

| Basic Information | |
|---|---|
| Species | Malus domestica |
| Cazyme ID | MDP0000235546 |
| Family | GH1 |
| Protein Properties | Length: 445 Molecular Weight: 50426 Isoelectric Point: 6.3798 |
| Chromosome | Chromosome/Scaffold: 000832270 Start: 8686 End: 11656 |
| Description | beta glucosidase 13 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH1 | 32 | 444 | 0 |
| RDDFPSDFYIGVGTAATQTEGAAEEEGXXPSVWDHRAKMLPGSIPNSDKFPFAIDGYKRYKEDIKIIKDLGVNGYRFSISWSRILPRIEPFVTIYHFDMP QALQVKYGGYLNSKFVQDFQDYSDLCFKLFGDRVKHWFTINEPSSLAVYGYEIGIAPPGRCSLPEGQCGLGAPPPGTCIVPAGPCFGGNSSTEPYIAAHN LILAHAAVEEQKGEVGIVLAATYFAPFSKSQEDKAAAKRLFDFTLGWFMEPLVFGDYPQSMRDLVKERLPTFSAEEKSLLSGSLGNFVGINYYSSAGAKH RPPPPTEXLRHSFDSWAEATGYTNTRNDSRSLHERLYDPHRASYIARHLYRLNXAMKNGANVKGYFYWALFDDVEWGLGFSLQYGLYYIDFGNNXKRYPK LSARWFRDFNNKN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 445 Download |
| MNVNHNEEEA XDAYFELRSH NIALIXELNI TRDDFPSDFY IGVGTAATQT EGAAEEEGXX 60 PSVWDHRAKM LPGSIPNSDK FPFAIDGYKR YKEDIKIIKD LGVNGYRFSI SWSRILPRIE 120 PFVTIYHFDM PQALQVKYGG YLNSKFVQDF QDYSDLCFKL FGDRVKHWFT INEPSSLAVY 180 GYEIGIAPPG RCSLPEGQCG LGAPPPGTCI VPAGPCFGGN SSTEPYIAAH NLILAHAAVE 240 EQKGEVGIVL AATYFAPFSK SQEDKAAAKR LFDFTLGWFM EPLVFGDYPQ SMRDLVKERL 300 PTFSAEEKSL LSGSLGNFVG INYYSSAGAK HRPPPPTEXL RHSFDSWAEA TGYTNTRNDS 360 RSLHERLYDP HRASYIARHL YRLNXAMKNG ANVKGYFYWA LFDDVEWGLG FSLQYGLYYI 420 DFGNNXKRYP KLSARWFRDF NNKNY |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02998 | PLN02998 | 3.0e-74 | 14 | 440 | 495 | + beta-glucosidase | ||
| PLN02814 | PLN02814 | 2.0e-77 | 31 | 442 | 481 | + beta-glucosidase | ||
| PLN02849 | PLN02849 | 1.0e-79 | 31 | 440 | 480 | + beta-glucosidase | ||
| COG2723 | BglB | 4.0e-80 | 35 | 439 | 480 | + Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism] | ||
| pfam00232 | Glyco_hydro_1 | 2.0e-100 | 31 | 444 | 483 | + Glycosyl hydrolase family 1. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | NP_001057511.1 | 0 | 30 | 440 | 130 | 577 | Os06g0320200 [Oryza sativa (japonica cultivar-group)] |
| Swiss-Prot | Q5Z9Z0 | 0 | 30 | 440 | 28 | 501 | BGL24_ORYSJ RecName: Full=Beta-glucosidase 24; Short=Os6bglu24; Flags: Precursor |
| RefSeq | XP_002330884.1 | 0 | 32 | 440 | 36 | 507 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002448027.1 | 0 | 30 | 443 | 36 | 512 | hypothetical protein SORBIDRAFT_06g019840 [Sorghum bicolor] |
| RefSeq | XP_002511218.1 | 0 | 27 | 389 | 48 | 460 | beta-glucosidase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1cbg_A | 0 | 29 | 443 | 13 | 490 | A Chain A, The Crystal Structure Of A Cyanogenic Beta-Glucosidase From White Clover (Trifolium Repens L.), A Family 1 Glycosyl-Hydrolase |
| PDB | 3ptq_B | 0 | 30 | 440 | 29 | 502 | A Chain A, The Crystal Structure Of A Cyanogenic Beta-Glucosidase From White Clover (Trifolium Repens L.), A Family 1 Glycosyl-Hydrolase |
| PDB | 3ptq_A | 0 | 30 | 440 | 29 | 502 | A Chain A, The Crystal Structure Of A Cyanogenic Beta-Glucosidase From White Clover (Trifolium Repens L.), A Family 1 Glycosyl-Hydrolase |
| PDB | 3ptm_B | 0 | 30 | 440 | 29 | 502 | A Chain A, The Crystal Structure Of A Cyanogenic Beta-Glucosidase From White Clover (Trifolium Repens L.), A Family 1 Glycosyl-Hydrolase |
| PDB | 3ptm_A | 0 | 30 | 440 | 29 | 502 | A Chain A, The Crystal Structure Of A Cyanogenic Beta-Glucosidase From White Clover (Trifolium Repens L.), A Family 1 Glycosyl-Hydrolase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| HO779715 | 440 | 61 | 432 | 0 |
| FG227815 | 488 | 30 | 440 | 0 |
| HO794849 | 469 | 32 | 432 | 0 |
| HO777739 | 438 | 73 | 443 | 0 |
| EE590590 | 434 | 87 | 440 | 0 |
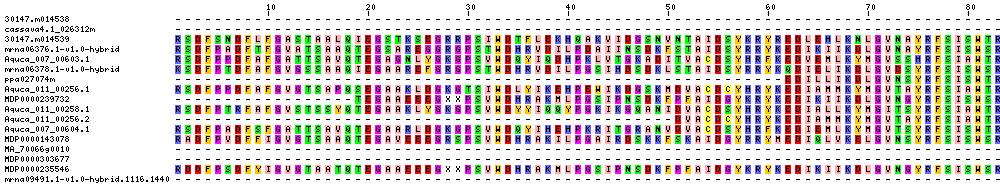
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |