

| Basic Information | |
|---|---|
| Species | Volvox carteri |
| Cazyme ID | Vocar20013124m |
| Family | GH20 |
| Protein Properties | Length: 887 Molecular Weight: 94627.7 Isoelectric Point: 9.3641 |
| Chromosome | Chromosome/Scaffold: 30 Start: 1422926 End: 1432603 |
| Description | beta-hexosaminidase 3 |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH20 | 219 | 300 | 8.1e-23 |
| DWPRFTHRGLLVDTARHFLPLSVLHTTLDAMAANKLNVLHWHATDDQSFPMEALVASEGGAVSDLATRGSFGDRMLYSREEV | |||
| Full Sequence |
|---|
| Protein Sequence Length: 887 Download |
| MSGLVLALVA LCAGVVTASA SLKTFAGIWP QPRSQRPVAT FYVGQHTGSL TTRHAATTAY 60 KAAVKPPSLA RGRHQGSRQY GRTPALTCAS RFYMVSIFPL LVLLVALFLP GSVPDLCFCQ 120 GQSPTPSSPG PKIVLQVDNQ DAQLGPAMDE SYQIQVPGRG SRGTRGEVLI QAATQVGAVH 180 ALETLAQLVV SYDHVKQQQP QQQLEGPGPL WLLAGNISDW PRFTHRGLLV DTARHFLPLS 240 VLHTTLDAMA ANKLNVLHWH ATDDQSFPME ALVASEGGAV SDLATRGSFG DRMLYSREEV 300 AGRPRTRRWV PYWVLLRRCS PIRSFTRGEM KLTWTAGGAT LLYGSGWSHT GLAGRSYSAL 360 IRTCCRSLTS PTAAHSLVVR LFPTCRTAVL PLLAAAYYMS RVLRGIVNGG SDSGGGSRGP 420 PRTPVVWQEA FDVAGGTGLP PGTIVQVWQS DVGPVGRTKI TAATGLVGGA SHGAGAAARA 480 ASGGEGVDLA SFDHNHGHMD LDDSDPNPYL DPYGDLGLED EEELMELLEA GEIAQEVAAG 540 RQLLFMEGLL GLLRRKEEPA VVAGGVGAVR GGGGGGGGGG GFDGGAEEAG GPRKRKKMKK 600 MKKMKKMKKM KKRAPGGGGR GGGGLGGGVG LRGGGGAHHA VGAGYDDADN DLYRRLRLGR 660 RLRYYQGKHE DDGVEYLQPY LHPVVRGIGR GTPSSSSSSS SSSSPSATAA AQQQQQQQQH 720 HHHHHHQDPD PELRSELQRL REAVAAADGA PGGGSSGSSA SRRTQAEELA AVTAAGFQAI 780 LSSGWYLDWI SWGEDWRRFY SQEPLGFEGS EEQKARVLGG EACMWGEYVD ATNLISRTWP 840 RASAVAERLW SDAAVRDEEE AGQRLRVHRC RMVRRRAEKR GECEGR* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
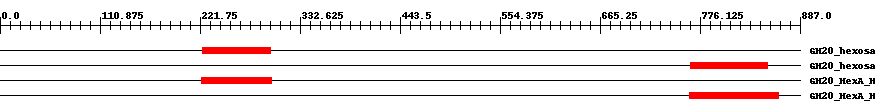 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd02742 | GH20_hexosaminidase | 1.0e-14 | 225 | 300 | 76 | + Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. | ||
| cd02742 | GH20_hexosaminidase | 2.0e-22 | 766 | 851 | 88 | + Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. | ||
| cd06562 | GH20_HexA_HexB-like | 3.0e-26 | 223 | 301 | 79 | + Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. | ||
| cd06562 | GH20_HexA_HexB-like | 2.0e-38 | 764 | 863 | 107 | + Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0004563 | beta-N-acetylhexosaminidase activity |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABQ22431.1 | 3.00004e-41 | 767 | 875 | 76 | 184 | beta-hexosaminidase beta chain precursor-like protein [Callithrix jacchus] |
| GenBank | EDM10130.1 | 9.80909e-45 | 768 | 875 | 93 | 200 | rCG44661, isoform CRA_a [Rattus norvegicus] |
| GenBank | EFB14981.1 | 4.99997e-41 | 770 | 875 | 335 | 440 | hypothetical protein PANDA_005458 [Ailuropoda melanoleuca] |
| RefSeq | XP_001513475.1 | 7e-24 | 133 | 473 | 146 | 473 | PREDICTED: similar to PSP94-like protein [Ornithorhynchus anatinus] |
| RefSeq | XP_001513475.1 | 1.99993e-41 | 766 | 875 | 446 | 555 | PREDICTED: similar to PSP94-like protein [Ornithorhynchus anatinus] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2gk1_H | 5e-39 | 767 | 875 | 382 | 490 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_H | 1e-26 | 141 | 463 | 89 | 390 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_F | 5e-39 | 767 | 875 | 382 | 490 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_F | 1e-26 | 141 | 463 | 89 | 390 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_D | 5e-39 | 767 | 875 | 382 | 490 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |