

| Basic Information | |
|---|---|
| Species | Zea mays |
| Cazyme ID | GRMZM2G128219_T01 |
| Family | CE10 |
| Protein Properties | Length: 333 Molecular Weight: 35303.5 Isoelectric Point: 5.9434 |
| Chromosome | Chromosome/Scaffold: 4 Start: 122677472 End: 122678792 |
| Description | alpha/beta-Hydrolases superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CE10 | 52 | 330 | 0 |
| AATGVQSKDVHLGSYSARLYLPPVADAGAKLPVVVFVHGGGFVAESAASPNYHLFLNRLAAACPALAVSVDYRLAPEHPLPAGYDDCLAALKWVLSAADP WVAAHGDLARVFVAGDSAGGNVCHYLAIHPDVVQAQQQGCPPPLKGAVLIHPWFWGSEAVGEEPRDPAVRTMGAGLWFFACPDANSMEDPRMNPMAPAAP GLHTLACERVMVCTAEGDFLRWRGRAYAEAVAAARGGRLGQAAGVELLETMGEGHVFFLFKPDCDKAKEMLDKMAAFIN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 333 Download |
| MASSKPLFPF PVPAPDPSDE VVREFGPLLR VYKSGRIERP LVAPPVEPGH DAATGVQSKD 60 VHLGSYSARL YLPPVADAGA KLPVVVFVHG GGFVAESAAS PNYHLFLNRL AAACPALAVS 120 VDYRLAPEHP LPAGYDDCLA ALKWVLSAAD PWVAAHGDLA RVFVAGDSAG GNVCHYLAIH 180 PDVVQAQQQG CPPPLKGAVL IHPWFWGSEA VGEEPRDPAV RTMGAGLWFF ACPDANSMED 240 PRMNPMAPAA PGLHTLACER VMVCTAEGDF LRWRGRAYAE AVAAARGGRL GQAAGVELLE 300 TMGEGHVFFL FKPDCDKAKE MLDKMAAFIN AP* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
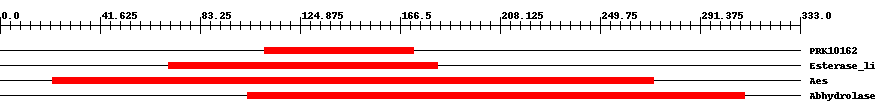 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK10162 | PRK10162 | 0.004 | 110 | 172 | 64 | + acetyl esterase; Provisional | ||
| cd00312 | Esterase_lipase | 4.0e-5 | 70 | 182 | 126 | + Esterases and lipases (includes fungal lipases, cholinesterases, etc.) These enzymes act on carboxylic esters (EC: 3.1.1.-). The catalytic apparatus involves three residues (catalytic triad): a serine, a glutamate or aspartate and a histidine.These catalytic residues are responsible for the nucleophilic attack on the carbonyl carbon atom of the ester bond. In contrast with other alpha/beta hydrolase fold family members, p-nitrobenzyl esterase and acetylcholine esterase have a Glu instead of Asp at the active site carboxylate. | ||
| COG0657 | Aes | 2.0e-20 | 22 | 272 | 261 | + Esterase/lipase [Lipid metabolism] | ||
| pfam07859 | Abhydrolase_3 | 9.0e-36 | 103 | 310 | 212 | + alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008152 | metabolic process |
| GO:0016787 | hydrolase activity |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | EAY86351.1 | 0 | 14 | 330 | 6 | 316 | hypothetical protein OsI_07729 [Oryza sativa Indica Group] |
| RefSeq | NP_001047179.1 | 0 | 14 | 330 | 6 | 313 | Os02g0567800 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | NP_001148459.1 | 0 | 1 | 332 | 1 | 334 | gibberellin receptor GID1L2 [Zea mays] |
| RefSeq | NP_001150025.1 | 0 | 1 | 332 | 1 | 332 | gibberellin receptor GID1L2 [Zea mays] |
| RefSeq | XP_002452312.1 | 0 | 1 | 332 | 1 | 341 | hypothetical protein SORBIDRAFT_04g023520 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2o7v_A | 2e-34 | 14 | 329 | 11 | 327 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2o7r_A | 2e-34 | 14 | 329 | 11 | 327 | A Chain A, Plant Carboxylesterase Aecxe1 From Actinidia Eriantha With Acyl Adduct |
| PDB | 2zsi_A | 4e-31 | 55 | 331 | 69 | 350 | A Chain A, Plant Carboxylesterase Aecxe1 From Actinidia Eriantha With Acyl Adduct |
| PDB | 2zsh_A | 4e-31 | 55 | 331 | 69 | 350 | B Chain B, Structural Basis Of Gibberellin(Ga3)-Induced Della Recognition By The Gibberellin Receptor |
| PDB | 3ed1_F | 2e-27 | 82 | 331 | 112 | 349 | B Chain B, Structural Basis Of Gibberellin(Ga3)-Induced Della Recognition By The Gibberellin Receptor |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| formononetin biosynthesis | RXN-3284 | EC-4.2.1.105 | 2-hydroxyisoflavanone dehydratase |
| isoflavonoid biosynthesis I | RXN-3284 | EC-4.2.1.105 | 2-hydroxyisoflavanone dehydratase |
| isoflavonoid biosynthesis II | RXN-3303 | EC-4.2.1.105 | 2-hydroxyisoflavanone dehydratase |