

| Basic Information | |
|---|---|
| Species | Physcomitrella patens |
| Cazyme ID | Pp1s54_189V6.2 |
| Family | GH13 |
| Protein Properties | Length: 776 Molecular Weight: 88383.8 Isoelectric Point: 6.3232 |
| Chromosome | Chromosome/Scaffold: 54 Start: 1157296 End: 1164372 |
| Description | starch branching enzyme 2.1 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH13 | 288 | 613 | 1.8e-26 |
| LPRIKANNYNTIQLMAIMEHAYYGCFGYHVTNFFAASSRSGTPEDLKYLIDKAHSMGLRVLMDVVHSHASTNAVDGLAGYNLDQTSQDSYFHSGARGYHK LWDSRLFNYGSWEVQRFLLSNLRWWMEEYMFDGFRFDGVTSMLYHHHGLNMCFTGNYHEYFSEATDVDAVVYLMLANELVHNLLRDATVIAEDVSGMPTL CRPVEEGGIGFDYRLAMAVPDKWIEYLKDRKDENWSMGDIVHTLTNRRYTEPCVGYAESHDQSMVGDKTFSFLLMDKEMYFNMSTQQPANLIVDRGIALH KMIHFITMALGGEGYLNFMGNEFGHP | |||
| Full Sequence |
|---|
| Protein Sequence Length: 776 Download |
| MLTVPACHGR STYPSLHPAS QIEGGSLASL DVLGLQRFPL KRSMSGIGLT RSLDFKAGSP 60 LVVKATGIKT QKTDRADNPA GWEDMSGMKK LGVMEVDPML TAHQDHLQYR FREYMKRKTE 120 IEKVEGSLEN FAKGFENFGF TRDGSCTVYR EWAPAAAAAQ LIGDFNNWDG SNHNLQRDEF 180 GVWSIRLPDE DGVSAVPHGS KVKFRMQKVD GTWVDRIPAW IKYAVVDPNV FAAYYDGVHW 240 DPPAAEKYQF KHARPEKPVA PIIYEAHVGM SSKEPVVTSY RKFADEVLPR IKANNYNTIQ 300 LMAIMEHAYY GCFGYHVTNF FAASSRSGTP EDLKYLIDKA HSMGLRVLMD VVHSHASTNA 360 VDGLAGYNLD QTSQDSYFHS GARGYHKLWD SRLFNYGSWE VQRFLLSNLR WWMEEYMFDG 420 FRFDGVTSML YHHHGLNMCF TGNYHEYFSE ATDVDAVVYL MLANELVHNL LRDATVIAED 480 VSGMPTLCRP VEEGGIGFDY RLAMAVPDKW IEYLKDRKDE NWSMGDIVHT LTNRRYTEPC 540 VGYAESHDQS MVGDKTFSFL LMDKEMYFNM STQQPANLIV DRGIALHKMI HFITMALGGE 600 GYLNFMGNEF GHPEWIDFPR DGNNWSFDKC RRRWDLLDNE QLRYKFMNNF NRAMIALEEE 660 FQFVSSSKQY ISCADESQKL IVFEKGDLVV VFNFHPTNTY SGLKVGCDVP GKYRICLDSD 720 AAEFGGHSRV DHKVDHFTSP EGEPGKPETN YNNRPHSFMI MAPSRSCQAY CRVPE* 780 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02960 | PLN02960 | 3.0e-11 | 39 | 189 | 162 | + alpha-amylase | ||
| PLN03244 | PLN03244 | 3.0e-139 | 193 | 737 | 548 | + alpha-amylase; Provisional | ||
| PLN02447 | PLN02447 | 0 | 87 | 775 | 689 | + 1,4-alpha-glucan-branching enzyme | ||
| cd11321 | AmyAc_bac_euk_BE | 0 | 246 | 652 | 407 | + Alpha amylase catalytic domain found in bacterial and eukaryotic branching enzymes. Branching enzymes (BEs) catalyze the formation of alpha-1,6 branch points in either glycogen or starch by cleavage of the alpha-1,4 glucosidic linkage yielding a non-reducing end oligosaccharide chain, and subsequent attachment to the alpha-1,6 position. By increasing the number of non-reducing ends, glycogen is more reactive to synthesis and digestion as well as being more soluble. This group includes bacterial and eukaryotic proteins. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. | ||
| PLN02960 | PLN02960 | 0 | 193 | 770 | 579 | + alpha-amylase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| GO:0043169 | cation binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI18866.1 | 0 | 63 | 775 | 70 | 780 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_001754772.1 | 0 | 88 | 775 | 1 | 688 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001762321.1 | 0 | 88 | 769 | 1 | 682 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001763855.1 | 0 | 88 | 775 | 1 | 688 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_002284841.1 | 0 | 63 | 775 | 47 | 757 | PREDICTED: hypothetical protein [Vitis vinifera] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3amk_A | 0 | 91 | 775 | 11 | 695 | A Chain A, Structure Of The Starch Branching Enzyme I (Bei) From Oryza Sativa L |
| PDB | 3aml_A | 0 | 91 | 775 | 11 | 695 | A Chain A, Structure Of The Starch Branching Enzyme I (Bei) From Oryza Sativa L |
| PDB | 3vu2_B | 0 | 91 | 775 | 11 | 695 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |
| PDB | 3vu2_A | 0 | 91 | 775 | 11 | 695 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |
| PDB | 1m7x_D | 0 | 141 | 726 | 20 | 577 | A Chain A, The X-Ray Crystallographic Structure Of Branching Enzyme |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| starch biosynthesis | RXN-7710 | EC-2.4.1.18 | 1,4-α-glucan branching enzyme |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
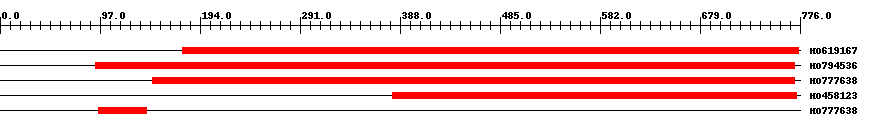 | ||||
| Hit | Length | Start | End | EValue |
| HO619167 | 600 | 177 | 775 | 0 |
| HO794536 | 691 | 93 | 771 | 0 |
| HO777638 | 636 | 148 | 771 | 0 |
| HO458123 | 404 | 381 | 773 | 0 |
| HO777638 | 47 | 96 | 142 | 0.00005 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |