RefSeq | NP_199295.2 | 0 | 375 | 714 | 18 | 364 | unknown protein [Arabidopsis thaliana] |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
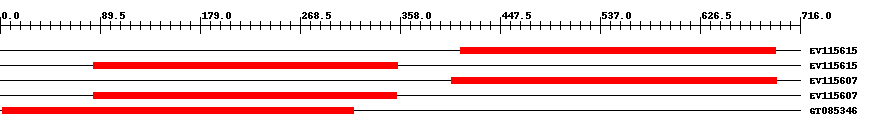 | ||||
| Hit | Length | Start | End | EValue |
| EV115615 | 284 | 412 | 694 | 0 |
| EV115615 | 273 | 84 | 356 | 0 |
| EV115607 | 293 | 404 | 695 | 0 |
| EV115607 | 272 | 84 | 355 | 0 |
| GT085346 | 315 | 2 | 316 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |