

| Basic Information | |
|---|---|
| Species | Gossypium raimondii |
| Cazyme ID | Gorai.002G186200.6 |
| Family | CBM45 |
| Protein Properties | Length: 1458 Molecular Weight: 163209 Isoelectric Point: 7.2783 |
| Chromosome | Chromosome/Scaffold: 02 Start: 49592453 End: 49606432 |
| Description | Pyruvate phosphate dikinase, PEP/pyruvate binding domain |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM45 | 451 | 530 | 3.6e-21 |
| LHWALSEKDGEWLAPPPAVLPPGSVSLEKAAESKFSTSTSGDLPKQVQCIEMEIADGNFKGMPFVLLSGGKWIKNNGSDF | |||
| CBM45 | 121 | 196 | 6.5e-24 |
| LHWGAIRGSNDKWVLPSRQPEGTRNHKNRALRTPFVKSGSSSYLKLEIDDPQIQAIEFLIFDEARNKWIKNNGQNF | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1458 Download |
| MSNSVGQNLI QQHFLRPTVL EHQSKLKGSS GIASNSLCAT ASLNQSLAQP RKYQISTKFY 60 GNSLSKRKHK LAMGSQRPLA FIPQAVLATD PASEVDASAP TSGSITNVNF RVMYTSDSLL 120 LHWGAIRGSN DKWVLPSRQP EGTRNHKNRA LRTPFVKSGS SSYLKLEIDD PQIQAIEFLI 180 FDEARNKWIK NNGQNFHVKL PQRKTLVSNI SVPEDLVQVQ AYLRWERKGK QMYTPEQEKE 240 EYEAARAELL EEISRGASVD DIRSKITKKS GQEYKETAIN EENNKIPDDL VQIQAYIRWE 300 KAGKPNYSPE QQLREFEEAR KELQSELEKG ASLDEIRKKI TKGEIKTKVA KQLQNKKYFS 360 PERIQRKQRD LMQLLNKHAV KVVEESISVE VEPKPSTAVE PFAKEKELDG SPVMNKKIYK 420 LGEKELLVLV TKPAGKIKIH LATDLEEPLT LHWALSEKDG EWLAPPPAVL PPGSVSLEKA 480 AESKFSTSTS GDLPKQVQCI EMEIADGNFK GMPFVLLSGG KWIKNNGSDF YVEFSQRFKQ 540 VQKDAGDGKG TSKVLLDRIA ALESEAQKSF MHRFNIASDL MDQAKNIGEL GLAGILVWMR 600 FMATRQLIWN RNYNVKPREI SKAQDRLTDL LQSIYTTHPQ HRELLRMIMS TIGRGGEGDV 660 GQRIRDEILV IQRNNDCKGG MMEEWHQKLH NNTSPDDVII CQALIDYIKS DFDINVYWKT 720 LNENGITKER LLSYDRAIHS EPSFKRDQKD GLLRDLGHYM RTLKAVHSGA DLESAISNCM 780 GYRAEGQGFM VGVQINPIPG LPSGFPDLLR FVLEHIEDRN VEALLEGLLE ARQELRPLLL 840 KSTGRLKDLL FLDIALDSTV RTAIERGYEE LNNARPEKIM HFITLVLENL ALSSDDNEDL 900 VYCLKGWHHS ISMCKSKSAH WALYAKSVLD RTRLALASKA ETYQRILQPS AEYLGSLLGV 960 DQWAINIFTE EIIRAGSAAT LSSLINRLDP VLRETAHLGS WQVISPVEVV GYVEVVDELL 1020 SVQNKSYDRP TILVAKSVKG EEEIPDGTIA VLTPDMPDVL SHVSVRARNC KVCFATCFDP 1080 NILADLQAKK GKLLRLKPSS ADVVYSEVKE GELADSSSSN LKGDGPSVTL VRKQFVGKYA 1140 ISAEEFTPEM VGAKSRNISY LKGKVPSWVG IPTSVALPFG VFEKVLADEA NKEVDQKLQI 1200 LKKKLGEGDF GALEEIRQTV LQLRAPSQLV QELKTKMLTS GMPWPGDEGE QRWEQAWTAI 1260 KKVWASKWNE RAYFSTRKVK LDHDYLCMAV LVQEVINADY AFVIHTTNPS SGDTSEIYAE 1320 VVKGLGETLV GAYPGRALSF VCKKNNLNSP EVLGYPSKPI GLFIRRSMIF RSDSNGEDLE 1380 GYAGAGLYDS VPMDKEEKVV VDYSSDPLIN DGKFQQAILS SIAGAGNAIE ELYGSPQDIE 1440 GVIRDGKVYV VQTRPQM* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
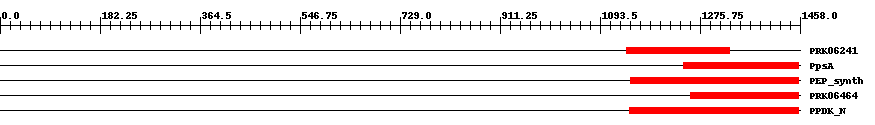 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK06241 | PRK06241 | 3.0e-7 | 1142 | 1330 | 224 | + phosphoenolpyruvate synthase; Validated | ||
| COG0574 | PpsA | 3.0e-10 | 1245 | 1455 | 213 | + Phosphoenolpyruvate synthase/pyruvate phosphate dikinase [Carbohydrate transport and metabolism] | ||
| TIGR01418 | PEP_synth | 1.0e-13 | 1149 | 1456 | 349 | + phosphoenolpyruvate synthase. Also called pyruvate,water dikinase and PEP synthase. The member from Methanococcus jannaschii contains a large intein. This enzyme generates phosphoenolpyruvate (PEP) from pyruvate, hydrolyzing ATP to AMP and releasing inorganic phosphate in the process. The enzyme shows extensive homology to other enzymes that use PEP as substrate or product. This enzyme may provide PEP for gluconeogenesis, for PTS-type carbohydrate transport systems, or for other processes [Energy metabolism, Glycolysis/gluconeogenesis]. | ||
| PRK06464 | PRK06464 | 2.0e-14 | 1259 | 1455 | 210 | + phosphoenolpyruvate synthase; Validated | ||
| pfam01326 | PPDK_N | 1.0e-33 | 1147 | 1456 | 336 | + Pyruvate phosphate dikinase, PEP/pyruvate binding domain. This enzyme catalyzes the reversible conversion of ATP to AMP, pyrophosphate and phosphoenolpyruvate (PEP). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005524 | ATP binding |
| GO:0016301 | kinase activity |
| GO:0016310 | phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ACG69788.1 | 0 | 1 | 1457 | 1 | 1465 | glucan water dikinase [Solanum lycopersicum] |
| Swiss-Prot | Q8LPT9 | 0 | 1 | 1457 | 1 | 1475 | GWD1_CITRE RecName: Full=Alpha-glucan water dikinase, chloroplastic; AltName: Full=Starch-related protein R1; Flags: Precursor |
| RefSeq | XP_002270485.1 | 0 | 1 | 1457 | 1 | 1470 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002315679.1 | 0 | 12 | 1457 | 17 | 1477 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002527902.1 | 0 | 1 | 1457 | 1 | 1469 | alpha-glucan water dikinase, chloroplast precursor, putative [Ricinus communis] |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| HO777523 | 898 | 563 | 1458 | 0 |
| HO794398 | 752 | 709 | 1458 | 0 |
| HO794398 | 142 | 568 | 709 | 0 |
| CO082451 | 294 | 1056 | 1349 | 0 |
| CO072750 | 291 | 1123 | 1413 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |