

| Basic Information | |
|---|---|
| Species | Glycine max |
| Cazyme ID | Glyma02g11653.1 |
| Family | GT1 |
| Protein Properties | Length: 514 Molecular Weight: 57834.5 Isoelectric Point: 6.6925 |
| Chromosome | Chromosome/Scaffold: 02 Start: 9854265 End: 9860748 |
| Description | UDP-glucosyl transferase 73B1 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT1 | 269 | 379 | 4.2e-24 |
| EHECLKWLDTQTTNSVVYVCFGSAVKFSDSQLLEIAMGLEASGQQFIWVVRKSIQEKGEKWLPEGFEKRMEGKGLIIRGWAPQVLILEHEAIGAFVTHCG WNSTLEAVSKG | |||
| GT1 | 385 | 490 | 3.5e-32 |
| WAPQVLILEHEAIGAFVTHCGWNSTLEAVSAGVPMITWPVGGEQFYNEKLVTEVLKIGVPVGVKKWTRFIGDDSVKWDALEKAVKMVMVEEMRNRAQVFK QMARRA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 514 Download |
| MGSNYGPLHI FFFPFMAHGH MIPLVDMAKL FAAKGVRTTI ITTPLNAPII SKTIEQTKTH 60 QSKEINIQTI KFPNVGVGLP EGCEHSDSVL STDLFPIFLK ATTLMQEPFE QLLLHQRPNC 120 VVADWFFPWT TDSAAKFGIP RLVFHGISFF SLCATKIMSL YKPYNNTCSD SELFVIPNFP 180 GEIKMTRLQV GNFHTKDNVG HNSFWNEAEE SEERSYGVVV NSFYELEKDY ADHYRNVHGR 240 KAWHIGPLSL CNRNKEEKIY RGKEASIDEH ECLKWLDTQT TNSVVYVCFG SAVKFSDSQL 300 LEIAMGLEAS GQQFIWVVRK SIQEKGEKWL PEGFEKRMEG KGLIIRGWAP QVLILEHEAI 360 GAFVTHCGWN STLEAVSKGL IIRGWAPQVL ILEHEAIGAF VTHCGWNSTL EAVSAGVPMI 420 TWPVGGEQFY NEKLVTEVLK IGVPVGVKKW TRFIGDDSVK WDALEKAVKM VMVEEMRNRA 480 QVFKQMARRA VEEGGSSDSN LDALVRELCS LSH* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN00164 | PLN00164 | 1.0e-54 | 217 | 509 | 316 | + glucosyltransferase; Provisional | ||
| PLN02448 | PLN02448 | 1.0e-62 | 9 | 509 | 527 | + UDP-glycosyltransferase family protein | ||
| PLN02863 | PLN02863 | 1.0e-95 | 9 | 511 | 522 | + UDP-glucoronosyl/UDP-glucosyl transferase family protein | ||
| PLN02534 | PLN02534 | 6.0e-154 | 8 | 508 | 519 | + UDP-glycosyltransferase | ||
| PLN03007 | PLN03007 | 0 | 8 | 510 | 515 | + UDP-glucosyltransferase family protein | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008152 | metabolic process |
| GO:0016758 | transferase activity, transferring hexosyl groups |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAN65903.1 | 0 | 1 | 508 | 1 | 475 | hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002298733.1 | 0 | 1 | 513 | 1 | 483 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002298734.1 | 0 | 1 | 508 | 1 | 480 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002313131.1 | 0 | 1 | 510 | 1 | 482 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002334266.1 | 0 | 1 | 510 | 1 | 483 | predicted protein [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2acv_B | 1.4013e-45 | 10 | 508 | 12 | 461 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2acv_A | 1.4013e-45 | 10 | 508 | 12 | 461 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2acw_B | 1.4013e-45 | 10 | 508 | 12 | 461 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2acw_A | 1.4013e-45 | 10 | 508 | 12 | 461 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2vg8_A | 1e-39 | 9 | 501 | 8 | 460 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
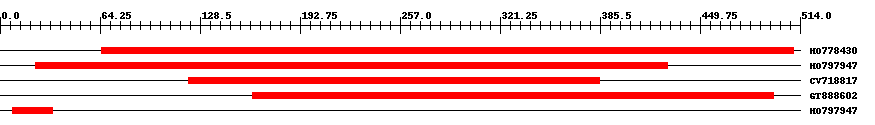 | ||||
| Hit | Length | Start | End | EValue |
| HO778430 | 453 | 65 | 510 | 0 |
| HO797947 | 410 | 23 | 429 | 0 |
| CV718817 | 272 | 121 | 385 | 0 |
| GT888602 | 341 | 162 | 497 | 0 |
| HO797947 | 27 | 8 | 34 | 0.72 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |