

| Basic Information | |
|---|---|
| Species | Populus trichocarpa |
| Cazyme ID | Potri.001G092200.5 |
| Family | CBM49 |
| Protein Properties | Length: 378 Molecular Weight: 41077.5 Isoelectric Point: 9.6502 |
| Chromosome | Chromosome/Scaffold: 01 Start: 7243607 End: 7245980 |
| Description | glycosyl hydrolase 9C2 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM49 | 285 | 362 | 1.3e-37 |
| VAIVQKTTATWIAKGKTYYRYSTIVTNKSAKELKDIKLSISKLYGPLWGLTKSGNFYAFPSWITSLAAGKSLEFVYVH | |||
| GH9 | 1 | 243 | 0 |
| MYQATNNQYYLNYLGNNGDSMGGTGWGMTEFGWDVKYAGVQTLVAKFLMQGKAGHHAPVFEKYQQKAEYFMCSCLGKGSRNVQKTPGGLIFRQRWNNMQF VTSASFLTTVYSDYLASAGRNLNCAAGNVAPTQLLAFAKSQVDYILGDNPRATSYMVGYGNNYPRQVHHRGSSIVSYKVDPTFVTCRGGYATWFSRKGSD PNLLTGAIVGGPDAYDNFADERDNYEQTEPATYNNAPLLGLLA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 378 Download |
| MYQATNNQYY LNYLGNNGDS MGGTGWGMTE FGWDVKYAGV QTLVAKFLMQ GKAGHHAPVF 60 EKYQQKAEYF MCSCLGKGSR NVQKTPGGLI FRQRWNNMQF VTSASFLTTV YSDYLASAGR 120 NLNCAAGNVA PTQLLAFAKS QVDYILGDNP RATSYMVGYG NNYPRQVHHR GSSIVSYKVD 180 PTFVTCRGGY ATWFSRKGSD PNLLTGAIVG GPDAYDNFAD ERDNYEQTEP ATYNNAPLLG 240 LLARLSGGHG GYNQLLPVVV PAPIEKKPAP QTKITPAPAS TSAPVAIVQK TTATWIAKGK 300 TYYRYSTIVT NKSAKELKDI KLSISKLYGP LWGLTKSGNF YAFPSWITSL AAGKSLEFVY 360 VHSASAADVS VSSYTLA* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam00759 | Glyco_hydro_9 | 1.0e-75 | 1 | 243 | 244 | + Glycosyl hydrolase family 9. | ||
| PLN00119 | PLN00119 | 7.0e-92 | 20 | 246 | 227 | + endoglucanase | ||
| PLN02420 | PLN02420 | 2.0e-96 | 1 | 245 | 247 | + endoglucanase | ||
| PLN02340 | PLN02340 | 1.0e-120 | 1 | 360 | 363 | + endoglucanase | ||
| PLN02171 | PLN02171 | 0 | 1 | 377 | 381 | + endoglucanase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005576 | extracellular region |
| GO:0005975 | carbohydrate metabolic process |
| GO:0030246 | carbohydrate binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAI68019.1 | 0 | 1 | 377 | 246 | 620 | endo-beta-1,4-glucanase [Prunus persica] |
| EMBL | CAI68020.1 | 0 | 1 | 377 | 246 | 620 | endo-beta-1,4-glucanase [Prunus persica] |
| RefSeq | XP_002270880.1 | 0 | 1 | 377 | 249 | 629 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002299478.1 | 0 | 1 | 377 | 246 | 622 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002303645.1 | 0 | 1 | 376 | 246 | 621 | glycosyl hydrolase family 9 [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1ksd_A | 4e-26 | 1 | 245 | 222 | 428 | B Chain B, Reassembly And Co-Crystallization Of A Family 9 Processive Endoglucanase From Separately Expressed Gh9 And Cbm3c Modules |
| PDB | 1ksc_A | 4e-26 | 1 | 245 | 222 | 428 | B Chain B, Reassembly And Co-Crystallization Of A Family 9 Processive Endoglucanase From Separately Expressed Gh9 And Cbm3c Modules |
| PDB | 1ks8_A | 4e-26 | 1 | 245 | 222 | 428 | A Chain A, The Structure Of Endoglucanase From Termite, Nasutitermes Takasagoensis, At Ph 2.5. |
| PDB | 1ia7_A | 3e-20 | 82 | 266 | 291 | 441 | A Chain A, The Structure Of Endoglucanase From Termite, Nasutitermes Takasagoensis, At Ph 2.5. |
| PDB | 1ia6_A | 3e-20 | 82 | 266 | 291 | 441 | A Chain A, Crystal Structure Of The Cellulase Cel9m Of C. Cellulolyticum |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
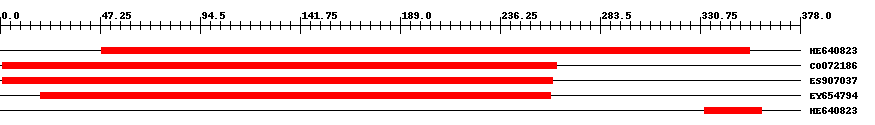 | ||||
| Hit | Length | Start | End | EValue |
| HE640823 | 308 | 48 | 354 | 0 |
| CO072186 | 263 | 1 | 263 | 0 |
| ES907037 | 261 | 1 | 261 | 0 |
| EY654794 | 242 | 19 | 260 | 0 |
| HE640823 | 28 | 333 | 360 | 2.5 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |