

| Basic Information | |
|---|---|
| Species | Gossypium raimondii |
| Cazyme ID | Gorai.002G186200.5 |
| Family | CBM45 |
| Protein Properties | Length: 1441 Molecular Weight: 161522 Isoelectric Point: 7.0033 |
| Chromosome | Chromosome/Scaffold: 02 Start: 49592453 End: 49606738 |
| Description | Pyruvate phosphate dikinase, PEP/pyruvate binding domain |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM45 | 465 | 544 | 3.5e-21 |
| LHWALSEKDGEWLAPPPAVLPPGSVSLEKAAESKFSTSTSGDLPKQVQCIEMEIADGNFKGMPFVLLSGGKWIKNNGSDF | |||
| CBM45 | 135 | 210 | 6.4e-24 |
| LHWGAIRGSNDKWVLPSRQPEGTRNHKNRALRTPFVKSGSSSYLKLEIDDPQIQAIEFLIFDEARNKWIKNNGQNF | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1441 Download |
| MSNSVGQNLI QQHFLRPTVL EHQSKLKGSS GIASNSLCAT ASLNQSLAQP RKYQISTKFY 60 GNSLSKRKHK LAMGSQRPLA FIPQAVLATD PASENLGKFN IDGNIELQVD ASAPTSGSIT 120 NVNFRVMYTS DSLLLHWGAI RGSNDKWVLP SRQPEGTRNH KNRALRTPFV KSGSSSYLKL 180 EIDDPQIQAI EFLIFDEARN KWIKNNGQNF HVKLPQRKTL VSNISVPEDL VQVQAYLRWE 240 RKGKQMYTPE QEKEEYEAAR AELLEEISRG ASVDDIRSKI TKKSGQEYKE TAINEENNKI 300 PDDLVQIQAY IRWEKAGKPN YSPEQQLREF EEARKELQSE LEKGASLDEI RKKITKGEIK 360 TKVAKQLQNK KYFSPERIQR KQRDLMQLLN KHAVKVVEES ISVEVEPKPS TAVEPFAKEK 420 ELDGSPVMNK KIYKLGEKEL LVLVTKPAGK IKIHLATDLE EPLTLHWALS EKDGEWLAPP 480 PAVLPPGSVS LEKAAESKFS TSTSGDLPKQ VQCIEMEIAD GNFKGMPFVL LSGGKWIKNN 540 GSDFYVEFSQ RFKQVQKDAG DGKGTSKVLL DRIAALESEA QKSFMHRFNI ASDLMDQAKN 600 IGELGLAGIL VWMRFMATRQ LIWNRNYNVK PREISKAQDR LTDLLQSIYT THPQHRELLR 660 MIMSTIGRGG EGDVGQRIRD EILVIQRNND CKGGMMEEWH QKLHNNTSPD DVIICQALID 720 YIKSDFDINV YWKTLNENGI TKERLLSYDR AIHSEPSFKR DQKDGLLRDL GHYMRTLKAV 780 HSGADLESAI SNCMGYRAEG QGFMVGVQIN PIPGLPSGFP DLLRFVLEHI EDRNVEALLE 840 GLLEARQELR PLLLKSTGRL KDLLFLDIAL DSTVRTAIER GYEELNNARP EKIMHFITLV 900 LENLALSSDD NEDLVYCLKG WHHSISMCKS KSAHWALYAK SVLDRTRLAL ASKAETYQRI 960 LQPSAEYLGS LLGVDQWAIN IFTEEIIRAG SAATLSSLIN RLDPVLRETA HLGSWQVISP 1020 VEVVGYVEVV DELLSVQNKS YDRPTILVAK SVKGEEEIPD GTIAVLTPDM PDVLSHVSVR 1080 ARNCKVCFAT CFDPNILADL QAKKGKLLRL KPSSADVVYS EVKEGELADS SSSNLKGDGP 1140 SVTLVRKQFV GKYAISAEEF TPEMVGAKSR NISYLKGKVP SWVGIPTSVA LPFGVFEKVL 1200 ADEANKEVDQ KLQILKKKLG EGDFGALEEI RQTVLQLRAP SQLVQELKTK MLTSGMPWPG 1260 DEGEQRWEQA WTAIKKVWAS KWNERAYFST RKVKLDHDYL CMAVLVQEVI NADYAFVIHT 1320 TNPSSGDTSE IYAEVLGYPS KPIGLFIRRS MIFRSDSNGE DLEGYAGAGL YDSVPMDKEE 1380 KVVVDYSSDP LINDGKFQQA ILSSIAGAGN AIEELYGSPQ DIEGVIRDGK VYVVQTRPQM 1440 * |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG0574 | PpsA | 2.0e-9 | 1266 | 1438 | 182 | + Phosphoenolpyruvate synthase/pyruvate phosphate dikinase [Carbohydrate transport and metabolism] | ||
| PRK06464 | PRK06464 | 2.0e-10 | 1273 | 1438 | 197 | + phosphoenolpyruvate synthase; Validated | ||
| TIGR01418 | PEP_synth | 1.0e-10 | 1163 | 1439 | 335 | + phosphoenolpyruvate synthase. Also called pyruvate,water dikinase and PEP synthase. The member from Methanococcus jannaschii contains a large intein. This enzyme generates phosphoenolpyruvate (PEP) from pyruvate, hydrolyzing ATP to AMP and releasing inorganic phosphate in the process. The enzyme shows extensive homology to other enzymes that use PEP as substrate or product. This enzyme may provide PEP for gluconeogenesis, for PTS-type carbohydrate transport systems, or for other processes [Energy metabolism, Glycolysis/gluconeogenesis]. | ||
| PRK06241 | PRK06241 | 2.0e-11 | 1156 | 1438 | 340 | + phosphoenolpyruvate synthase; Validated | ||
| pfam01326 | PPDK_N | 4.0e-29 | 1161 | 1439 | 311 | + Pyruvate phosphate dikinase, PEP/pyruvate binding domain. This enzyme catalyzes the reversible conversion of ATP to AMP, pyrophosphate and phosphoenolpyruvate (PEP). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005524 | ATP binding |
| GO:0016301 | kinase activity |
| GO:0016310 | phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAK11735.1 | 0 | 1 | 1440 | 1 | 1464 | starch associated protein R1 [Solanum tuberosum] |
| Swiss-Prot | Q8LPT9 | 0 | 1 | 1440 | 1 | 1475 | GWD1_CITRE RecName: Full=Alpha-glucan water dikinase, chloroplastic; AltName: Full=Starch-related protein R1; Flags: Precursor |
| RefSeq | XP_002270485.1 | 0 | 1 | 1440 | 1 | 1470 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002315679.1 | 0 | 12 | 1440 | 17 | 1477 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002527902.1 | 0 | 1 | 1440 | 1 | 1469 | alpha-glucan water dikinase, chloroplast precursor, putative [Ricinus communis] |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
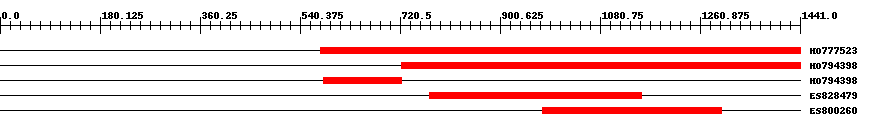 | ||||
| Hit | Length | Start | End | EValue |
| HO777523 | 898 | 577 | 1441 | 0 |
| HO794398 | 752 | 723 | 1441 | 0 |
| HO794398 | 142 | 582 | 723 | 0 |
| ES828479 | 385 | 774 | 1155 | 0 |
| ES800260 | 324 | 978 | 1300 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |