

| Basic Information | |
|---|---|
| Species | Setaria italica |
| Cazyme ID | Si015393m |
| Family | CBM53 |
| Protein Properties | Length: 1508 Molecular Weight: 171171 Isoelectric Point: 5.2146 |
| Chromosome | Chromosome/Scaffold: 6 Start: 2776414 End: 2785871 |
| Description | starch synthase 3 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM53 | 625 | 708 | 1.5e-32 |
| STIDLYFNRDLSTLANEHDILIKGAFNGWKWRFFTEKLHMSEQGGDWWCCKLYIPKQAYRLDFVFFNGRTVYENNGNKDFMIQI | |||
| CBM53 | 961 | 1049 | 3.4e-33 |
| GTTVDVLYNPSNTVLTGKLEVWFRCSFNRWMHPGGVLTPQKMVKVENGSHLKATVNVPRDAYMMDFVFSEFEEGGIYDNRNGLDYHIPV | |||
| CBM53 | 801 | 887 | 1.6e-33 |
| AVRLYYNRSSRPLVHSTEIWMHGGYNNWIDGLSFSERLVHLDDKDGDWWYADVVLPERTFVLDWVFADGPPGNARNYDNNAWQDFHA | |||
| GT5 | 1060 | 1502 | 0 |
| HIVHIAVEMAPIAKVGGLADVVTSLSRAVQDLGHNVEVILPKHDCLNLSNVKNLHMHQSFSFGGSEIKVWCGLVEDLCVYFLEPQNGLFGVGCVYGRNDD LRFGFFCHSALEFLLQSGSSPHILHCHDWSSAPVAWLYKEHYAQSSLTNARVVFTIHNLEFGAHYIGKAMRYCDKATTVSNTYSREVSGHGAIAPHLGKF HGILNGIDPDIWDPYNDNFIPVHYTSENVVEGKSAAKKALQQKLGLQQNDVPIVGIVTRLTAQKGIHLIKHAIQRTLEWNGQVVLLGSAPDPRIQGDFVN FANTLHGVNYGRVRLCLTYDEPLSHLIYAGSDFILVPSIFEPCGLTQLVAMRYGAIPIVRKTGGLYDTVFDVDSDKERAWARGLEPNGFSFDGADSRGVD YALNRAISAWFDARSWFHSLCKRVMEQDWSWNRPALDYIELYH | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1508 Download |
| MEIALRPQSL LCSRSHQPLV IQPAFEGGGP VQSLLRTSRC TRSRIIRCTV ASSGVGTILL 60 KAYFSVGCFT DLMFSCTAYA DCSNRKSRRL VYPKIKVNAF RGYAPKVTIQ SSTEKSEHHD 120 CNEETIGTYN GQLSTDLAES TNGRDAENAK EDQAHNASSC SAFRETDVIE EVGLDLFKAE 180 FPGNALINIS LAEVEALDEA EVEEDKFEVD FSGIALSSAA VWALDSKDEA KAKEDTFVVD 240 LSRVAPYCAA AGELGDKQKQ LTSGLPEQDL CIVQFSEQNH AIVGSPKQDV GLLEQKQAIV 300 GSYEQDQSIV SSHEQDRSAD GSHSQDESIV GAPEQIRSIF GYHKSDQSIV GSYRQDKYTV 360 GLPENFKSIV GYYKSDEYVV VPHKQDEPIV GVPEQTQPVI SHRTPDRSIV GFPKHHHSIV 420 HIPEQKQSIV GFHIQDLSIV DSSMESQTKQ LAVVRTHDSL LTKKVEAKDG DYTSQKTDGD 480 TLNAKFDVDN LLQKYKEGFT EEAAEMTTST RIDEERLDMI EEKKGIRVDE WIVTEEGISM 540 SEADFLPLLC EKESSWVEDE VEITEDEEQY EVDEIPMFAE QDIQELPHHD VDQQALQRML 600 QELTIKNYSL GSKLFVFPEV LKADSTIDLY FNRDLSTLAN EHDILIKGAF NGWKWRFFTE 660 KLHMSEQGGD WWCCKLYIPK QAYRLDFVFF NGRTVYENNG NKDFMIQIKS TMDEHLFEDF 720 LVEDKKRELE RLAIEEAERR RQAEEQWRRE EEREADEADK VQTKVEVEIK KKKLHNVLGL 780 ARPSIDNLWY IEPVVTRQRA AVRLYYNRSS RPLVHSTEIW MHGGYNNWID GLSFSERLVH 840 LDDKDGDWWY ADVVLPERTF VLDWVFADGP PGNARNYDNN AWQDFHAILL NNMTEEEYWV 900 EEEQQIYRRL QQERRERKDS IKRKAERTAK MKDEMKKKTT KMFLLSQKHI VYTEPLEIRA 960 GTTVDVLYNP SNTVLTGKLE VWFRCSFNRW MHPGGVLTPQ KMVKVENGSH LKATVNVPRD 1020 AYMMDFVFSE FEEGGIYDNR NGLDYHIPVF GSTAKEPPIH IVHIAVEMAP IAKVGGLADV 1080 VTSLSRAVQD LGHNVEVILP KHDCLNLSNV KNLHMHQSFS FGGSEIKVWC GLVEDLCVYF 1140 LEPQNGLFGV GCVYGRNDDL RFGFFCHSAL EFLLQSGSSP HILHCHDWSS APVAWLYKEH 1200 YAQSSLTNAR VVFTIHNLEF GAHYIGKAMR YCDKATTVSN TYSREVSGHG AIAPHLGKFH 1260 GILNGIDPDI WDPYNDNFIP VHYTSENVVE GKSAAKKALQ QKLGLQQNDV PIVGIVTRLT 1320 AQKGIHLIKH AIQRTLEWNG QVVLLGSAPD PRIQGDFVNF ANTLHGVNYG RVRLCLTYDE 1380 PLSHLIYAGS DFILVPSIFE PCGLTQLVAM RYGAIPIVRK TGGLYDTVFD VDSDKERAWA 1440 RGLEPNGFSF DGADSRGVDY ALNRAISAWF DARSWFHSLC KRVMEQDWSW NRPALDYIEL 1500 YHSASKL* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02316 | PLN02316 | 7.0e-5 | 93 | 173 | 81 | + synthase/transferase | ||
| PLN02316 | PLN02316 | 5.0e-21 | 761 | 1056 | 322 | + synthase/transferase | ||
| PRK00654 | glgA | 8.0e-149 | 1060 | 1504 | 483 | + glycogen synthase; Provisional | ||
| cd03791 | GT1_Glycogen_synthase_DULL1_like | 8.0e-159 | 1060 | 1503 | 490 | + This family is most closely related to the GT1 family of glycosyltransferases. Glycogen synthase catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. | ||
| PLN02316 | PLN02316 | 0 | 593 | 1506 | 915 | + synthase/transferase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0009058 | biosynthetic process |
| GO:2001070 | starch binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ACC86846.1 | 0 | 1 | 448 | 1 | 450 | starch synthase IIIa precursor [Sorghum bicolor] |
| GenBank | ACC86846.1 | 0 | 256 | 1507 | 358 | 1684 | starch synthase IIIa precursor [Sorghum bicolor] |
| RefSeq | NP_001104881.1 | 1.99993e-41 | 1 | 457 | 1 | 425 | starch synthase DULL1 [Zea mays] |
| RefSeq | NP_001104881.1 | 0 | 161 | 1507 | 231 | 1674 | starch synthase DULL1 [Zea mays] |
| RefSeq | XP_002443985.1 | 0 | 1 | 1507 | 1 | 1664 | hypothetical protein SORBIDRAFT_07g005400 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3d1j_A | 0 | 1059 | 1502 | 1 | 474 | A Chain A, Crystal Structure Of E.Coli Gs Mutant Dmgs(C7s;c40 |
| PDB | 3guh_A | 0 | 1059 | 1502 | 1 | 474 | A Chain A, Crystal Structure Of E.Coli Gs Mutant Dmgs(C7s;c40 |
| PDB | 2r4u_A | 0 | 1059 | 1502 | 1 | 474 | A Chain A, Crystal Structure Of E.Coli Gs Mutant Dmgs(C7s;c40 |
| PDB | 2r4t_A | 0 | 1059 | 1502 | 1 | 474 | A Chain A, Crystal Structure Of E.Coli Gs Mutant Dmgs(C7s;c40 |
| PDB | 2qzs_A | 0 | 1059 | 1502 | 1 | 474 | A Chain A, Crystal Structure Of Wild-Type E.Coli Gs In Complex With Adp And Glucose(Wtgsb) |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| starch biosynthesis | GLYCOGENSYN-RXN | EC-2.4.1.21 | starch synthase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
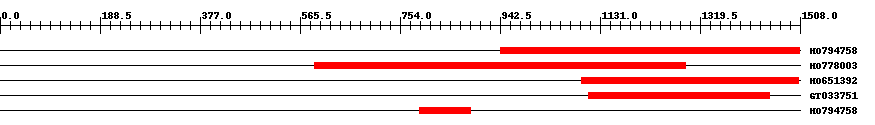 | ||||
| Hit | Length | Start | End | EValue |
| HO794758 | 566 | 943 | 1507 | 0 |
| HO778003 | 701 | 593 | 1292 | 0 |
| HO651392 | 412 | 1096 | 1506 | 0 |
| GT033751 | 342 | 1110 | 1451 | 0 |
| HO794758 | 99 | 790 | 886 | 8.8 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |