

| Basic Information | |
|---|---|
| Species | Aquilegia coerulea |
| Cazyme ID | Aquca_027_00219.2 |
| Family | PL1 |
| Protein Properties | Length: 636 Molecular Weight: 71569.5 Isoelectric Point: 8.181 |
| Chromosome | Chromosome/Scaffold: 27 Start: 1972294 End: 1977656 |
| Description | Pectin lyase-like superfamily protein |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| PL1 | 123 | 302 | 0 |
| IVFEVSGTIHLSSYLNVSSYKTIDGRGQRIKFMGKGLRLKECEHVIICNLEFESGRGHDVDGIQIKPNSKHIWIDRCSLRDYDDGLIDITRESTDITISR CHFSGHDKTMLIGADPTHIGDRCIRVTIHHCFFDGTRQRHPRVRYGKVHLYNNYTRDWGIYAVCASVEAQIYSQCNIYEA | |||
| PL1 | 467 | 633 | 0 |
| SGTIPLSSYLNVSSYKTIDGRGQRIKLTGKGLRLKECEHVIICNLEFEVGRGPDVDAIQIKPNSKHIWIDRCSLRDYEDGLIDITRESTDITVSRCHFSG HNKTMLIGGDPSHIGDRCIRVTIHHCFFDGTRQRHPRVRYAKVHLYNNYTRDWGIYAVCASVEAQVQ | |||
| Full Sequence |
|---|
| Protein Sequence Length: 636 Download |
| MGTVHGRYTT DKRNSRKHNN NNPSSSVSNQ QQQHHHLHNH NNNNKPTAFQ NNPPNTNTNT 60 KKMCSLPYSH VDSCLRGLAA QAEGFGRLAI GGLHGPIYHV TTLADDGPGS LRDGCRRKEP 120 LWIVFEVSGT IHLSSYLNVS SYKTIDGRGQ RIKFMGKGLR LKECEHVIIC NLEFESGRGH 180 DVDGIQIKPN SKHIWIDRCS LRDYDDGLID ITRESTDITI SRCHFSGHDK TMLIGADPTH 240 IGDRCIRVTI HHCFFDGTRQ RHPRVRYGKV HLYNNYTRDW GIYAVCASVE AQIYSQCNIY 300 EAGQKKGTFK YLPEKAADKE ETSSGWVISE GDIYLNGAQA CLPKEAINGC LFHPSEFYPT 360 WTLESPSESL KEVLQHCTGW QSVPRPTDQI VGNHHSNHNN NMSPVPYAHV DSNLRALAGQ 420 AEGFGRFAIG GLHGPLYHVT TLSDDGPGSL RYGCRLKEPL WIVFDISGTI PLSSYLNVSS 480 YKTIDGRGQR IKLTGKGLRL KECEHVIICN LEFEVGRGPD VDAIQIKPNS KHIWIDRCSL 540 RDYEDGLIDI TRESTDITVS RCHFSGHNKT MLIGGDPSHI GDRCIRVTIH HCFFDGTRQR 600 HPRVRYAKVH LYNNYTRDWG IYAVCASVEA QVQFF* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG3866 | PelB | 3.0e-37 | 430 | 624 | 212 | + Pectate lyase [Carbohydrate transport and metabolism] | ||
| pfam00544 | Pec_lyase_C | 3.0e-37 | 118 | 298 | 201 | + Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. | ||
| COG3866 | PelB | 2.0e-43 | 91 | 367 | 296 | + Pectate lyase [Carbohydrate transport and metabolism] | ||
| smart00656 | Amb_all | 8.0e-58 | 476 | 632 | 169 | + Amb_all domain. | ||
| smart00656 | Amb_all | 1.0e-63 | 137 | 303 | 179 | + Amb_all domain. | ||
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABK93648.1 | 0 | 46 | 389 | 37 | 381 | unknown [Populus trichocarpa] |
| GenBank | ABK93648.1 | 0 | 405 | 632 | 58 | 285 | unknown [Populus trichocarpa] |
| GenBank | ABK96132.1 | 0 | 54 | 388 | 48 | 383 | unknown [Populus trichocarpa] |
| GenBank | ABK96132.1 | 0 | 353 | 632 | 9 | 288 | unknown [Populus trichocarpa] |
| RefSeq | XP_002277576.1 | 0 | 47 | 389 | 31 | 369 | PREDICTED: hypothetical protein [Vitis vinifera] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1pxz_B | 9.99995e-41 | 71 | 338 | 4 | 307 | A Chain A, 1.7 Angstrom Crystal Structure Of Jun A 1, The Major Allergen From Cedar Pollen |
| PDB | 1pxz_B | 6e-37 | 411 | 627 | 15 | 256 | A Chain A, 1.7 Angstrom Crystal Structure Of Jun A 1, The Major Allergen From Cedar Pollen |
| PDB | 1pxz_A | 9.99995e-41 | 71 | 338 | 4 | 307 | A Chain A, 1.7 Angstrom Crystal Structure Of Jun A 1, The Major Allergen From Cedar Pollen |
| PDB | 1pxz_A | 6e-37 | 411 | 627 | 15 | 256 | A Chain A, 1.7 Angstrom Crystal Structure Of Jun A 1, The Major Allergen From Cedar Pollen |
| PDB | 3zsc_A | 4e-24 | 90 | 379 | 22 | 327 | A Chain A, Catalytic Function And Substrate Recognition Of The Pectate Lyase From Thermotoga Maritima |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
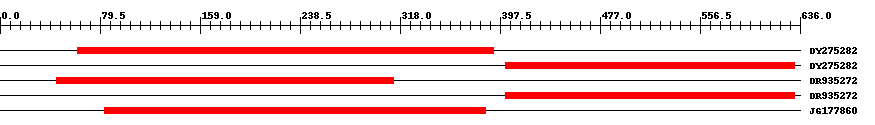 | ||||
| Hit | Length | Start | End | EValue |
| DY275282 | 332 | 62 | 392 | 0 |
| DY275282 | 231 | 402 | 632 | 0 |
| DR935272 | 269 | 45 | 313 | 0 |
| DR935272 | 231 | 402 | 632 | 0 |
| JG177860 | 304 | 83 | 386 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |