

| Basic Information | |
|---|---|
| Species | Gossypium raimondii |
| Cazyme ID | Gorai.010G194200.1 |
| Family | AA1 |
| Protein Properties | Length: 414 Molecular Weight: 46045.6 Isoelectric Point: 8.8298 |
| Chromosome | Chromosome/Scaffold: 10 Start: 55212590 End: 55214647 |
| Description | laccase 3 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 4 | 279 | 0 |
| IQATKVKRLCKTHNTITVNGMFPGPTLEIKNGDTLEVQVVNKARYNVTIHWHGVRQMGTGWADGPEFVTQCPIRPGGSYTYRFTVQGQEGTLWWHAHSSW LRATVYGALIIRPREGKSYPFPKPKRETPILLGEWWDANPIDVVREATRTGAAPNVSDAYTINAQPGDLYKCSSKETTVVSVDSGETNLLRVINAALNQP LFFKVANHKLTVVGADASYTKPFTTSVLMLGPGQTTDVLIRGDQPPSRYYMAARAYQSAQNAPFDNTTTTAILEYN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 414 Download |
| MKQIQATKVK RLCKTHNTIT VNGMFPGPTL EIKNGDTLEV QVVNKARYNV TIHWHGVRQM 60 GTGWADGPEF VTQCPIRPGG SYTYRFTVQG QEGTLWWHAH SSWLRATVYG ALIIRPREGK 120 SYPFPKPKRE TPILLGEWWD ANPIDVVREA TRTGAAPNVS DAYTINAQPG DLYKCSSKET 180 TVVSVDSGET NLLRVINAAL NQPLFFKVAN HKLTVVGADA SYTKPFTTSV LMLGPGQTTD 240 VLIRGDQPPS RYYMAARAYQ SAQNAPFDNT TTTAILEYNR SLFQPVPGTK LYKLKYGSRV 300 QIVLQDTSIV TPENHPIHLH GYDFYIIAEG FGNFDPKRDT SKFNLVDPPM RNTVGVPVNG 360 WAVIRFVADN PVWIMHCHLD VHINWGLAMA FLVDNGVGEL QTIQPPPPDL PIC* 420 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| TIGR03388 | ascorbase | 6.0e-30 | 248 | 391 | 148 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03388 | ascorbase | 1.0e-47 | 13 | 278 | 292 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| pfam07732 | Cu-oxidase_3 | 2.0e-54 | 3 | 119 | 119 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| TIGR03389 | laccase | 1.0e-74 | 280 | 413 | 135 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| TIGR03389 | laccase | 1.0e-164 | 4 | 287 | 284 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABK92474.1 | 0 | 4 | 290 | 15 | 300 | unknown [Populus trichocarpa] |
| GenBank | ABK92474.1 | 0 | 279 | 413 | 415 | 550 | unknown [Populus trichocarpa] |
| RefSeq | XP_002312186.1 | 0 | 4 | 290 | 39 | 324 | laccase 90a [Populus trichocarpa] |
| RefSeq | XP_002312186.1 | 0 | 279 | 413 | 439 | 574 | laccase 90a [Populus trichocarpa] |
| RefSeq | XP_002315131.1 | 0 | 4 | 282 | 40 | 318 | laccase 90c [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1asq_B | 2e-37 | 13 | 253 | 19 | 276 | A Chain A, Native Xylanase10c From Cellvibrio Japonicus |
| PDB | 1asq_B | 2e-21 | 288 | 391 | 415 | 521 | A Chain A, Native Xylanase10c From Cellvibrio Japonicus |
| PDB | 1asq_A | 2e-37 | 13 | 253 | 19 | 276 | A Chain A, Native Xylanase10c From Cellvibrio Japonicus |
| PDB | 1asq_A | 2e-21 | 288 | 391 | 415 | 521 | A Chain A, Native Xylanase10c From Cellvibrio Japonicus |
| PDB | 1asp_B | 2e-37 | 13 | 253 | 19 | 276 | A Chain A, Native Xylanase10c From Cellvibrio Japonicus |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
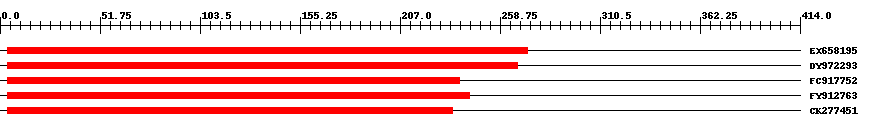 | ||||
| Hit | Length | Start | End | EValue |
| EX658195 | 270 | 4 | 273 | 0 |
| DY972293 | 265 | 4 | 268 | 0 |
| FC917752 | 235 | 4 | 238 | 0 |
| FY912763 | 240 | 4 | 243 | 0 |
| CK277451 | 231 | 4 | 234 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |