

| Basic Information | |
|---|---|
| Species | Malus domestica |
| Cazyme ID | MDP0000273111 |
| Family | CBM43 |
| Protein Properties | Length: 2383 Molecular Weight: 264441 Isoelectric Point: 9.3904 |
| Chromosome | Chromosome/Scaffold: 00513076 Start: 2099 End: 19965 |
| Description | Sugar isomerase (SIS) family protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM43 | 1462 | 1537 | 7.3e-28 |
| CVPKAGVSDSQLQANIDYVCGSGFDCTPIQPGGSCFEPNNVKSHAAYAMNLLYQTTGGDTQNCDFSQTATLSSNNP | |||
| GH17 | 1100 | 1417 | 0 |
| IGVNYGQVADNLPPPSSTAXLLQSTSIQKVRLYGSDPAIIKALANTGVGIVIGAANGDIPGLASDPSFAKSWVGANVSPFYPASNIILITVGNEVLTSGD QGLISQLVPAIQNVQNALNSASLGGKIKVSTVHSMAVLKQSEPPSSGSFDPGFGDVMKGLLGFNNATGSPFSINPYPYFAYRGDPRPETLAFCLFQPNEG RLDTNTNIKYMNMFDAQVDAIRSALDSMGFKGVEIVVAETGWPYKGDDNEVGPSVENAKAYNGNLIAHLRSMLGTPLMPGKSVDTYLFALYDENLKPGPG SERAFGLFKPDLTMAYDV | |||
| Full Sequence |
|---|
| Protein Sequence Length: 2383 Download |
| MAKRSDFAQK LLDDLRVRKE KMGASQSSNR SNSMAIDAYA YSKQAYRGRG DARTSGTIGS 60 RGSSRTPIIQ DGSNQIVAYG RGGRSSQQMG DLSMALAFAL ENGGKLGRAD TSSRSSMLGF 120 LNQIGRGAME FSKMERKGST NRYFGSSNQF PNLSHLHIEE ISRGAHKLNQ ILKACSNGLN 180 VNGFSIEVGK ELXKGAMDLE ESLRMLVNLQ ETSDYMVRSQ RKNRITLLDE EEDDEDNAVK 240 IDEQKQIDLP RFSFDKGSRH GRKXQEXGRT GFMQKMXAAL TYTTEGSSFN REKQGKITSS 300 PSVTQRQSVS YSSGVKNPSA EYKESKPEKE RIPNVIAKLM GLNELPKNES MKHKAHEDLS 360 SKSKTESRTR EQTMRERSKI SGVKTKDPLM HNTTFVLQAE KNMLPNNVSL ELVVHDGTSQ 420 WKDPEGFKPV ARSEKATIKS DKQQKTTQSV KPNKAVVHNN SEARLATQGK TEFKENRVQT 480 EKQNGIKIPL RNQHKSPNNH ELQQQSMFLK SETLEDKRRR ETKEQQSPQP KLPARKQRGG 540 EIISRTKSTG AVDLQKKQTL XDQARLHRKN TREVGGAMQP KGVSNGRFHE NPVRSKSXSD 600 LNFATNXSSP KYLDPEPSKE XFGTPLAMEE RPVHAAPPLQ KAKSRRVNKS EMPRRIDEVA 660 TRRSXEKLGS QNAXEKVRAS RLKQAEPRIV KSNKSRASIQ SVDSTQNLHI EAKLEAPTLC 720 DPNEVHLLPN ESYDQQYQAP VFGDDECITA STTXNGKRRT SLQRGMSLWQ YFKPITHCHV 780 NKWQYLXXSQ XHLVAGNREX GLDICYXEVR EHHKAFNLRK QEPLTESENR LKQVVIKSHH 840 FLNTAEALFK LEIPFGILHD SGRDCSPDVD IKLTLECGYE VMKRKGRRHE LNVHPNCTKT 900 SISFVKIHSL DELVRQLHKD FETLKFYGRK GKYECEVEAY LPQMLESDMH NWEPDVNCMW 960 DMGWDETMFA VIQVDEVVKD LERLLLNGLV DELARDLRRT GFIPLIKIVI TKFIFVLDLC 1020 MSSLRSHSSL YRSNFARDNS NSARVKAPPE LVLVTFVAVN IIAMLCHIPS TLSSAMAITF 1080 LHAVSLFVLS VNLANCQSFI GVNYGQVADN LPPPSSTAXL LQSTSIQKVR LYGSDPAIIK 1140 ALANTGVGIV IGAANGDIPG LASDPSFAKS WVGANVSPFY PASNIILITV GNEVLTSGDQ 1200 GLISQLVPAI QNVQNALNSA SLGGKIKVST VHSMAVLKQS EPPSSGSFDP GFGDVMKGLL 1260 GFNNATGSPF SINPYPYFAY RGDPRPETLA FCLFQPNEGR LDTNTNIKYM NMFDAQVDAI 1320 RSALDSMGFK GVEIVVAETG WPYKGDDNEV GPSVENAKAY NGNLIAHLRS MLGTPLMPGK 1380 SVDTYLFALY DENLKPGPGS ERAFGLFKPD LTMAYDVGLS KSSXTPTTPI TPTTPSASPM 1440 PSAVPKTPVN PSGPNPKKES YCVPKAGVSD SQLQANIDYV CGSGFDCTPI QPGGSCFEPN 1500 NVKSHAAYAM NLLYQTTGGD TQNCDFSQTA TLSSNNPIIV VRVFLNNTKG SRDAEAKQQW 1560 RPEREKGKGI SSSSSQLLPF PKPEEIPNRP PQQTPHLPPP PIPIIKPAAR FSSNFIFLLT 1620 FCIRSIYDRN ISYHIMRMKC GNIFLAFSKV IQSWASKKFM TGCVILLPIA ITFYVTWAFI 1680 KFVDGFFSPI YNHLGINIFG LGFATSLTFI FLVGIFMQSW LGTSVLTLWE WFIKKMPLIS 1740 YIYAASKQIS IAISPDQSSK AFKEVAIIXH PRIGEYALGF ITSTVVLSQR NVGEELCCVY 1800 IPTNHLYLGD IFLISPKDIL RPNLSVREGI AVEGFEGNRG STSLKSRSLK AHVEDINKTH 1860 LRDLLNDVKR SXAMMVEFDG LLLDYSRQCA TVETLQKLLK LAEAASLKEK INRMFAGERI 1920 NSTENRAVLH VALRAXRDAV IQCDGKNVVP DVWEVLDKIQ KFSESIRSGS WVGATGKALK 1980 DVIAVGIGGS FLGPLFVHTA LQTDPEAMGA AKGRQLHFLA NVDPIDVARN IAGFNPETTL 2040 VVVVSKTFTT AETMLNARTL REWISSSLGP AAVAKHMVAV STNLTLVEKF GIDPKNAFAF 2100 WDWVGGRYSV CSAVGVLPLS LQYGFXVVEK FLKGASSIDQ HFYSTPFEKN IPVLLGLLSI 2160 WNVSFLGYPA RVSMESNGKG VSIDGVILPF ETGEIDFGEP XTNGQHSFYQ LIHQGRVIPC 2220 EFIGVMKSQQ PVYLKGEVVS NHDELMSNFF AQPDALAYGK HPEQLKKENV PEHLITHKTF 2280 SGNRPSLSLL LPSLNAYNIG QLLAIYEHRI AVEGFVWGIN SFDQWGVELG KSLASQVRKQ 2340 LNASRTKGQP IEGFNYSTTT LLTKYLEATK DIPADLPTLL PQI |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
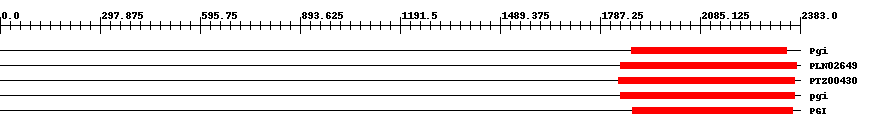 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG0166 | Pgi | 6.0e-125 | 1880 | 2342 | 483 | + Glucose-6-phosphate isomerase [Carbohydrate transport and metabolism] | ||
| PLN02649 | PLN02649 | 0 | 1847 | 2373 | 546 | + glucose-6-phosphate isomerase | ||
| PTZ00430 | PTZ00430 | 0 | 1842 | 2367 | 549 | + glucose-6-phosphate isomerase; Provisional | ||
| PRK00179 | pgi | 0 | 1847 | 2368 | 551 | + glucose-6-phosphate isomerase; Reviewed | ||
| pfam00342 | PGI | 0 | 1884 | 2362 | 499 | + Phosphoglucose isomerase. Phosphoglucose isomerase catalyzes the interconversion of glucose-6-phosphate and fructose-6-phosphate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004347 | glucose-6-phosphate isomerase activity |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| GO:0006094 | gluconeogenesis |
| GO:0006096 | glycolysis |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAU00726.1 | 0 | 1847 | 2383 | 14 | 568 | glucose-6-phosphate isomerase [Solanum tuberosum] |
| GenBank | ACU20878.1 | 0 | 1841 | 2383 | 8 | 568 | unknown [Glycine max] |
| Swiss-Prot | P54237 | 0 | 1847 | 2383 | 14 | 568 | G6PI1_CLAMI RecName: Full=Glucose-6-phosphate isomerase, cytosolic 1; Short=GPI; AltName: Full=Phosphoglucose isomerase; Short=PGI; AltName: Full=Phosphohexose isomerase; Short=PHI |
| RefSeq | XP_002282774.1 | 0 | 1847 | 2383 | 14 | 568 | PREDICTED: similar to glucose-6-phosphate isomerase [Vitis vinifera] |
| RefSeq | XP_002510371.1 | 0 | 1847 | 2383 | 14 | 568 | glucose-6-phosphate isomerase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3ujh_B | 0 | 1856 | 2370 | 26 | 566 | A Chain A, Crystal Structure Of Substrate-Bound Glucose-6-Phosphate Isomerase From Toxoplasma Gondii |
| PDB | 3ujh_A | 0 | 1856 | 2370 | 26 | 566 | A Chain A, Crystal Structure Of Substrate-Bound Glucose-6-Phosphate Isomerase From Toxoplasma Gondii |
| PDB | 3qki_C | 0 | 1825 | 2366 | 9 | 595 | A Chain A, Crystal Structure Of Substrate-Bound Glucose-6-Phosphate Isomerase From Toxoplasma Gondii |
| PDB | 3qki_B | 0 | 1825 | 2366 | 9 | 595 | A Chain A, Crystal Structure Of Substrate-Bound Glucose-6-Phosphate Isomerase From Toxoplasma Gondii |
| PDB | 3qki_A | 0 | 1825 | 2366 | 9 | 595 | A Chain A, Crystal Structure Of Substrate-Bound Glucose-6-Phosphate Isomerase From Toxoplasma Gondii |