

| Basic Information | |
|---|---|
| Species | Glycine max |
| Cazyme ID | Glyma08g39110.1 |
| Family | CBM45 |
| Protein Properties | Length: 1460 Molecular Weight: 163762 Isoelectric Point: 6.8286 |
| Chromosome | Chromosome/Scaffold: 08 Start: 38392238 End: 38409081 |
| Description | Pyruvate phosphate dikinase, PEP/pyruvate binding domain |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM45 | 452 | 531 | 1.9e-21 |
| LHWALSRTSEEWLVPPATALPPGSVTMNEAAETPFKAGSSSHPSYEVQSLDIEVDDDTFKGIPFVILSDGEWIKNNGSNF | |||
| CBM45 | 123 | 198 | 3.2e-24 |
| LHWGVVRDQPGKWVLPSHHPDGTKNYKNRALRTPFVKSDSGSFLKIEIDDPAAQAIEFLILDEAKNKWFKNKGENF | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1460 Download |
| MSQSIFHQTV LCQTQTVAEH RSKVSSLSVS ANKGKKNLFL APTNFRGNRL CVRKRKLAMG 60 RHHHRHVDAV PRAVLTTNPA SELSGKFNLD GNIELQVAVS SSEPGAARQV DIKVSYNSDS 120 LFLHWGVVRD QPGKWVLPSH HPDGTKNYKN RALRTPFVKS DSGSFLKIEI DDPAAQAIEF 180 LILDEAKNKW FKNKGENFHI KLPVKSKLSQ EVSVPEDLVQ IQAYLRWERK GKQMYTPEQE 240 KEEYEAARNE LFEEVARGTS VQDLRAKLTK KTKAAEVKEP SVSETKTIPD ELVQIQAFIR 300 WEKAGKPNYS QEQQLMEFEE ARKELLAELE KGASLDEIRK KITKGEIQTK VAKQLKTKKY 360 FRAERIQRKK RDLVQLINRN VAENIVEQVI DAPKALTVIE HYANAREEYE SGPVLNKTIY 420 KLGDNDLLVL VTKDAGKIKV HLATDSKKPF TLHWALSRTS EEWLVPPATA LPPGSVTMNE 480 AAETPFKAGS SSHPSYEVQS LDIEVDDDTF KGIPFVILSD GEWIKNNGSN FYIEFGGKKQ 540 IQKDFGDGKG TAKFLLNKIA EMESEAQKSF MHRFNIASDL IDEAKNAGQQ GLAGILVWMR 600 FMATRQLIWN KNYNVKPREI SKAQDRLTDL LQDVYASYPQ YREIVRMILS TVGRGGEGDV 660 GQRIRDEILV IQRNNDCKGG MMEEWHQKLH NNTSPDDVVI CQALIDYINS DFDIGVYWKT 720 LNANGITKER LLSYDRAIHS EPNFRRDQKE GLLRDLGNYM RTLKAVHSGA DLESAISNCM 780 GYKSEGQGFM VGVQINPVPG LPNGFPELLE FVAEHVEEKN VEPLLEGLLE ARQELQPSLS 840 KSQSRLKDLI FLDVALDSTV RTAVERSYEE LNNAGPEKIM YFISLVLENL ALSSDDNEDL 900 IYCLKGWDVA LSMCKSKDTH WALYAKSVLD RTRLALTNKA HLYQEILQPS AEYLGSLLGV 960 DRWAVEIFTE EIIRAGSAAS LSTLLNRLDP VLRKTAHLGS WQVISPVETV GYVEVIDELL 1020 AVQNKSYERP TILIAKSVRG EEEIPDGTVA VLTPDMPDVL SHVSVRARNS KVCFATCFDP 1080 NILANLQENK GKLLRLKPTS ADVVYSEVKE GELIDDKSTQ LKDVGSVSPI SLARKKFSGR 1140 YAVSSEEFTG EMVGAKSRNI SYLKGKVASW IGIPTSVAIP FGVFEHVLSD KPNQAVAERV 1200 NNLKKKLIEG DFSVLKEIRE TVLQLNAPSH LVEELKTKMK SSGMPWPGDE GEQRWEQAWI 1260 AIKKVWGSKW NERAYFSTRK VKLDHEYLSM AVLVQEVINA DYAFVIHTTN PASGDSSEIY 1320 AEVVKGLGET LVGAYPGRAL SFICKKRDLN SPQVLGYPSK PVGLFIRRSI IFRSDSNGED 1380 LEGYAGAGLY DSVPMDEAEK VVLDYSSDKL ILDGSFRQSI LSSIARAGNE IEELYGTPQD 1440 IEGVIKDGKV YVVQTRPQM* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
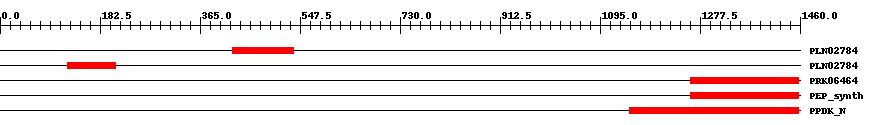 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02784 | PLN02784 | 1.0e-8 | 424 | 535 | 116 | + alpha-amylase | ||
| PLN02784 | PLN02784 | 6.0e-10 | 123 | 211 | 92 | + alpha-amylase | ||
| PRK06464 | PRK06464 | 3.0e-14 | 1261 | 1457 | 212 | + phosphoenolpyruvate synthase; Validated | ||
| TIGR01418 | PEP_synth | 1.0e-14 | 1261 | 1458 | 208 | + phosphoenolpyruvate synthase. Also called pyruvate,water dikinase and PEP synthase. The member from Methanococcus jannaschii contains a large intein. This enzyme generates phosphoenolpyruvate (PEP) from pyruvate, hydrolyzing ATP to AMP and releasing inorganic phosphate in the process. The enzyme shows extensive homology to other enzymes that use PEP as substrate or product. This enzyme may provide PEP for gluconeogenesis, for PTS-type carbohydrate transport systems, or for other processes [Energy metabolism, Glycolysis/gluconeogenesis]. | ||
| pfam01326 | PPDK_N | 4.0e-32 | 1149 | 1458 | 334 | + Pyruvate phosphate dikinase, PEP/pyruvate binding domain. This enzyme catalyzes the reversible conversion of ATP to AMP, pyrophosphate and phosphoenolpyruvate (PEP). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005524 | ATP binding |
| GO:0016301 | kinase activity |
| GO:0016310 | phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAK11735.1 | 0 | 1 | 1459 | 5 | 1464 | starch associated protein R1 [Solanum tuberosum] |
| Swiss-Prot | Q8LPT9 | 0 | 1 | 1459 | 5 | 1475 | GWD1_CITRE RecName: Full=Alpha-glucan water dikinase, chloroplastic; AltName: Full=Starch-related protein R1; Flags: Precursor |
| RefSeq | XP_002270485.1 | 0 | 1 | 1459 | 5 | 1470 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002315679.1 | 0 | 5 | 1459 | 13 | 1477 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002527902.1 | 0 | 1 | 1459 | 1 | 1469 | alpha-glucan water dikinase, chloroplast precursor, putative [Ricinus communis] |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| starch degradation II | RXN-12203 | EC-2.7.9.4 | α-glucan, water dikinase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
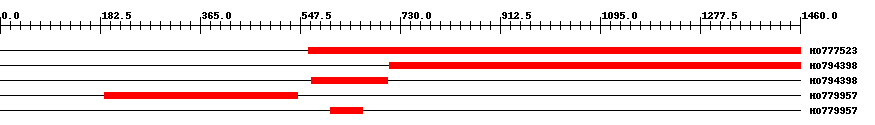 | ||||
| Hit | Length | Start | End | EValue |
| HO777523 | 898 | 563 | 1460 | 0 |
| HO794398 | 751 | 710 | 1460 | 0 |
| HO794398 | 141 | 568 | 708 | 0 |
| HO779957 | 353 | 191 | 543 | 0 |
| HO779957 | 60 | 603 | 662 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |