

| Basic Information | |
|---|---|
| Species | Medicago truncatula |
| Cazyme ID | Medtr3g101640.1 |
| Family | AA1 |
| Protein Properties | Length: 1114 Molecular Weight: 125452 Isoelectric Point: 9.4151 |
| Chromosome | Chromosome/Scaffold: 3 Start: 35839122 End: 35850733 |
| Description | Laccase/Diphenol oxidase family protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 30 | 558 | 0 |
| SHYNFVVKEARYTRLCCTKNILTVNGQFPGPTIRVHKGDTIYVNVYNKGKYNITIHWHGVKQPRNPWTDGPEYITQCPIKPGGKFRQKVIFSEEEGTLWW HAHSDWARATVHGAINIYPGINSTYPFPKPYGEIPIIFGEWWKNDVNKVLTESLESGGAPNSSDAITINGQPGDLYLCSKSETFKLRVEQGRTYLLRVVN AAMNLILFFSISKHKLTVVGADAMYTKPMTRDYICISPGQTMDVLLHANQEPSHYYMAARAYSSAAGLEFDNTTATGIVQYSWSHTPLSTPSLPHLPNYN DTQAAFDFIRSIRGLPKKYPHEVPTKITTHVITTVSINTFPCPNGRQTCEGPNGTILSSSMNNISFQTPNKFDILEAYYYHINGVFSKGFPSFPPLIFNF TADSFPLTLNTPKRGTKVKVLKYGSTVELVFQGTNLVGGGLDHPMHLHGFSFYVVGYGFGNFNKSKHPMNYNLIDPPLLNTLMVPKSGWAAVRFLASNPG VWFLHCHLERHLTWGMETVFIVKNGKSLN | |||
| AA1 | 573 | 1100 | 0 |
| YEHKKMDVQEARYTRLCSTKSILTVNGKFPGPTIRVHKGDTIYVNVYNKGKYNITIHWHGVMQPRNPWTDGPEYITQCPIQPGGKFRQKVVFSDEEGTLW WHAHSDWARATVHGAINIYPIINSTYPFPKPYGEIPIIFGEWWKNDVNKVFTEFLESGGAPNSSDAITINGQPGDLYPCSKSETFKLRVEQGRTYLLRVV NAAMNLILFFSISKHKLTVVGADAMYTKPMTRDYICISPGQTMDVLLHANQEPSHYYMAARAYSTDNARVEFDNTTTTAIVQYRESYTPISTSSLPFLPN YNDTQAAFNFIRSISGLHKYSNDVPKKIKTRIITTVSINTFPCPNGRSCEGPNGTILSASMNNISFQTPNKFDILEAYYYHINGVFRKGFPRFPPFIFNF NGNSLPMTLNTPRRGTKVKVLKYGSAVEVVFQGTNLVAPLDHPMHLHGFSFYVVGYGFGNFNKSKDTKNYNLIDPPLLNTVLVPVNGWAAIRFVATNPGV WFLHCHLERHLSWGMETVFIVKNGKSLN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1114 Download |
| MSFYKELFLQ ILLIFSLIAL SSQTRYQTKS HYNFVVKEAR YTRLCCTKNI LTVNGQFPGP 60 TIRVHKGDTI YVNVYNKGKY NITIHWHGVK QPRNPWTDGP EYITQCPIKP GGKFRQKVIF 120 SEEEGTLWWH AHSDWARATV HGAINIYPGI NSTYPFPKPY GEIPIIFGEW WKNDVNKVLT 180 ESLESGGAPN SSDAITINGQ PGDLYLCSKS ETFKLRVEQG RTYLLRVVNA AMNLILFFSI 240 SKHKLTVVGA DAMYTKPMTR DYICISPGQT MDVLLHANQE PSHYYMAARA YSSAAGLEFD 300 NTTATGIVQY SWSHTPLSTP SLPHLPNYND TQAAFDFIRS IRGLPKKYPH EVPTKITTHV 360 ITTVSINTFP CPNGRQTCEG PNGTILSSSM NNISFQTPNK FDILEAYYYH INGVFSKGFP 420 SFPPLIFNFT ADSFPLTLNT PKRGTKVKVL KYGSTVELVF QGTNLVGGGL DHPMHLHGFS 480 FYVVGYGFGN FNKSKHPMNY NLIDPPLLNT LMVPKSGWAA VRFLASNPGV WFLHCHLERH 540 LTWGMETVFI VKNGKSLNET LPPPPPDMTC GKYEHKKMDV QEARYTRLCS TKSILTVNGK 600 FPGPTIRVHK GDTIYVNVYN KGKYNITIHW HGVMQPRNPW TDGPEYITQC PIQPGGKFRQ 660 KVVFSDEEGT LWWHAHSDWA RATVHGAINI YPIINSTYPF PKPYGEIPII FGEWWKNDVN 720 KVFTEFLESG GAPNSSDAIT INGQPGDLYP CSKSETFKLR VEQGRTYLLR VVNAAMNLIL 780 FFSISKHKLT VVGADAMYTK PMTRDYICIS PGQTMDVLLH ANQEPSHYYM AARAYSTDNA 840 RVEFDNTTTT AIVQYRESYT PISTSSLPFL PNYNDTQAAF NFIRSISGLH KYSNDVPKKI 900 KTRIITTVSI NTFPCPNGRS CEGPNGTILS ASMNNISFQT PNKFDILEAY YYHINGVFRK 960 GFPRFPPFIF NFNGNSLPMT LNTPRRGTKV KVLKYGSAVE VVFQGTNLVA PLDHPMHLHG 1020 FSFYVVGYGF GNFNKSKDTK NYNLIDPPLL NTVLVPVNGW AAIRFVATNP GVWFLHCHLE 1080 RHLSWGMETV FIVKNGKSLN ATLPPPPPDM PPC* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
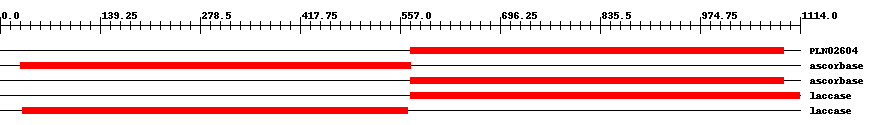 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02604 | PLN02604 | 8.0e-62 | 572 | 1091 | 550 | + oxidoreductase | ||
| TIGR03388 | ascorbase | 6.0e-65 | 29 | 572 | 579 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03388 | ascorbase | 9.0e-71 | 572 | 1091 | 551 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03389 | laccase | 0 | 571 | 1113 | 545 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| TIGR03389 | laccase | 0 | 31 | 568 | 539 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ACU17753.1 | 0 | 5 | 570 | 8 | 566 | unknown [Glycine max] |
| GenBank | ACU17753.1 | 0 | 580 | 1113 | 35 | 566 | unknown [Glycine max] |
| EMBL | CBI31651.1 | 0 | 4 | 1113 | 2 | 1094 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002264394.1 | 0 | 1 | 568 | 1 | 564 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002264394.1 | 0 | 580 | 1113 | 35 | 567 | PREDICTED: hypothetical protein [Vitis vinifera] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1asq_B | 0 | 572 | 1091 | 2 | 521 | A Chain A, Crystal Structure Of The Nasturtium Seedling Mutant Xyloglucanase Isoform Nxg1-Delta-Yniig |
| PDB | 1asq_B | 0 | 31 | 549 | 5 | 521 | A Chain A, Crystal Structure Of The Nasturtium Seedling Mutant Xyloglucanase Isoform Nxg1-Delta-Yniig |
| PDB | 1asq_A | 0 | 572 | 1091 | 2 | 521 | A Chain A, Crystal Structure Of The Nasturtium Seedling Mutant Xyloglucanase Isoform Nxg1-Delta-Yniig |
| PDB | 1asq_A | 0 | 31 | 549 | 5 | 521 | A Chain A, Crystal Structure Of The Nasturtium Seedling Mutant Xyloglucanase Isoform Nxg1-Delta-Yniig |
| PDB | 1asp_B | 0 | 572 | 1091 | 2 | 521 | A Chain A, Crystal Structure Of The Nasturtium Seedling Mutant Xyloglucanase Isoform Nxg1-Delta-Yniig |