

| Basic Information | |
|---|---|
| Species | Glycine max |
| Cazyme ID | Glyma08g19190.2 |
| Family | AA2 |
| Protein Properties | Length: 157 Molecular Weight: 17149.7 Isoelectric Point: 6.4954 |
| Chromosome | Chromosome/Scaffold: 08 Start: 14481104 End: 14482506 |
| Description | Peroxidase superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA2 | 2 | 76 | 5.3e-23 |
| AAGLLRIHFDDCFVQGCDASVLIAGDATERTAFANLGLRGYEVIDDAKTQLEAACPGVVSCADILALAARDSVSL | |||
| Full Sequence |
|---|
| Protein Sequence Length: 157 Download |
| MAAGLLRIHF DDCFVQGCDA SVLIAGDATE RTAFANLGLR GYEVIDDAKT QLEAACPGVV 60 SCADILALAA RDSVSLWRFK SGGQNRFDIS YLANLRIGHG ILQSDQALWN DPSTKSFVQR 120 YLQDGFKGLL FNVEFAKSMF CHQLAIVHVK INLYMA* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd00693 | secretory_peroxidase | 4.0e-13 | 50 | 139 | 90 | + Horseradish peroxidase and related secretory plant peroxidases. Secretory peroxidases belong to class III of the plant heme-dependent peroxidase superfamily. All members of the superfamily share a heme prosthetic group and catalyze a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. Class III peroxidases are found in the extracellular space or in the vacuole in plants where they have been implicated in hydrogen peroxide detoxification, auxin catabolism and lignin biosynthesis, and stress response. Class III peroxidases contain four conserved disulphide bridges and two conserved calcium binding sites. | ||
| PLN03030 | PLN03030 | 3.0e-20 | 50 | 139 | 91 | + cationic peroxidase; Provisional | ||
| pfam00141 | peroxidase | 1.0e-40 | 1 | 124 | 167 | + Peroxidase. | ||
| PLN03030 | PLN03030 | 8.0e-41 | 1 | 82 | 87 | + cationic peroxidase; Provisional | ||
| cd00693 | secretory_peroxidase | 2.0e-48 | 1 | 83 | 86 | + Horseradish peroxidase and related secretory plant peroxidases. Secretory peroxidases belong to class III of the plant heme-dependent peroxidase superfamily. All members of the superfamily share a heme prosthetic group and catalyze a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. Class III peroxidases are found in the extracellular space or in the vacuole in plants where they have been implicated in hydrogen peroxide detoxification, auxin catabolism and lignin biosynthesis, and stress response. Class III peroxidases contain four conserved disulphide bridges and two conserved calcium binding sites. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004601 | peroxidase activity |
| GO:0006979 | response to oxidative stress |
| GO:0020037 | heme binding |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAD37429.2 | 5e-37 | 1 | 83 | 27 | 105 | AF149279_1 peroxidase 4 precursor [Phaseolus vulgaris] |
| GenBank | AAD37429.2 | 0.0000000003 | 81 | 139 | 211 | 271 | AF149279_1 peroxidase 4 precursor [Phaseolus vulgaris] |
| GenBank | ACU20183.1 | 3e-38 | 1 | 74 | 57 | 130 | unknown [Glycine max] |
| GenBank | ACU20183.1 | 0.000000000002 | 51 | 140 | 222 | 302 | unknown [Glycine max] |
| Swiss-Prot | P22196 | 7e-37 | 1 | 82 | 62 | 148 | PER2_ARAHY RecName: Full=Cationic peroxidase 2; AltName: Full=PNPC2; Flags: Precursor |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1qgj_B | 4e-25 | 1 | 82 | 32 | 119 | A Chain A, Arabidopsis Thaliana Peroxidase N |
| PDB | 1qgj_A | 4e-25 | 1 | 82 | 32 | 119 | A Chain A, Arabidopsis Thaliana Peroxidase N |
| PDB | 1sch_B | 1e-24 | 1 | 74 | 32 | 109 | A Chain A, Peanut Peroxidase |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| betanidin degradation | RXN-8635 | EC-1.11.1.7 | peroxidase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
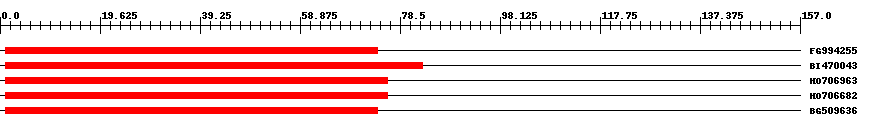 | ||||
| Hit | Length | Start | End | EValue |
| FG994255 | 74 | 1 | 74 | 6e-40 |
| BI470043 | 83 | 1 | 83 | 2e-39 |
| HO706963 | 76 | 1 | 76 | 2e-39 |
| HO706682 | 76 | 1 | 76 | 2e-39 |
| BG509636 | 74 | 1 | 74 | 2e-39 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |