

| Basic Information | |
|---|---|
| Species | Vitis vinifera |
| Cazyme ID | GSVIVT01016490001 |
| Family | AA1 |
| Protein Properties | Length: 1051 Molecular Weight: 115984 Isoelectric Point: 9.8536 |
| Chromosome | Chromosome/Scaffold: 13 Start: 3444003 End: 3452360 |
| Description | laccase 17 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 35 | 454 | 0 |
| TRHYKFNIELQNVTRLCHTKSIVSVNGQFPGPPIIAREGDRVVVKVVNHVQNNVTIHWHGVRQLRSGWADGPAYVTQCPIQTGQTYVYNFTITGQRGTLF WHAHISWLRSTLYGPIIIFPKKNVPYPFAKPYKEVPIIFGEWWNTDPEAVITQALQNGGGPNVSDGYTINGLPGPLYNCSAQDTFKLKVKPGKTYLLRLI NAALNDELFFSIANHTLTVVDVDAIYVKPFKTDTILITPGQTTNLLLKAKPHFPNATFLMAARPYATGSGTFDNSTVAGILHYEPPPKSSPSAPSFENLP LFKPTLPPLNDTSFAANFTRKLRSLANTQFPANVPQKVDKRFFFTVGLGTNPCPQNQTCQGPNNSTMFSASVNNVSFVLPTTAILQAHFFQQSNGVYTTD FPATPIFPFNYTGTPPNNTM | |||
| AA1 | 501 | 1038 | 0 |
| TRHYKFNIKLQNVTRLCHTKSIVSVNGQFPGPHIIAREGDRVVVKVVNHVQNNVTIHWHGVRQLRSGWADGPAYVTQCPIQTGQTYVYNFTITGQRGTLF WHAHISWLRSTLYGPIIILPKKNVPYPFVKPYKEVPIIFGEWWNADPEAVITQALQNGGGPNVSDAYTINGLPGPLYNCSAQDTFKLKVKPGKTYLLRLI NAALNDELFFSIANHTLTVVDVDAIYVKPFKTDTILITPGQTTNLLLKAKPHFPNATFLMAARPYATGRGTFDNSTVAGILHYEPPPKSSPSASSFKNLP LFKPTLPPLNDTSFAANFTGKLRSLANTQFPANVPQKVDQRFFFTVGLGTNPCPQNQTCQGPTNITMFSASVNNVSFVLPTTAILQAHFFQQSKGVYTTD FPTTPIFPFNYTGTPPNNTMVSNGTKVVVLPFNTSVELVLQDTSILGIESHPLHLHGFNFFVVGQGFGNFDSKKDPPKFNLVDPVERNTVGVPSGGWVAI RFLADNPGVWFMHCHLEVHTSWGLKMAWIVMDGKLPNQ | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1051 Download |
| MGASLIPSTT IFKVLFLVLN SLWYLPELAN GSGITRHYKF NIELQNVTRL CHTKSIVSVN 60 GQFPGPPIIA REGDRVVVKV VNHVQNNVTI HWHGVRQLRS GWADGPAYVT QCPIQTGQTY 120 VYNFTITGQR GTLFWHAHIS WLRSTLYGPI IIFPKKNVPY PFAKPYKEVP IIFGEWWNTD 180 PEAVITQALQ NGGGPNVSDG YTINGLPGPL YNCSAQDTFK LKVKPGKTYL LRLINAALND 240 ELFFSIANHT LTVVDVDAIY VKPFKTDTIL ITPGQTTNLL LKAKPHFPNA TFLMAARPYA 300 TGSGTFDNST VAGILHYEPP PKSSPSAPSF ENLPLFKPTL PPLNDTSFAA NFTRKLRSLA 360 NTQFPANVPQ KVDKRFFFTV GLGTNPCPQN QTCQGPNNST MFSASVNNVS FVLPTTAILQ 420 AHFFQQSNGV YTTDFPATPI FPFNYTGTPP NNTMVSNGTK RRFSGEMGAS LLSSTAIFRV 480 LFVVLNTLWL FPELASSAGI TRHYKFNIKL QNVTRLCHTK SIVSVNGQFP GPHIIAREGD 540 RVVVKVVNHV QNNVTIHWHG VRQLRSGWAD GPAYVTQCPI QTGQTYVYNF TITGQRGTLF 600 WHAHISWLRS TLYGPIIILP KKNVPYPFVK PYKEVPIIFG EWWNADPEAV ITQALQNGGG 660 PNVSDAYTIN GLPGPLYNCS AQDTFKLKVK PGKTYLLRLI NAALNDELFF SIANHTLTVV 720 DVDAIYVKPF KTDTILITPG QTTNLLLKAK PHFPNATFLM AARPYATGRG TFDNSTVAGI 780 LHYEPPPKSS PSASSFKNLP LFKPTLPPLN DTSFAANFTG KLRSLANTQF PANVPQKVDQ 840 RFFFTVGLGT NPCPQNQTCQ GPTNITMFSA SVNNVSFVLP TTAILQAHFF QQSKGVYTTD 900 FPTTPIFPFN YTGTPPNNTM VSNGTKVVVL PFNTSVELVL QDTSILGIES HPLHLHGFNF 960 FVVGQGFGNF DSKKDPPKFN LVDPVERNTV GVPSGGWVAI RFLADNPGVW FMHCHLEVHT 1020 SWGLKMAWIV MDGKLPNQKL PPPPSDLPKC * |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
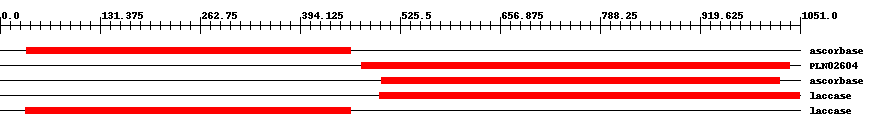 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| TIGR03388 | ascorbase | 6.0e-50 | 35 | 461 | 457 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| PLN02604 | PLN02604 | 4.0e-66 | 475 | 1037 | 597 | + oxidoreductase | ||
| TIGR03388 | ascorbase | 6.0e-79 | 501 | 1024 | 556 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03389 | laccase | 0 | 499 | 1050 | 553 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| TIGR03389 | laccase | 0 | 33 | 460 | 429 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI31631.1 | 0 | 1 | 1050 | 1 | 1050 | unnamed protein product [Vitis vinifera] |
| EMBL | CBI31651.1 | 0 | 1 | 460 | 511 | 970 | unnamed protein product [Vitis vinifera] |
| EMBL | CBI31651.1 | 0 | 11 | 1050 | 1 | 1094 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002282823.1 | 0 | 1 | 460 | 1 | 460 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002282823.1 | 0 | 467 | 1050 | 1 | 584 | PREDICTED: hypothetical protein [Vitis vinifera] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1asq_B | 0 | 502 | 1040 | 4 | 536 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 1asq_B | 4.99983e-42 | 36 | 479 | 4 | 438 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 1asq_A | 0 | 502 | 1040 | 4 | 536 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 1asq_A | 4.99983e-42 | 36 | 479 | 4 | 438 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 1asp_B | 0 | 502 | 1040 | 4 | 536 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |