

| Basic Information | |
|---|---|
| Species | Malus domestica |
| Cazyme ID | MDP0000174496 |
| Family | GH1 |
| Protein Properties | Length: 446 Molecular Weight: 49425.5 Isoelectric Point: 6.4084 |
| Chromosome | Chromosome/Scaffold: 005483475 Start: 7056 End: 13257 |
| Description | beta glucosidase 13 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH1 | 348 | 446 | 1.2e-31 |
| ALDENQTWNVEDIKLIKDAGLNSYRFSISWSRILPKGSVSGGINQEGIDYYNSLIDELIKNGEKATSFYASHLFVTIYHFDMPLALEEKYGGLLNRSFV | |||
| Full Sequence |
|---|
| Protein Sequence Length: 446 Download |
| MARGLIAICL ITLLACSSVE GRGLELAHLN GAEHQSVGDH VHGVVENVTV GGFNLQLNLS 60 NPKLVEELKI KRSDFPSDFM FGAATSAAQS EGSAKEGGRG PSGWDHRMET LQARNVLDSQ 120 LDGAQVVEAL QLFRNFTHTL GRCQGNNYHH SSACVMQFKG KKLKERKNXT IENEKMIIEN 180 SIAMLGKFAD FFDGSDTTTH DNIPVTPFFH KDSENNPQGE SLMDTFPLPT PGKIHANLPN 240 PIINSNIDEV VSEEVPSAPT PQPQAATPQS VVAPRRNPAR DQHPPLGLQE YVTYFARYPI 300 SEAITYQKLS ASHALFLNQL SNNLEPRSFQ KAVCTPVWQK AMHKELKALD ENQTWNVEDI 360 KLIKDAGLNS YRFSISWSRI LPKGSVSGGI NQEGIDYYNS LIDELIKNGE KATSFYASHL 420 FVTIYHFDMP LALEEKYGGL LNRSFV |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
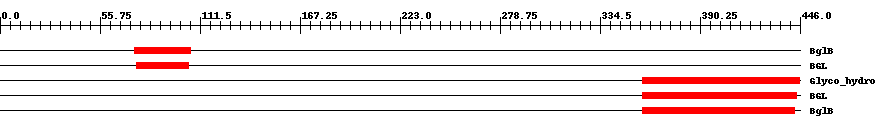 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG2723 | BglB | 8.0e-8 | 75 | 106 | 32 | + Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism] | ||
| TIGR03356 | BGL | 1.0e-8 | 76 | 105 | 30 | + beta-galactosidase. | ||
| pfam00232 | Glyco_hydro_1 | 3.0e-27 | 358 | 446 | 89 | + Glycosyl hydrolase family 1. | ||
| TIGR03356 | BGL | 2.0e-27 | 358 | 444 | 87 | + beta-galactosidase. | ||
| COG2723 | BglB | 4.0e-30 | 358 | 443 | 86 | + Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAL27856.1 | 1e-24 | 358 | 446 | 17 | 97 | raucaffricine-O-beta-D-glucosidase-like protein [Davidia involucrata] |
| GenBank | ABK60303.2 | 0.002 | 70 | 105 | 30 | 65 | glycosylhydrolase family 1 [Leucaena leucocephala] |
| GenBank | ABK60303.2 | 8e-24 | 337 | 446 | 63 | 173 | glycosylhydrolase family 1 [Leucaena leucocephala] |
| RefSeq | XP_002512137.1 | 0.009 | 72 | 105 | 45 | 78 | beta-glucosidase, putative [Ricinus communis] |
| RefSeq | XP_002512137.1 | 6e-25 | 358 | 446 | 106 | 186 | beta-glucosidase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 4ek7_B | 1e-23 | 358 | 446 | 80 | 160 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 4ek7_B | 0.00000001 | 66 | 163 | 13 | 108 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 4ek7_A | 1e-23 | 358 | 446 | 80 | 160 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 4ek7_A | 0.00000001 | 66 | 163 | 13 | 108 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 3u5y_B | 1e-23 | 358 | 446 | 80 | 160 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |