

| Basic Information | |
|---|---|
| Species | Volvox carteri |
| Cazyme ID | Vocar20012246m |
| Family | GT47 |
| Protein Properties | Length: 1512 Molecular Weight: 170372 Isoelectric Point: 7.0215 |
| Chromosome | Chromosome/Scaffold: 47 Start: 335983 End: 364785 |
| Description | exostosin family protein |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT47 | 1032 | 1298 | 9.10844e-44 |
| AQNRTRFSEWCYGAESALHEYLLLSEHRTFDPEEADFFYVPYYGTCMIWPVLHWADFPYFHTTGGPRILQVINMLIDTVDWINKMYPFWGRRGGRDHIFL FPHDEGACWAPNVLVNATWLTHWGRTDMIHESKTSFDADNYTRDYVGWRQPGGFVNLIRGHPCYDPVKIYRLAKENNWQDKHNILIGDAADVPGDYSDLL SRSLFCLVATGDGWSARTEDAVLHGCIPVIIIDGVHIKFETVFSVDEFSIRIPEANASRILEILKEI | |||
| GT47 | 303 | 655 | 0 |
| KRPLIYVYDLEPLYQSKILQYRISPPWCVHRRHDLPGNQTVWSDGWVYAADTLLHELLLISEHRTFDPEEADFFYVPHSASCLPFPMGSWADYPWFLGPG GPRIRQMVNMLREVVDWIDKTYPFWRRRGGRDHIWLFTHDEGACWAPKVLENSTWLTHWGRMGLEHRSGTAFLADKYDIDFVSPHQPEGFLTHIKGHPCY DSTKDLVVPAFKQPRHYRSSPLLGSATKQRDIFLFFRGDVGKHRMAHYSRGVRQKLYKLSVENNWKSKNVLIGGTHEVRGEYSDLLSRSQFCLVAAGDGW SARLEDAVLHGCIPVIVIDEVHVVFESILNVDSFAVRIDEQQLPQILDILAAI | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1512 Download |
| MYQRWLAIVL VAAFVSWHGA RALSEGTHSW QHAHVHSKER TFPLTEHAPL ALRCANTNGD 60 WCSKFLSQEP VPWKPAPRGS KDCPDKCNGV GRCNYDTGYC DCPAGDHEDP GTEPLSHIGP 120 DKRDLDWTEG GVTYSRCAGI CDDDTAICYC DGPLGRLLPP PGSAPGTPPI RRGRPLVTFH 180 MAPSTTWDGR KAFGEQPYNN VYGPQGYCNA TQPIWTPVCS LDDLGGPTCD DPIQAFCPGA 240 CSGHGTCNLG FCVCDEGYYG HDCARRRAGL PLLPSAVPTT PWLASVIREP PAAMEPPPHA 300 TRKRPLIYVY DLEPLYQSKI LQYRISPPWC VHRRHDLPGN QTVWSDGWVY AADTLLHELL 360 LISEHRTFDP EEADFFYVPH SASCLPFPMG SWADYPWFLG PGGPRIRQMV NMLREVVDWI 420 DKTYPFWRRR GGRDHIWLFT HDEGACWAPK VLENSTWLTH WGRMGLEHRS GTAFLADKYD 480 IDFVSPHQPE GFLTHIKGHP CYDSTKDLVV PAFKQPRHYR SSPLLGSATK QRDIFLFFRG 540 DVGKHRMAHY SRGVRQKLYK LSVENNWKSK NVLIGGTHEV RGEYSDLLSR SQFCLVAAGD 600 GWSARLEDAV LHGCIPVIVI DEVHVVFESI LNVDSFAVRI DEQQLPQILD ILAAIPERKI 660 RAKQAHLGHV WHRFRYGSLP GLASEIQSLT EANRAHQHQQ ELDIGQDQHA QGRRLVANDR 720 HALNTPHEGG GDPEFGRGAG HGEAADVQYP RPFRGDPAVD DAFATIMQWL YSRIPHTRTS 780 QIELPNLWAS LGSRCVYVTC FATRHLQVPL FLVSIGDTGT AFGQLSHERK VQLTDDAPLK 840 LRCSSIKGSW CMDFHQQQQG LSRQVQRRGQ VQLRHRLLRL SRGVDGAGVY DTAKEALYQR 900 FSVSVTMTWG SATVTDRWDA SRHLTTRRPT YDGKPVPGGQ QPYDNVYGPE GFCNATKPKW 960 APGGCGGPED LNGPYCDEPT ESFCPGACSG HGTCNLGFCV CDEGYYGHDC ARRRAGLPLL 1020 PSRLGELPWV AAQNRTRFSE WCYGAESALH EYLLLSEHRT FDPEEADFFY VPYYGTCMIW 1080 PVLHWADFPY FHTTGGPRIL QVINMLIDTV DWINKMYPFW GRRGGRDHIF LFPHDEGACW 1140 APNVLVNATW LTHWGRTDMI HESKTSFDAD NYTRDYVGWR QPGGFVNLIR GHPCYDPVKI 1200 YRLAKENNWQ DKHNILIGDA ADVPGDYSDL LSRSLFCLVA TGDGWSARTE DAVLHGCIPV 1260 IIIDGVHIKF ETVFSVDEFS IRIPEANASR ILEILKEIPK TKIRSIQAHL GRVWHRYRYA 1320 NLPGLASELR RLMVSNTADP LIREAAQLSA SEEVRLPRPF RGDPAVDDAF ATIMQWLYSR 1380 IPHTRQFQGR VDIGDSDAPY CGISVRNTKV AEELVWELFT QAGPVGKSLT IRDLRDCFKP 1440 PQHHAMICPR AVNVYMPKDR VTSQHQGYGF VEFKGEEDAD YRRTPFSSQP CISPHPSRFT 1500 LLQTFHSMDT L* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
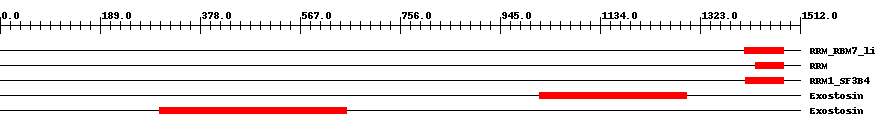
| cd12336 | RRM_RBM7_like | 9.0e-6 | 1407 | 1481 | 75 | + RNA recognition motif in RNA-binding protein 7 (RBM7) and similar proteins. This subfamily corresponds to the RRM of RBM7, RBM11 and their eukaryotic homologous. RBM7 is an ubiquitously expressed pre-mRNA splicing factor that enhances messenger RNA (mRNA) splicing in a cell-specific manner or in a certain developmental process, such as spermatogenesis. It interacts with splicing factors SAP145 (the spliceosomal splicing factor 3b subunit 2) and SRp20, and may play a more specific role in meiosis entry and progression. Together with additional testis-specific RNA-binding proteins, RBM7 may regulate the splicing of specific pre-mRNA species that are important in the meiotic cell cycle. RBM11 is a novel tissue-specific splicing regulator that is selectively expressed in brain, cerebellum and testis, and to a lower extent in kidney. It is localized in the nucleoplasm and enriched in SRSF2-containing splicing speckles. It may play a role in the modulation of alternative splicing during neuron and germ cell differentiation. Both, RBM7 and RBM11, contain an N-terminal RNA recognition motif (RRM), also termed RBD (RNA binding domain) or RNP (ribonucleoprotein domain), and a region lacking known homology at the C-terminus. The RRM is responsible for RNA binding, whereas the C-terminal region permits nuclear localization and homodimerization. | ||
| smart00360 | RRM | 3.0e-6 | 1427 | 1480 | 54 | + RNA recognition motif. | ||
| cd12334 | RRM1_SF3B4 | 3.0e-22 | 1409 | 1481 | 73 | + RNA recognition motif 1 in splicing factor 3B subunit 4 (SF3B4) and similar proteins. This subfamily corresponds to the RRM1 of SF3B4, also termed pre-mRNA-splicing factor SF3b 49 kDa (SF3b50), or spliceosome-associated protein 49 (SAP 49). SF3B4 a component of the multiprotein complex splicing factor 3b (SF3B), an integral part of the U2 small nuclear ribonucleoprotein (snRNP) and the U11/U12 di-snRNP. SF3B is essential for the accurate excision of introns from pre-messenger RNA, and is involved in the recognition of the pre-mRNA's branch site within the major and minor spliceosomes. SF3B4 functions to tether U2 snRNP with pre-mRNA at the branch site during spliceosome assembly. It is an evolutionarily highly conserved protein with orthologs across diverse species. SF3B4 contains two closely adjacent N-terminal RNA recognition motifs (RRMs), also termed RBDs (RNA binding domains) or RNPs (ribonucleoprotein domains). It binds directly to pre-mRNA and also interacts directly and highly specifically with another SF3B subunit called SAP 145. | ||
| pfam03016 | Exostosin | 1.0e-33 | 1020 | 1298 | 300 | + Exostosin family. The EXT family is a family of tumour suppressor genes. Mutations of EXT1 on 8q24.1, EXT2 on 11p11-13, and EXT3 on 19p have been associated with the autosomal dominant disorder known as hereditary multiple exostoses (HME). This is the most common known skeletal dysplasia. The chromosomal locations of other EXT genes suggest association with other forms of neoplasia. EXT1 and EXT2 have both been shown to encode a heparan sulphate polymerase with both D-glucuronyl (GlcA) and N-acetyl-D-glucosaminoglycan (GlcNAC) transferase activities. The nature of the defect in heparan sulphate biosynthesis in HME is unclear. | ||
| pfam03016 | Exostosin | 2.0e-44 | 302 | 655 | 355 | + Exostosin family. The EXT family is a family of tumour suppressor genes. Mutations of EXT1 on 8q24.1, EXT2 on 11p11-13, and EXT3 on 19p have been associated with the autosomal dominant disorder known as hereditary multiple exostoses (HME). This is the most common known skeletal dysplasia. The chromosomal locations of other EXT genes suggest association with other forms of neoplasia. EXT1 and EXT2 have both been shown to encode a heparan sulphate polymerase with both D-glucuronyl (GlcA) and N-acetyl-D-glucosaminoglycan (GlcNAC) transferase activities. The nature of the defect in heparan sulphate biosynthesis in HME is unclear. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0016020 | membrane |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_001693194.1 | 0 | 70 | 778 | 6 | 661 | exostosin-like glycosyltransferase [Chlamydomonas reinhardtii] |
| RefSeq | XP_001693194.1 | 0 | 928 | 1385 | 146 | 661 | exostosin-like glycosyltransferase [Chlamydomonas reinhardtii] |
| RefSeq | XP_001697525.1 | 0 | 105 | 673 | 5 | 593 | exostosin-like glycosyltransferase [Chlamydomonas reinhardtii] |
| RefSeq | XP_001700890.1 | 0 | 70 | 778 | 12 | 703 | exostosin-like glycosyltransferase [Chlamydomonas reinhardtii] |
| RefSeq | XP_001700890.1 | 0 | 928 | 1385 | 145 | 703 | exostosin-like glycosyltransferase [Chlamydomonas reinhardtii] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1x5u_A | 0.00000000000006 | 1407 | 1481 | 24 | 72 | A Chain A, Solution Structure Of Rrm Domain In Splicing Factor 3b |
| PDB | 2ygq_A | 0.005 | 951 | 1012 | 159 | 212 | A Chain A, Wif Domain-Epidermal Growth Factor (Egf)-Like Domains 1-3 Of Human Wnt Inhibitory Factor 1 In Complex With 1,2-Dipalmitoylphosphatidylcholine |