

| Basic Information | |
|---|---|
| Species | Zea mays |
| Cazyme ID | GRMZM2G025302_T01 |
| Family | GT43 |
| Protein Properties | Length: 913 Molecular Weight: 102688 Isoelectric Point: 8.7822 |
| Chromosome | Chromosome/Scaffold: 1 Start: 194522655 End: 194533077 |
| Description | ethylene induced calmodulin binding protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT43 | 216 | 310 | 1.2e-36 |
| LAHVLKAVQAPLLWLVVEWPEQSYETAEILRSSGVMYRHLICRKNTTSVRKIAVCQRNNAIYHVKRHHLDGIMHFADEERSYSADVFEEMQKIRH | |||
| Full Sequence |
|---|
| Protein Sequence Length: 913 Download |
| MVSSRRNSGI ILREGSVRDW SEFNNPSPSP KLLYSQSYVA MRGLLASVIS LDFFLLSSKL 60 KSVCAAMTSQ RHSRSQERSK SKGLTCRRVA VHLLFFFMVG IFIGFMPLFS VDVYKKIVSE 120 NERLPFHDGV IEVEMMGTKV KELETVVVEK EVELIDEPQV QESPPVPAML DDEADFAESS 180 PALPAIEESD IPVKKLLIIV TITSVRPQQA YYLNRLAHVL KAVQAPLLWL VVEWPEQSYE 240 TAEILRSSGV MYRHLICRKN TTSVRKIAVC QRNNAIYHVK RHHLDGIMHF ADEERSYSAD 300 VFEEMQKIRH KNTSRSRRTP NAGFSFTSPI IFFPSTLIHA RSSVIMVFAA GGTSRFVGGG 360 EPREISRVLP VLRDHDVHAD AHLYNALMKA NVAADDPGAV LRVFRQVSVT GTFLVNKEHV 420 ESHRWSCMFG DVEVPAEVLT DGTLRCYAPA HQSGRVPFYV TCSNMVACSE VREFEYRDSE 480 AHYMETSRSQ ANGVNEMHLH IRLEKLLTLG PDDHQMLAIN SLMLDGKWSN QESSVKEVVS 540 TARVQSLKKL VKEKLHQWLI CKHWGYDWAI RPIIIAGVNV NFRDAHGWTT LHWVASLGRE 600 RTVSVLIANG HKIPMRQWRT ATGAQRHRAS CEPMVEHLQT RTTGYGSSNP LFGGASSNKN 660 NRIFFFLHAI PGMLLALVLF FDNRKFKDVN FPPAMSPPKR WDHVCCTRCW DLVLVSSTAL 720 LYLVMSMSTG LANMSLKHNS ILTGLLSFMM DDALTTGSIK TSDAEKRRLA KASLAYNCES 780 KNCPHFRKLF PEYVEKYNQQ LQLENTAAEP VPQENPAPAP SPAAQQAPTV ANRAQPLAEA 840 RRDKNQKKAV PFWMVLSAKT NLNVEQVFFS IARDIKQRLS ETDSKPEDRT IKIKAEGEAD 900 AAEAQKSACC DS* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
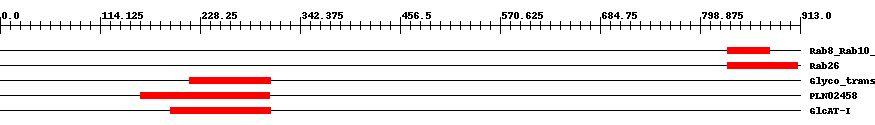 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd01867 | Rab8_Rab10_Rab13_like | 0.0002 | 830 | 878 | 56 | + Rab GTPase families 8, 10, 13 (Rab8, Rab10, Rab13). Rab8/Sec4/Ypt2 are known or suspected to be involved in post-Golgi transport to the plasma membrane. It is likely that these Rabs have functions that are specific to the mammalian lineage and have no orthologs in plants. Rab8 modulates polarized membrane transport through reorganization of actin and microtubules, induces the formation of new surface extensions, and has an important role in directed membrane transport to cell surfaces. The Ypt2 gene of the fission yeast Schizosaccharomyces pombe encodes a member of the Ypt/Rab family of small GTP-binding proteins, related in sequence to Sec4p of Saccharomyces cerevisiae but closer to mammalian Rab8. GTPase activating proteins (GAPs) interact with GTP-bound Rab and accelerate the hydrolysis of GTP to GDP. Guanine nucleotide exchange factors (GEFs) interact with GDP-bound Rabs to promote the formation of the GTP-bound state. Rabs are further regulated by guanine nucleotide dissociation inhibitors (GDIs), which facilitate Rab recycling by masking C-terminal lipid binding and promoting cytosolic localization. Most Rab GTPases contain a lipid modification site at the C-terminus, with sequence motifs CC, CXC, or CCX. Lipid binding is essential for membrane attachment, a key feature of most Rab proteins. Due to the presence of truncated sequences in this CD, the lipid modification site is not available for annotation. | ||
| cd04112 | Rab26 | 8.0e-5 | 830 | 910 | 87 | + Rab GTPase family 26 (Rab26). Rab26 subfamily. First identified in rat pancreatic acinar cells, Rab26 is believed to play a role in recruiting mature granules to the plasma membrane upon beta-adrenergic stimulation. Rab26 belongs to the Rab functional group III, which are considered key regulators of intracellular vesicle transport during exocytosis. GTPase activating proteins (GAPs) interact with GTP-bound Rab and accelerate the hydrolysis of GTP to GDP. Guanine nucleotide exchange factors (GEFs) interact with GDP-bound Rabs to promote the formation of the GTP-bound state. Rabs are further regulated by guanine nucleotide dissociation inhibitors (GDIs), which facilitate Rab recycling by masking C-terminal lipid binding and promoting cytosolic localization. Most Rab GTPases contain a lipid modification site at the C-terminus, with sequence motifs CC, CXC, or CCX. Lipid binding is essential for membrane attachment, a key feature of most Rab proteins. | ||
| pfam03360 | Glyco_transf_43 | 3.0e-17 | 216 | 309 | 103 | + Glycosyltransferase family 43. | ||
| PLN02458 | PLN02458 | 2.0e-25 | 160 | 308 | 153 | + transferase, transferring glycosyl groups | ||
| cd00218 | GlcAT-I | 5.0e-43 | 195 | 309 | 121 | + Beta1,3-glucuronyltransferase I (GlcAT-I) is involved in the initial steps of proteoglycan synthesis. Beta1,3-glucuronyltransferase I (GlcAT-I) domain; GlcAT-I is a Key enzyme involved in the initial steps of proteoglycan synthesis. GlcAT-I catalyzes the transfer of a glucuronic acid moiety from the uridine diphosphate-glucuronic acid (UDP-GlcUA) to the common linkage region of trisaccharide Gal-beta-(1-3)-Gal-beta-(1-4)-Xyl of proteoglycans. The enzyme has two subdomains that bind the donor and acceptor substrate separately. The active site is located at the cleft between both subdomains in which the trisaccharide molecule is oriented perpendicular to the UDP. This family has been classified as Glycosyltransferase family 43 (GT-43). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity |
| GO:0016020 | membrane |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ACL54619.1 | 0 | 1 | 309 | 1 | 299 | unknown [Zea mays] |
| GenBank | ACN26728.1 | 0 | 1 | 309 | 1 | 309 | unknown [Zea mays] |
| EMBL | CAI96159.1 | 0 | 1 | 309 | 1 | 309 | glycosyltransferase [Saccharum officinarum] |
| RefSeq | NP_001152042.1 | 0 | 1 | 309 | 1 | 299 | LOC100285679 [Zea mays] |
| RefSeq | XP_002440234.1 | 0 | 1 | 309 | 1 | 309 | hypothetical protein SORBIDRAFT_09g028220 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2f4w_B | 0.000000002 | 739 | 805 | 114 | 177 | A Chain A, Human Ubiquitin-Conjugating Enzyme E2 J2 |
| PDB | 2f4w_A | 0.000000002 | 739 | 805 | 114 | 177 | A Chain A, Human Ubiquitin-Conjugating Enzyme E2 J2 |
| PDB | 2cxk_E | 0.0000004 | 422 | 477 | 35 | 89 | A Chain A, Crystal Structure Of The Tig Domain Of Human Calmodulin- Binding Transcription Activator 1 (Camta1) |
| PDB | 2cxk_D | 0.0000004 | 422 | 477 | 35 | 89 | A Chain A, Crystal Structure Of The Tig Domain Of Human Calmodulin- Binding Transcription Activator 1 (Camta1) |
| PDB | 2cxk_C | 0.0000004 | 422 | 477 | 35 | 89 | A Chain A, Crystal Structure Of The Tig Domain Of Human Calmodulin- Binding Transcription Activator 1 (Camta1) |