

| Basic Information | |
|---|---|
| Species | Oryza sativa |
| Cazyme ID | LOC_Os04g33040.1 |
| Family | GH13 |
| Protein Properties | Length: 413 Molecular Weight: 46951.6 Isoelectric Point: 5.9313 |
| Chromosome | Chromosome/Scaffold: 4 Start: 20006128 End: 20009276 |
| Description | alpha-amylase-like 2 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH13 | 48 | 333 | 8.5e-34 |
| DLAQSGFTSAWLPPPTQSLSPEGYLPQNLYCLDSCYGSLHDLQALLRKMKEHNVRAMADVVINHRVGTTQGSNGMYNRYDGIPVSWDEHAVTSCSGGKGN ESTGDNFDGVPNIDHTQPFVRKDIIDWLIWLRESIGFQDFRFDFTKGYAAKFVKEYIEQSKPLFAVGEYWDSCEYSPPDYRLNYNQDKHRQRIINWMDST GGLCAAFDFTTKGILQEAVKGELWRLRDPEGKPPGVMGWWPSRSVTFVENHDTGSTQGHWPFPSDHIMEGYAYILTHPGIPTVFYD | |||
| Full Sequence |
|---|
| Protein Sequence Length: 413 Download |
| MVSDENFEEQ AARNGGIIKH GREILFQAFN WESHKHNWWS NLEEKVVDLA QSGFTSAWLP 60 PPTQSLSPEG YLPQNLYCLD SCYGSLHDLQ ALLRKMKEHN VRAMADVVIN HRVGTTQGSN 120 GMYNRYDGIP VSWDEHAVTS CSGGKGNEST GDNFDGVPNI DHTQPFVRKD IIDWLIWLRE 180 SIGFQDFRFD FTKGYAAKFV KEYIEQSKPL FAVGEYWDSC EYSPPDYRLN YNQDKHRQRI 240 INWMDSTGGL CAAFDFTTKG ILQEAVKGEL WRLRDPEGKP PGVMGWWPSR SVTFVENHDT 300 GSTQGHWPFP SDHIMEGYAY ILTHPGIPTV FYDHFYGKDD SFHGGIAKLM EIRKCQDIHS 360 RSAVKILEAS SDLYSAIVDD KLCMKIGDGS WCPSGPEWKL AASGDRYAVW HK* 420 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK09441 | PRK09441 | 1.0e-45 | 23 | 354 | 415 | + cytoplasmic alpha-amylase; Reviewed | ||
| PLN00196 | PLN00196 | 3.0e-138 | 23 | 412 | 408 | + alpha-amylase; Provisional | ||
| cd11314 | AmyAc_arch_bac_plant_AmyA | 3.0e-157 | 24 | 364 | 345 | + Alpha amylase catalytic domain found in archaeal, bacterial, and plant Alpha-amylases (also called 1,4-alpha-D-glucan-4-glucanohydrolase). AmyA (EC 3.2.1.1) catalyzes the hydrolysis of alpha-(1,4) glycosidic linkages of glycogen, starch, related polysaccharides, and some oligosaccharides. This group includes AmyA from bacteria, archaea, water fleas, and plants. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. | ||
| PLN02784 | PLN02784 | 7.0e-165 | 21 | 412 | 399 | + alpha-amylase | ||
| PLN02361 | PLN02361 | 0 | 12 | 412 | 401 | + alpha-amylase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0004556 | alpha-amylase activity |
| GO:0005488 | binding |
| GO:0005509 | calcium ion binding |
| GO:0005576 | extracellular region |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAE02023.2 | 0 | 1 | 412 | 1 | 412 | OSJNBb0118P14.5 [Oryza sativa (japonica cultivar-group)] |
| EMBL | CAX51375.1 | 0 | 1 | 412 | 4 | 415 | alpha-amylase [Hordeum vulgare subsp. vulgare] |
| RefSeq | NP_001052697.1 | 0 | 2 | 412 | 34 | 444 | Os04g0403300 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | NP_001141482.1 | 0 | 1 | 412 | 4 | 415 | hypothetical protein LOC100273593 [Zea mays] |
| RefSeq | XP_002446386.1 | 0 | 1 | 412 | 4 | 415 | hypothetical protein SORBIDRAFT_06g015110 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1bg9_A | 0 | 23 | 412 | 1 | 402 | A Chain A, The Structure Of Endoglucanase From Termite, Nasutitermes Takasagoensis, At Ph 2.5. |
| PDB | 1ava_B | 0 | 23 | 412 | 1 | 402 | A Chain A, Amy2BASI PROTEIN-Protein Complex From Barley Seed |
| PDB | 1ava_A | 0 | 23 | 412 | 1 | 402 | A Chain A, Amy2BASI PROTEIN-Protein Complex From Barley Seed |
| PDB | 1amy_A | 0 | 23 | 412 | 1 | 402 | A Chain A, Amy2BASI PROTEIN-Protein Complex From Barley Seed |
| PDB | 3bsg_A | 0 | 23 | 412 | 2 | 404 | A Chain A, Barley Alpha-Amylase Isozyme 1 (Amy1) H395a Mutant |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| starch degradation I | RXN-1823 | EC-3.2.1.1 | α-amylase |
| starch degradation I | RXN-1825 | EC-3.2.1.1 | α-amylase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
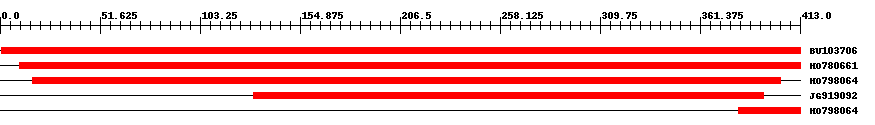 | ||||
| Hit | Length | Start | End | EValue |
| BU103706 | 413 | 1 | 413 | 0 |
| HO780661 | 404 | 10 | 413 | 0 |
| HO798064 | 389 | 17 | 403 | 0 |
| JG919092 | 264 | 131 | 394 | 0 |
| HO798064 | 33 | 381 | 413 | 0.000003 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |