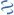MultiGeneBlast hits

Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028287 : Acidovorax avenae subsp. avenae strain AA78_5 chromosome. Total score: 1.0 Cumulative Blast bit score: 195
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8241_01345
C8241_01350
branched-chain amino acid transaminase
Accession: AVS60532
Location: 354758-355696
NCBI BlastP on this gene
Accession: AVS60532
Location: 354758-355696
NCBI BlastP on this gene
C8241_01355
zinc-finger domain-containing protein
Accession: AVS60533
Location: 355738-355944
NCBI BlastP on this gene
Accession: AVS60533
Location: 355738-355944
NCBI BlastP on this gene
C8241_01360
capsular biosynthesis protein
Accession: AVS63922
Location: 355983-356870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 97
Sequence coverage: 67 %
E-value: 9e-21
NCBI BlastP on this gene
Accession: AVS63922
Location: 355983-356870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 97
Sequence coverage: 67 %
E-value: 9e-21
NCBI BlastP on this gene
C8241_01365
C8241_01370
C8241_01375
C8241_01380
C8241_01385
mannosyltransferase
Accession: AVS60536
Location: 360670-361701
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVS60536
Location: 360670-361701
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8241_01390
C8241_01395
LysR family transcriptional regulator
Accession: AVS60538
Location: 362816-363700
NCBI BlastP on this gene
Accession: AVS60538
Location: 362816-363700
NCBI BlastP on this gene
C8241_01400
C8241_01405
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028290 : Acidovorax cattleyae strain CAT98_1 chromosome. Total score: 1.0 Cumulative Blast bit score: 194
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8240_01065
C8240_01070
branched-chain amino acid transaminase
Accession: AVS72831
Location: 348314-349252
NCBI BlastP on this gene
Accession: AVS72831
Location: 348314-349252
NCBI BlastP on this gene
C8240_01075
zinc-finger domain-containing protein
Accession: AVS72832
Location: 349290-349496
NCBI BlastP on this gene
Accession: AVS72832
Location: 349290-349496
NCBI BlastP on this gene
C8240_01080
capsular biosynthesis protein
Accession: AVS76217
Location: 349537-350427
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS76217
Location: 349537-350427
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8240_01085
C8240_01090
C8240_01095
C8240_01100
C8240_01105
C8240_01110
mannosyltransferase
Accession: AVS72838
Location: 354376-355401
BlastP hit with GL636894_1983_2612_-
Percentage identity: 36 %
BlastP bit score: 100
Sequence coverage: 70 %
E-value: 9e-22
NCBI BlastP on this gene
Accession: AVS72838
Location: 354376-355401
BlastP hit with GL636894_1983_2612_-
Percentage identity: 36 %
BlastP bit score: 100
Sequence coverage: 70 %
E-value: 9e-22
NCBI BlastP on this gene
C8240_01115
LysR family transcriptional regulator
Accession: AVS72839
Location: 357086-357406
NCBI BlastP on this gene
Accession: AVS72839
Location: 357086-357406
NCBI BlastP on this gene
C8240_01120
C8240_01125
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028302 : Acidovorax avenae subsp. avenae isolate SF12 chromosome. Total score: 1.0 Cumulative Blast bit score: 193
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8244_01120
C8244_01125
branched-chain amino acid transaminase
Accession: AVT14961
Location: 360944-361882
NCBI BlastP on this gene
Accession: AVT14961
Location: 360944-361882
NCBI BlastP on this gene
C8244_01130
zinc-finger domain-containing protein
Accession: AVT14962
Location: 361924-362130
NCBI BlastP on this gene
Accession: AVT14962
Location: 361924-362130
NCBI BlastP on this gene
C8244_01135
capsular biosynthesis protein
Accession: AVT18179
Location: 362168-363058
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
Accession: AVT18179
Location: 362168-363058
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
C8244_01140
C8244_01145
C8244_01150
C8244_01155
mannosyltransferase
Accession: AVT14966
Location: 366862-367893
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVT14966
Location: 366862-367893
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8244_01160
C8244_01165
LysR family transcriptional regulator
Accession: AVT14968
Location: 369008-369892
NCBI BlastP on this gene
Accession: AVT14968
Location: 369008-369892
NCBI BlastP on this gene
C8244_01170
C8244_01175
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028297 : Acidovorax avenae subsp. avenae strain MOR chromosome. Total score: 1.0 Cumulative Blast bit score: 193
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8236_01480
C8236_01485
branched chain amino acid aminotransferase
Accession: AVS97630
Location: 365975-366913
NCBI BlastP on this gene
Accession: AVS97630
Location: 365975-366913
NCBI BlastP on this gene
C8236_01490
zinc-finger domain-containing protein
Accession: AVS97631
Location: 366955-367161
NCBI BlastP on this gene
Accession: AVS97631
Location: 366955-367161
NCBI BlastP on this gene
C8236_01495
capsular biosynthesis protein
Accession: AVT00759
Location: 367200-368087
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 6e-20
NCBI BlastP on this gene
Accession: AVT00759
Location: 367200-368087
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 6e-20
NCBI BlastP on this gene
C8236_01500
C8236_01505
C8236_01510
C8236_01515
mannosyltransferase
Accession: AVS97635
Location: 371891-372922
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVS97635
Location: 371891-372922
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C8236_01520
C8236_01525
LysR family transcriptional regulator
Accession: AVS97637
Location: 374037-374921
NCBI BlastP on this gene
Accession: AVS97637
Location: 374037-374921
NCBI BlastP on this gene
C8236_01530
C8236_01535
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028292 : Acidovorax avenae subsp. avenae strain INDB2 chromosome. Total score: 1.0 Cumulative Blast bit score: 193
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8237_01140
C8237_01145
branched-chain amino acid transaminase
Accession: AVS79845
Location: 370722-371660
NCBI BlastP on this gene
Accession: AVS79845
Location: 370722-371660
NCBI BlastP on this gene
C8237_01150
zinc-finger domain-containing protein
Accession: AVS79846
Location: 371702-371908
NCBI BlastP on this gene
Accession: AVS79846
Location: 371702-371908
NCBI BlastP on this gene
C8237_01155
capsular biosynthesis protein
Accession: AVS83051
Location: 371946-372836
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
Accession: AVS83051
Location: 371946-372836
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
C8237_01160
C8237_01165
C8237_01170
C8237_01175
C8237_01180
mannosyltransferase
Accession: AVS79851
Location: 376884-377915
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVS79851
Location: 376884-377915
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8237_01185
C8237_01190
LysR family transcriptional regulator
Accession: AVS79853
Location: 379030-379914
NCBI BlastP on this gene
Accession: AVS79853
Location: 379030-379914
NCBI BlastP on this gene
C8237_01195
C8237_01200
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028303 : Acidovorax avenae subsp. avenae strain SH7 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C7Y68_01460
C7Y68_01465
branched-chain amino acid transaminase
Accession: AVT18817
Location: 379755-380693
NCBI BlastP on this gene
Accession: AVT18817
Location: 379755-380693
NCBI BlastP on this gene
C7Y68_01470
zinc-finger domain-containing protein
Accession: AVT18818
Location: 380735-380941
NCBI BlastP on this gene
Accession: AVT18818
Location: 380735-380941
NCBI BlastP on this gene
C7Y68_01475
capsular biosynthesis protein
Accession: AVT22261
Location: 380980-381870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVT22261
Location: 380980-381870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C7Y68_01480
C7Y68_01485
C7Y68_01490
C7Y68_01495
mannosyltransferase
Accession: AVT18822
Location: 385674-386705
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVT18822
Location: 385674-386705
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C7Y68_01500
C7Y68_01505
LysR family transcriptional regulator
Accession: AVT18824
Location: 387820-388704
NCBI BlastP on this gene
Accession: AVT18824
Location: 387820-388704
NCBI BlastP on this gene
C7Y68_01510
C7Y68_01515
C7Y68_01520
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028300 : Acidovorax avenae subsp. avenae strain QHB1 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8242_01295
C8242_01300
branched-chain amino acid transaminase
Accession: AVT08274
Location: 372258-373196
NCBI BlastP on this gene
Accession: AVT08274
Location: 372258-373196
NCBI BlastP on this gene
C8242_01305
zinc-finger domain-containing protein
Accession: AVT08275
Location: 373238-373444
NCBI BlastP on this gene
Accession: AVT08275
Location: 373238-373444
NCBI BlastP on this gene
C8242_01310
capsular biosynthesis protein
Accession: AVT11131
Location: 373483-374373
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVT11131
Location: 373483-374373
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8242_01315
C8242_01320
C8242_01325
C8242_01330
mannosyltransferase
Accession: AVT08279
Location: 378176-379207
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVT08279
Location: 378176-379207
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C8242_01335
C8242_01340
LysR family transcriptional regulator
Accession: AVT08281
Location: 380322-381206
NCBI BlastP on this gene
Accession: AVT08281
Location: 380322-381206
NCBI BlastP on this gene
C8242_01345
C8242_01350
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028298 : Acidovorax avenae subsp. avenae strain NCT3 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8243_00950
C8243_00955
branched-chain amino acid transaminase
Accession: AVT01204
Location: 364567-365505
NCBI BlastP on this gene
Accession: AVT01204
Location: 364567-365505
NCBI BlastP on this gene
C8243_00960
zinc-finger domain-containing protein
Accession: AVT01205
Location: 365547-365753
NCBI BlastP on this gene
Accession: AVT01205
Location: 365547-365753
NCBI BlastP on this gene
C8243_00965
capsular biosynthesis protein
Accession: AVT04260
Location: 365792-366682
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVT04260
Location: 365792-366682
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8243_00970
C8243_00975
C8243_00980
C8243_00985
mannosyltransferase
Accession: AVT01209
Location: 370485-371516
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVT01209
Location: 370485-371516
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8243_00990
C8243_00995
LysR family transcriptional regulator
Accession: AVT01211
Location: 372631-373515
NCBI BlastP on this gene
Accession: AVT01211
Location: 372631-373515
NCBI BlastP on this gene
C8243_01000
C8243_01005
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028296 : Acidovorax avenae subsp. avenae strain MDB1 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8232_00180
C8232_00185
branched-chain amino acid transaminase
Accession: AVS94857
Location: 362617-363555
NCBI BlastP on this gene
Accession: AVS94857
Location: 362617-363555
NCBI BlastP on this gene
C8232_00190
zinc-finger domain-containing protein
Accession: AVS94858
Location: 363597-363803
NCBI BlastP on this gene
Accession: AVS94858
Location: 363597-363803
NCBI BlastP on this gene
C8232_00195
capsular biosynthesis protein
Accession: AVS97109
Location: 363842-364732
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS97109
Location: 363842-364732
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8232_00200
C8232_00205
C8232_00210
C8232_00215
mannosyltransferase
Accession: AVS94862
Location: 368535-369566
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVS94862
Location: 368535-369566
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C8232_00220
C8232_00225
LysR family transcriptional regulator
Accession: AVS94864
Location: 370681-371565
NCBI BlastP on this gene
Accession: AVS94864
Location: 370681-371565
NCBI BlastP on this gene
C8232_00230
C8232_00235
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028288 : Acidovorax avenae subsp. avenae strain AA81_1 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8245_04500
LysR family transcriptional regulator
Accession: AVS65061
Location: 998017-998901
NCBI BlastP on this gene
Accession: AVS65061
Location: 998017-998901
NCBI BlastP on this gene
C8245_04505
C8245_04510
mannosyltransferase
Accession: AVS65063
Location: 1000016-1001047
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 99
Sequence coverage: 66 %
E-value: 4e-21
NCBI BlastP on this gene
Accession: AVS65063
Location: 1000016-1001047
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 99
Sequence coverage: 66 %
E-value: 4e-21
NCBI BlastP on this gene
C8245_04515
glycosyl transferase family 2
Accession: AVS65064
Location: 1001044-1002105
NCBI BlastP on this gene
Accession: AVS65064
Location: 1001044-1002105
NCBI BlastP on this gene
C8245_04520
C8245_04525
C8245_04530
capsular biosynthesis protein
Accession: AVS68648
Location: 1004851-1005738
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS68648
Location: 1004851-1005738
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8245_04535
zinc-finger domain-containing protein
Accession: AVS65067
Location: 1005777-1005983
NCBI BlastP on this gene
Accession: AVS65067
Location: 1005777-1005983
NCBI BlastP on this gene
C8245_04540
branched-chain amino acid transaminase
Accession: AVS65068
Location: 1006025-1006963
NCBI BlastP on this gene
Accession: AVS65068
Location: 1006025-1006963
NCBI BlastP on this gene
C8245_04545
C8245_04550
C8245_04555
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP002521 : Acidovorax avenae subsp. avenae ATCC 19860 Total score: 1.0 Cumulative Blast bit score: 192
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
Acav_0319
Acav_0320
branched-chain amino acid aminotransferase
Accession: ADX44244
Location: 360935-361873
NCBI BlastP on this gene
Accession: ADX44244
Location: 360935-361873
NCBI BlastP on this gene
Acav_0321
Acav_0322
Capsular polysaccharide synthesis
Accession: ADX44246
Location: 362160-363149
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: ADX44246
Location: 362160-363149
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Acav_0323
Acav_0324
Acav_0325
Acav_0326
Capsular polysaccharide synthesis
Accession: ADX44250
Location: 366851-367882
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: ADX44250
Location: 366851-367882
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Acav_0327
Acav_0328
transcriptional regulator, LysR family
Accession: ADX44252
Location: 368997-369881
NCBI BlastP on this gene
Accession: ADX44252
Location: 368997-369881
NCBI BlastP on this gene
Acav_0329
protein of unknown function DUF1228
Accession: ADX44253
Location: 369951-371225
NCBI BlastP on this gene
Accession: ADX44253
Location: 369951-371225
NCBI BlastP on this gene
Acav_0330
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028295 : Acidovorax avenae subsp. avenae strain MD5 chromosome. Total score: 1.0 Cumulative Blast bit score: 191
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8246_13775
LysR family transcriptional regulator
Accession: AVS92686
Location: 3063597-3064481
NCBI BlastP on this gene
Accession: AVS92686
Location: 3063597-3064481
NCBI BlastP on this gene
C8246_13780
C8246_13785
mannosyltransferase
Accession: AVS92688
Location: 3065596-3066633
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVS92688
Location: 3065596-3066633
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
C8246_13790
glycosyl transferase family 2
Accession: AVS92689
Location: 3066630-3067691
NCBI BlastP on this gene
Accession: AVS92689
Location: 3066630-3067691
NCBI BlastP on this gene
C8246_13795
C8246_13800
C8246_13805
capsular biosynthesis protein
Accession: AVS94633
Location: 3070437-3071327
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS94633
Location: 3070437-3071327
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8246_13810
zinc-finger domain-containing protein
Accession: AVS92692
Location: 3071366-3071572
NCBI BlastP on this gene
Accession: AVS92692
Location: 3071366-3071572
NCBI BlastP on this gene
C8246_13815
branched-chain amino acid transaminase
Accession: AVS92693
Location: 3071614-3072552
NCBI BlastP on this gene
Accession: AVS92693
Location: 3071614-3072552
NCBI BlastP on this gene
C8246_13820
C8246_13825
C8246_13830
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028294 : Acidovorax avenae subsp. avenae strain KL3 chromosome. Total score: 1.0 Cumulative Blast bit score: 191
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8238_01105
C8238_01110
branched-chain amino acid transaminase
Accession: AVS87029
Location: 353399-354337
NCBI BlastP on this gene
Accession: AVS87029
Location: 353399-354337
NCBI BlastP on this gene
C8238_01115
zinc-finger domain-containing protein
Accession: AVS87030
Location: 354379-354585
NCBI BlastP on this gene
Accession: AVS87030
Location: 354379-354585
NCBI BlastP on this gene
C8238_01120
capsular biosynthesis protein
Accession: AVS90195
Location: 354624-355514
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS90195
Location: 354624-355514
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8238_01125
C8238_01130
C8238_01135
C8238_01140
mannosyltransferase
Accession: AVS87034
Location: 359317-360354
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVS87034
Location: 359317-360354
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
C8238_01145
C8238_01150
LysR family transcriptional regulator
Accession: AVS87036
Location: 361469-362353
NCBI BlastP on this gene
Accession: AVS87036
Location: 361469-362353
NCBI BlastP on this gene
C8238_01155
C8238_01160
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028293 : Acidovorax avenae subsp. avenae strain INV chromosome. Total score: 1.0 Cumulative Blast bit score: 191
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8239_01575
C8239_01580
branched-chain amino acid transaminase
Accession: AVS83605
Location: 354178-355116
NCBI BlastP on this gene
Accession: AVS83605
Location: 354178-355116
NCBI BlastP on this gene
C8239_01585
zinc-finger domain-containing protein
Accession: AVS83606
Location: 355158-355364
NCBI BlastP on this gene
Accession: AVS83606
Location: 355158-355364
NCBI BlastP on this gene
C8239_01590
capsular biosynthesis protein
Accession: AVS86546
Location: 355403-356293
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS86546
Location: 355403-356293
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8239_01595
C8239_01600
C8239_01605
C8239_01610
mannosyltransferase
Accession: AVS83610
Location: 360097-361134
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVS83610
Location: 360097-361134
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
C8239_01615
C8239_01620
LysR family transcriptional regulator
Accession: AVS83612
Location: 362249-363133
NCBI BlastP on this gene
Accession: AVS83612
Location: 362249-363133
NCBI BlastP on this gene
C8239_01625
C8239_01630
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP028289 : Acidovorax avenae subsp. avenae strain AA99_2 chromosome. Total score: 1.0 Cumulative Blast bit score: 191
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8247_01030
C8247_01035
branched-chain amino acid transaminase
Accession: AVS69168
Location: 354803-355741
NCBI BlastP on this gene
Accession: AVS69168
Location: 354803-355741
NCBI BlastP on this gene
C8247_01040
zinc-finger domain-containing protein
Accession: AVS69169
Location: 355783-355989
NCBI BlastP on this gene
Accession: AVS69169
Location: 355783-355989
NCBI BlastP on this gene
C8247_01045
capsular biosynthesis protein
Accession: AVS72347
Location: 356028-356918
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 3e-19
NCBI BlastP on this gene
Accession: AVS72347
Location: 356028-356918
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 3e-19
NCBI BlastP on this gene
C8247_01050
C8247_01055
C8247_01060
C8247_01065
mannosyltransferase
Accession: AVS69173
Location: 360722-361753
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 6e-21
NCBI BlastP on this gene
Accession: AVS69173
Location: 360722-361753
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 6e-21
NCBI BlastP on this gene
C8247_01070
C8247_01075
LysR family transcriptional regulator
Accession: AVS69175
Location: 362868-363752
NCBI BlastP on this gene
Accession: AVS69175
Location: 362868-363752
NCBI BlastP on this gene
C8247_01080
C8247_01085
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP044016 : Arachidicoccus sp. B3-10 chromosome Total score: 1.0 Cumulative Blast bit score: 174
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
glycerol-3-phosphate dehydrogenase
Accession: QES89184
Location: 2575778-2576725
NCBI BlastP on this gene
Accession: QES89184
Location: 2575778-2576725
NCBI BlastP on this gene
E0W69_011090
NAD-dependent epimerase/dehydratase family protein
Accession: E0W69_011095
Location: 2576739-2577721
NCBI BlastP on this gene
Accession: E0W69_011095
Location: 2576739-2577721
NCBI BlastP on this gene
E0W69_011095
E0W69_011100
capsular biosynthesis protein
Accession: QES89186
Location: 2579014-2579943
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 69
Sequence coverage: 48 %
E-value: 6e-11
NCBI BlastP on this gene
Accession: QES89186
Location: 2579014-2579943
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 69
Sequence coverage: 48 %
E-value: 6e-11
NCBI BlastP on this gene
E0W69_011105
glycosyltransferase family 4 protein
Accession: QES89187
Location: 2579991-2581187
NCBI BlastP on this gene
Accession: QES89187
Location: 2579991-2581187
NCBI BlastP on this gene
E0W69_011110
glycosyltransferase family 4 protein
Accession: QES89188
Location: 2581205-2582293
NCBI BlastP on this gene
Accession: QES89188
Location: 2581205-2582293
NCBI BlastP on this gene
E0W69_011115
E0W69_011120
hypothetical protein
Accession: QES89190
Location: 2583330-2584277
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 105
Sequence coverage: 67 %
E-value: 2e-23
NCBI BlastP on this gene
Accession: QES89190
Location: 2583330-2584277
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 105
Sequence coverage: 67 %
E-value: 2e-23
NCBI BlastP on this gene
E0W69_011125
E0W69_011130
glycosyltransferase family 2 protein
Accession: QES89192
Location: 2585610-2586629
NCBI BlastP on this gene
Accession: QES89192
Location: 2585610-2586629
NCBI BlastP on this gene
E0W69_011135
beta-1,6-N-acetylglucosaminyltransferase
Accession: QES89193
Location: 2586640-2587530
NCBI BlastP on this gene
Accession: QES89193
Location: 2586640-2587530
NCBI BlastP on this gene
E0W69_011140
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP049868 : Pedobacter sp. HDW13 chromosome Total score: 1.0 Cumulative Blast bit score: 172
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
G7074_10860
G7074_10865
glycosyltransferase family 4 protein
Accession: QIL39724
Location: 2563276-2564181
NCBI BlastP on this gene
Accession: QIL39724
Location: 2563276-2564181
NCBI BlastP on this gene
G7074_10870
G7074_10875
G7074_10880
G7074_10885
hypothetical protein
Accession: QIL39728
Location: 2567273-2567776
BlastP hit with GL636894_880_1674_-
Percentage identity: 36 %
BlastP bit score: 83
Sequence coverage: 44 %
E-value: 4e-16
NCBI BlastP on this gene
Accession: QIL39728
Location: 2567273-2567776
BlastP hit with GL636894_880_1674_-
Percentage identity: 36 %
BlastP bit score: 83
Sequence coverage: 44 %
E-value: 4e-16
NCBI BlastP on this gene
G7074_10890
hypothetical protein
Accession: QIL39729
Location: 2567746-2568003
BlastP hit with GL636894_880_1674_-
Percentage identity: 50 %
BlastP bit score: 89
Sequence coverage: 31 %
E-value: 4e-19
NCBI BlastP on this gene
Accession: QIL39729
Location: 2567746-2568003
BlastP hit with GL636894_880_1674_-
Percentage identity: 50 %
BlastP bit score: 89
Sequence coverage: 31 %
E-value: 4e-19
NCBI BlastP on this gene
G7074_10895
ABC transporter ATP-binding protein
Accession: G7074_10900
Location: 2568109-2569943
NCBI BlastP on this gene
Accession: G7074_10900
Location: 2568109-2569943
NCBI BlastP on this gene
G7074_10900
glycosyltransferase family 2 protein
Accession: QIL39730
Location: 2570072-2571058
NCBI BlastP on this gene
Accession: QIL39730
Location: 2570072-2571058
NCBI BlastP on this gene
G7074_10905
glycosyltransferase family 2 protein
Accession: G7074_10910
Location: 2571059-2571933
NCBI BlastP on this gene
Accession: G7074_10910
Location: 2571059-2571933
NCBI BlastP on this gene
G7074_10910
methylmalonyl Co-A mutase-associated GTPase MeaB
Accession: QIL39731
Location: 2571993-2572889
NCBI BlastP on this gene
Accession: QIL39731
Location: 2571993-2572889
NCBI BlastP on this gene
meaB
aminopeptidase P family protein
Accession: QIL39732
Location: 2572941-2574230
NCBI BlastP on this gene
Accession: QIL39732
Location: 2572941-2574230
NCBI BlastP on this gene
G7074_10920
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP018259 : Acinetobacter bereziniae strain XH901 Total score: 1.0 Cumulative Blast bit score: 167
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
BSR55_03865
BSR55_03860
lipid A biosynthesis acyltransferase
Accession: ATZ62537
Location: 828310-829176
NCBI BlastP on this gene
Accession: ATZ62537
Location: 828310-829176
NCBI BlastP on this gene
BSR55_03855
capsular biosynthesis protein
Accession: ATZ62536
Location: 827288-828295
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 73
Sequence coverage: 61 %
E-value: 2e-12
NCBI BlastP on this gene
Accession: ATZ62536
Location: 827288-828295
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 73
Sequence coverage: 61 %
E-value: 2e-12
NCBI BlastP on this gene
BSR55_03850
BSR55_03845
BSR55_03840
BSR55_03835
BSR55_03830
hypothetical protein
Accession: ATZ62531
Location: 822890-823804
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 94
Sequence coverage: 67 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: ATZ62531
Location: 822890-823804
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 94
Sequence coverage: 67 %
E-value: 2e-19
NCBI BlastP on this gene
BSR55_03825
BSR55_03820
capsular polysaccharide biosynthesis protein
Accession: ATZ62529
Location: 821027-822034
NCBI BlastP on this gene
Accession: ATZ62529
Location: 821027-822034
NCBI BlastP on this gene
BSR55_03815
BSR55_03810
BSR55_03805
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP037934 : Marinobacter sp. JH2 chromosome Total score: 1.0 Cumulative Blast bit score: 144
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
hypothetical protein
Accession: QBM18131
Location: 2476210-2477229
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 102 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QBM18131
Location: 2476210-2477229
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 102 %
E-value: 2e-36
NCBI BlastP on this gene
MARI_22600
MARI_22590
MARI_22580
lipid A biosynthesis lauroyltransferase
Accession: QBM18128
Location: 2473173-2474132
NCBI BlastP on this gene
Accession: QBM18128
Location: 2473173-2474132
NCBI BlastP on this gene
lpxL_1
3-deoxy-D-manno-octulosonic acid transferase
Accession: QBM18127
Location: 2471817-2473070
NCBI BlastP on this gene
Accession: QBM18127
Location: 2471817-2473070
NCBI BlastP on this gene
waaA
tolC
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP002637 : Selenomonas sputigena ATCC 35185 Total score: 1.0 Cumulative Blast bit score: 140
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
Selsp_1174
polysaccharide pyruvyl transferase CsaB
Accession: AEC00133
Location: 1314550-1315668
NCBI BlastP on this gene
Accession: AEC00133
Location: 1314550-1315668
NCBI BlastP on this gene
Selsp_1173
UspA domain-containing protein
Accession: AEC00132
Location: 1313992-1314372
NCBI BlastP on this gene
Accession: AEC00132
Location: 1313992-1314372
NCBI BlastP on this gene
Selsp_1172
Selsp_1171
prolipoprotein diacylglyceryl transferase
Accession: AEC00130
Location: 1312190-1313005
NCBI BlastP on this gene
Accession: AEC00130
Location: 1312190-1313005
NCBI BlastP on this gene
Selsp_1170
polysaccharide deacetylase
Accession: AEC00129
Location: 1311206-1312054
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 140
Sequence coverage: 99 %
E-value: 6e-36
NCBI BlastP on this gene
Accession: AEC00129
Location: 1311206-1312054
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 140
Sequence coverage: 99 %
E-value: 6e-36
NCBI BlastP on this gene
Selsp_1169
Selsp_1168
Selsp_1167
biotin/lipoyl attachment domain-containing protein
Accession: AEC00126
Location: 1305941-1306927
NCBI BlastP on this gene
Accession: AEC00126
Location: 1305941-1306927
NCBI BlastP on this gene
Selsp_1166
Selsp_1165
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP010086 : Clostridium beijerinckii strain NCIMB 14988 Total score: 1.0 Cumulative Blast bit score: 136
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
LF65_00314
LF65_00317
LF65_00318
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AJG96988
Location: 318116-318595
NCBI BlastP on this gene
Accession: AJG96988
Location: 318116-318595
NCBI BlastP on this gene
LF65_00319
LF65_00321
polysaccharide deacetylase
Accession: AJG96991
Location: 320024-320983
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 136
Sequence coverage: 82 %
E-value: 5e-34
NCBI BlastP on this gene
Accession: AJG96991
Location: 320024-320983
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 136
Sequence coverage: 82 %
E-value: 5e-34
NCBI BlastP on this gene
LF65_00322
LF65_00323
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: AJG96993
Location: 322387-323433
NCBI BlastP on this gene
Accession: AJG96993
Location: 322387-323433
NCBI BlastP on this gene
LF65_00324
MarR family transcriptional regulator
Accession: AJG96994
Location: 323646-324080
NCBI BlastP on this gene
Accession: AJG96994
Location: 323646-324080
NCBI BlastP on this gene
LF65_00325
LF65_00326
LF65_00327
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: AJG96997
Location: 325224-327806
NCBI BlastP on this gene
Accession: AJG96997
Location: 325224-327806
NCBI BlastP on this gene
LF65_00328
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP016090 : Clostridium beijerinckii strain BAS/B3/I/124 Total score: 1.0 Cumulative Blast bit score: 135
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
methyl-accepting chemotaxis protein McpB
Accession: AQS02926
Location: 312480-314957
NCBI BlastP on this gene
Accession: AQS02926
Location: 312480-314957
NCBI BlastP on this gene
mcpB_1
CLBIJ_03080
CLBIJ_03090
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AQS02929
Location: 316870-317349
NCBI BlastP on this gene
Accession: AQS02929
Location: 316870-317349
NCBI BlastP on this gene
ispF
CLBIJ_03110
RNA polymerase sigma factor SigI
Accession: AQS02931
Location: 317731-318762
NCBI BlastP on this gene
Accession: AQS02931
Location: 317731-318762
NCBI BlastP on this gene
sigI_1
poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase precursor
Accession: AQS02932
Location: 318778-319737
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 135
Sequence coverage: 82 %
E-value: 8e-34
NCBI BlastP on this gene
Accession: AQS02932
Location: 318778-319737
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 135
Sequence coverage: 82 %
E-value: 8e-34
NCBI BlastP on this gene
icaB
rsiV
bioB_1
CLBIJ_03160
CLBIJ_03170
quaternary ammonium compound-resistance protein SugE
Accession: AQS02937
Location: 323352-323678
NCBI BlastP on this gene
Accession: AQS02937
Location: 323352-323678
NCBI BlastP on this gene
sugE_1
adhE_1
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
LN908213 : Clostridium beijerinckii isolate C. beijerinckii DSM 6423 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 134
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
Methyl-accepting chemotaxis protein
Accession: CUU45382
Location: 313057-315534
NCBI BlastP on this gene
Accession: CUU45382
Location: 313057-315534
NCBI BlastP on this gene
CIBE_0297
conserved protein of unknown function
Accession: CUU45383
Location: 315909-316313
NCBI BlastP on this gene
Accession: CUU45383
Location: 315909-316313
NCBI BlastP on this gene
CIBE_0298
conserved protein of unknown function
Accession: CUU45384
Location: 316584-317339
NCBI BlastP on this gene
Accession: CUU45384
Location: 316584-317339
NCBI BlastP on this gene
CIBE_0299
2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase
Accession: CUU45385
Location: 317443-317919
NCBI BlastP on this gene
Accession: CUU45385
Location: 317443-317919
NCBI BlastP on this gene
ispF
conserved protein of unknown function
Accession: CUU45386
Location: 317993-318163
NCBI BlastP on this gene
Accession: CUU45386
Location: 317993-318163
NCBI BlastP on this gene
CIBE_0301
conserved protein of unknown function
Accession: CUU45387
Location: 318300-319331
NCBI BlastP on this gene
Accession: CUU45387
Location: 318300-319331
NCBI BlastP on this gene
CIBE_0302
Polysaccharide deacetylase
Accession: CUU45388
Location: 319347-320405
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 4e-33
NCBI BlastP on this gene
Accession: CUU45388
Location: 319347-320405
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 4e-33
NCBI BlastP on this gene
CIBE_0303
conserved protein of unknown function
Accession: CUU45389
Location: 320709-321464
NCBI BlastP on this gene
Accession: CUU45389
Location: 320709-321464
NCBI BlastP on this gene
CIBE_0304
CIBE_0305
fused acetaldehyde-CoA dehydrogenase;
Accession: CUU45391
Location: 323285-325867
NCBI BlastP on this gene
Accession: CUU45391
Location: 323285-325867
NCBI BlastP on this gene
adhE
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP043998 : Clostridium diolis strain DSM 15410 chromosome Total score: 1.0 Cumulative Blast bit score: 134
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
methyl-accepting chemotaxis protein
Accession: QES71561
Location: 296925-299402
NCBI BlastP on this gene
Accession: QES71561
Location: 296925-299402
NCBI BlastP on this gene
F3K33_01475
F3K33_01490
F3K33_01495
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: QES71564
Location: 301311-301787
NCBI BlastP on this gene
Accession: QES71564
Location: 301311-301787
NCBI BlastP on this gene
F3K33_01500
anti-sigma factor domain-containing protein
Accession: QES71565
Location: 302169-303200
NCBI BlastP on this gene
Accession: QES71565
Location: 302169-303200
NCBI BlastP on this gene
F3K33_01505
polysaccharide deacetylase family protein
Accession: QES71566
Location: 303216-304175
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
Accession: QES71566
Location: 303216-304175
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
F3K33_01510
DUF3298 and DUF4163 domain-containing protein
Accession: QES71567
Location: 304582-305337
NCBI BlastP on this gene
Accession: QES71567
Location: 304582-305337
NCBI BlastP on this gene
F3K33_01515
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: QES71568
Location: 305579-306625
NCBI BlastP on this gene
Accession: QES71568
Location: 305579-306625
NCBI BlastP on this gene
hydE
MarR family transcriptional regulator
Accession: QES71569
Location: 306838-307272
NCBI BlastP on this gene
Accession: QES71569
Location: 306838-307272
NCBI BlastP on this gene
F3K33_01525
F3K33_01530
multidrug efflux SMR transporter
Accession: QES71571
Location: 307791-308117
NCBI BlastP on this gene
Accession: QES71571
Location: 307791-308117
NCBI BlastP on this gene
F3K33_01535
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: QES71572
Location: 308416-311010
NCBI BlastP on this gene
Accession: QES71572
Location: 308416-311010
NCBI BlastP on this gene
adhE
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP014331 : Clostridium sp. MF28, genome. Total score: 1.0 Cumulative Blast bit score: 134
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
AXY43_15660
AXY43_15675
AXY43_15680
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AVK49323
Location: 3657982-3658458
NCBI BlastP on this gene
Accession: AVK49323
Location: 3657982-3658458
NCBI BlastP on this gene
AXY43_15685
AXY43_15690
polysaccharide deacetylase
Accession: AVK49325
Location: 3659887-3660846
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
Accession: AVK49325
Location: 3659887-3660846
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
AXY43_15695
AXY43_15700
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: AVK49327
Location: 3662250-3663296
NCBI BlastP on this gene
Accession: AVK49327
Location: 3662250-3663296
NCBI BlastP on this gene
AXY43_15705
MarR family transcriptional regulator
Accession: AVK49328
Location: 3663509-3663943
NCBI BlastP on this gene
Accession: AVK49328
Location: 3663509-3663943
NCBI BlastP on this gene
AXY43_15710
AXY43_15715
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: AVK49329
Location: 3665086-3667680
NCBI BlastP on this gene
Accession: AVK49329
Location: 3665086-3667680
NCBI BlastP on this gene
AXY43_15720
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP011966 : Clostridium beijerinckii NRRL B-598 chromosome Total score: 1.0 Cumulative Blast bit score: 134
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
methyl-accepting chemotaxis protein
Accession: ALB48351
Location: 301218-303695
NCBI BlastP on this gene
Accession: ALB48351
Location: 301218-303695
NCBI BlastP on this gene
X276_25350
X276_25335
X276_25330
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: ALB48349
Location: 305604-306080
NCBI BlastP on this gene
Accession: ALB48349
Location: 305604-306080
NCBI BlastP on this gene
X276_25325
anti-sigma factor domain-containing protein
Accession: ALB48348
Location: 306462-307493
NCBI BlastP on this gene
Accession: ALB48348
Location: 306462-307493
NCBI BlastP on this gene
X276_25320
polysaccharide deacetylase family protein
Accession: ALB48347
Location: 307509-308468
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
Accession: ALB48347
Location: 307509-308468
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
X276_25315
DUF3298/DUF4163 domain-containing protein
Accession: ALB48346
Location: 308875-309630
NCBI BlastP on this gene
Accession: ALB48346
Location: 308875-309630
NCBI BlastP on this gene
X276_25310
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: ALB48345
Location: 309872-310918
NCBI BlastP on this gene
Accession: ALB48345
Location: 309872-310918
NCBI BlastP on this gene
hydE
QacE family quaternary ammonium compound efflux SMR transporter
Accession: X276_27430
Location: 311030-311143
NCBI BlastP on this gene
Accession: X276_27430
Location: 311030-311143
NCBI BlastP on this gene
X276_27430
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: ALB48344
Location: 311442-314036
NCBI BlastP on this gene
Accession: ALB48344
Location: 311442-314036
NCBI BlastP on this gene
X276_25300
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP006777 : Clostridium beijerinckii ATCC 35702 Total score: 1.0 Cumulative Blast bit score: 134
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
methyl-accepting chemotaxis sensory transducer
Accession: AIU02043
Location: 352392-354869
NCBI BlastP on this gene
Accession: AIU02043
Location: 352392-354869
NCBI BlastP on this gene
Cbs_0294
Cbs_0295
Cbs_0296
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AIU00312
Location: 356778-357254
NCBI BlastP on this gene
Accession: AIU00312
Location: 356778-357254
NCBI BlastP on this gene
ispF
Cbs_0298
polysaccharide deacetylase
Accession: AIU03183
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: AIU03183
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Cbs_0299
Cbs_0300
Cbs_0301
MarR family transcriptional regulator
Accession: AIU03625
Location: 362304-362738
NCBI BlastP on this gene
Accession: AIU03625
Location: 362304-362738
NCBI BlastP on this gene
Cbs_0302
Cbs_0303
small multidrug resistance protein
Accession: AIU03858
Location: 363257-363583
NCBI BlastP on this gene
Accession: AIU03858
Location: 363257-363583
NCBI BlastP on this gene
Cbs_0304
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: AIU02688
Location: 363882-366476
NCBI BlastP on this gene
Accession: AIU02688
Location: 363882-366476
NCBI BlastP on this gene
Cbs_0305
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP000721 : Clostridium beijerinckii NCIMB 8052 Total score: 1.0 Cumulative Blast bit score: 134
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
methyl-accepting chemotaxis sensory transducer
Accession: ABR32482
Location: 352392-354869
NCBI BlastP on this gene
Accession: ABR32482
Location: 352392-354869
NCBI BlastP on this gene
Cbei_0294
Cbei_0295
Cbei_0296
2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: ABR32485
Location: 356778-357254
NCBI BlastP on this gene
Accession: ABR32485
Location: 356778-357254
NCBI BlastP on this gene
Cbei_0297
Cbei_0298
polysaccharide deacetylase
Accession: ABR32487
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: ABR32487
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Cbei_0299
Cbei_0300
Cbei_0301
transcriptional regulator, MarR family
Accession: ABR32490
Location: 362304-362738
NCBI BlastP on this gene
Accession: ABR32490
Location: 362304-362738
NCBI BlastP on this gene
Cbei_0302
Cbei_0303
small multidrug resistance protein
Accession: ABR32492
Location: 363257-363583
NCBI BlastP on this gene
Accession: ABR32492
Location: 363257-363583
NCBI BlastP on this gene
Cbei_0304
iron-containing alcohol dehydrogenase
Accession: ABR32493
Location: 363882-366476
NCBI BlastP on this gene
Accession: ABR32493
Location: 363882-366476
NCBI BlastP on this gene
Cbei_0305
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP002637 : Selenomonas sputigena ATCC 35185 Total score: 1.0 Cumulative Blast bit score: 130
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
phospho-N-acetylmuramoyl-pentapeptide- transferase
Accession: AEC00777
Location: 2004406-2005380
NCBI BlastP on this gene
Accession: AEC00777
Location: 2004406-2005380
NCBI BlastP on this gene
Selsp_1822
UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-
Accession: AEC00778
Location: 2005377-2008277
NCBI BlastP on this gene
Accession: AEC00778
Location: 2005377-2008277
NCBI BlastP on this gene
Selsp_1823
Selsp_1824
S-adenosyl-methyltransferase MraW
Accession: AEC00780
Location: 2008764-2009699
NCBI BlastP on this gene
Accession: AEC00780
Location: 2008764-2009699
NCBI BlastP on this gene
Selsp_1825
Selsp_1826
polysaccharide deacetylase
Accession: AEC00782
Location: 2010387-2011193
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 130
Sequence coverage: 82 %
E-value: 1e-32
NCBI BlastP on this gene
Accession: AEC00782
Location: 2010387-2011193
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 130
Sequence coverage: 82 %
E-value: 1e-32
NCBI BlastP on this gene
Selsp_1827
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP014331 : Clostridium sp. MF28, genome. Total score: 1.0 Cumulative Blast bit score: 127
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase
Accession: AVK51319
Location: 6105005-6105919
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 9e-31
NCBI BlastP on this gene
Accession: AVK51319
Location: 6105005-6105919
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 9e-31
NCBI BlastP on this gene
AXY43_26710
AXY43_26705
AXY43_26700
AXY43_26695
AXY43_26690
AXY43_26685
AXY43_26680
AXY43_26675
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP010086 : Clostridium beijerinckii strain NCIMB 14988 Total score: 1.0 Cumulative Blast bit score: 127
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
LF65_03181
YgiQ family radical SAM protein
Accession: AJG99747
Location: 3511379-3513358
NCBI BlastP on this gene
Accession: AJG99747
Location: 3511379-3513358
NCBI BlastP on this gene
LF65_03182
LF65_03184
LF65_03185
polysaccharide deacetylase
Accession: AJG99751
Location: 3515874-3516788
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 1e-30
NCBI BlastP on this gene
Accession: AJG99751
Location: 3515874-3516788
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 1e-30
NCBI BlastP on this gene
LF65_03186
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP016090 : Clostridium beijerinckii strain BAS/B3/I/124 Total score: 1.0 Cumulative Blast bit score: 126
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase precursor
Accession: AQS05090
Location: 2750134-2751048
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 126
Sequence coverage: 85 %
E-value: 2e-30
NCBI BlastP on this gene
Accession: AQS05090
Location: 2750134-2751048
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 126
Sequence coverage: 85 %
E-value: 2e-30
NCBI BlastP on this gene
pgaB
CLBIJ_25200
CLBIJ_25190
CLBIJ_25180
CLBIJ_25170
CLBIJ_25160
small, acid-soluble spore protein beta
Accession: AQS05084
Location: 2746964-2747170
NCBI BlastP on this gene
Accession: AQS05084
Location: 2746964-2747170
NCBI BlastP on this gene
CLBIJ_25150
CLBIJ_25140
CLBIJ_25130
CLBIJ_25120
CLBIJ_25110
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP043998 : Clostridium diolis strain DSM 15410 chromosome Total score: 1.0 Cumulative Blast bit score: 125
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase family protein
Accession: QES73502
Location: 2638497-2639411
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: QES73502
Location: 2638497-2639411
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
F3K33_12030
F3K33_12025
F3K33_12020
F3K33_12015
alpha/beta-type small acid-soluble spore protein
Accession: QES73498
Location: 2635135-2635341
NCBI BlastP on this gene
Accession: QES73498
Location: 2635135-2635341
NCBI BlastP on this gene
F3K33_12010
F3K33_12005
F3K33_12000
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP011966 : Clostridium beijerinckii NRRL B-598 chromosome Total score: 1.0 Cumulative Blast bit score: 125
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase family protein
Accession: ALB46438
Location: 2662157-2663071
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: ALB46438
Location: 2662157-2663071
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
X276_14905
X276_14910
X276_14915
X276_14920
alpha/beta-type small acid-soluble spore protein
Accession: ALB46442
Location: 2658795-2659001
NCBI BlastP on this gene
Accession: ALB46442
Location: 2658795-2659001
NCBI BlastP on this gene
X276_14925
X276_14930
X276_14935
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP006777 : Clostridium beijerinckii ATCC 35702 Total score: 1.0 Cumulative Blast bit score: 125
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase
Accession: AIU03179
Location: 2712562-2713476
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: AIU03179
Location: 2712562-2713476
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Cbs_2350
Cbs_2349
Cbs_2348
Cbs_2347
Cbs_2346
small acid-soluble spore protein, alpha/beta type
Accession: AIU03854
Location: 2709191-2709397
NCBI BlastP on this gene
Accession: AIU03854
Location: 2709191-2709397
NCBI BlastP on this gene
Cbs_2345
Cbs_2344
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP000721 : Clostridium beijerinckii NCIMB 8052 Total score: 1.0 Cumulative Blast bit score: 125
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase
Accession: ABR34510
Location: 2712567-2713481
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: ABR34510
Location: 2712567-2713481
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Cbei_2350
Cbei_2349
Cbei_2348
Cbei_2347
Cbei_2346
small acid-soluble spore protein, alpha/beta type
Accession: ABR34505
Location: 2709196-2709402
NCBI BlastP on this gene
Accession: ABR34505
Location: 2709196-2709402
NCBI BlastP on this gene
Cbei_2345
Cbei_2344
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
LN908213 : Clostridium beijerinckii isolate C. beijerinckii DSM 6423 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 123
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
conserved exported protein of unknown function
Accession: CUU47882
Location: 2805018-2805932
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 123
Sequence coverage: 85 %
E-value: 2e-29
NCBI BlastP on this gene
Accession: CUU47882
Location: 2805018-2805932
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 123
Sequence coverage: 85 %
E-value: 2e-29
NCBI BlastP on this gene
CIBE_2781
conserved protein of unknown function
Accession: CUU47881
Location: 2804007-2804312
NCBI BlastP on this gene
Accession: CUU47881
Location: 2804007-2804312
NCBI BlastP on this gene
CIBE_2780
conserved protein of unknown function
Accession: CUU47880
Location: 2803182-2803688
NCBI BlastP on this gene
Accession: CUU47880
Location: 2803182-2803688
NCBI BlastP on this gene
CIBE_2779
conserved protein of unknown function
Accession: CUU47879
Location: 2801031-2803007
NCBI BlastP on this gene
Accession: CUU47879
Location: 2801031-2803007
NCBI BlastP on this gene
CIBE_2778
conserved protein of unknown function
Accession: CUU47878
Location: 2800724-2800954
NCBI BlastP on this gene
Accession: CUU47878
Location: 2800724-2800954
NCBI BlastP on this gene
CIBE_2777
conserved protein of unknown function
Accession: CUU47877
Location: 2800408-2800569
NCBI BlastP on this gene
Accession: CUU47877
Location: 2800408-2800569
NCBI BlastP on this gene
CIBE_2776
Small, acid-soluble spore protein beta
Accession: CUU47876
Location: 2799941-2800147
NCBI BlastP on this gene
Accession: CUU47876
Location: 2799941-2800147
NCBI BlastP on this gene
CIBE_2775
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
LT906477 : Clostridium cochlearium strain NCTC13027 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 122
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
tpl_1
metal-binding protein of ferredoxin fold
Accession: SNV63480
Location: 70754-70957
NCBI BlastP on this gene
Accession: SNV63480
Location: 70754-70957
NCBI BlastP on this gene
SAMEA4530647_00079
SAMEA4530647_00080
SAMEA4530647_00081
corrin/porphyrin methyltransferase
Accession: SNV63526
Location: 72087-72935
NCBI BlastP on this gene
Accession: SNV63526
Location: 72087-72935
NCBI BlastP on this gene
rsmI
iap
transition state regulatory protein AbrB
Accession: SNV63556
Location: 74352-74594
NCBI BlastP on this gene
Accession: SNV63556
Location: 74352-74594
NCBI BlastP on this gene
abrB
tal
polysaccharide deacetylase
Accession: SNV63583
Location: 75801-76688
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 122
Sequence coverage: 86 %
E-value: 4e-29
NCBI BlastP on this gene
Accession: SNV63583
Location: 75801-76688
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 122
Sequence coverage: 86 %
E-value: 4e-29
NCBI BlastP on this gene
icaB
glcK
SAMEA4530647_00088
hexA_1
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: SNV63641
Location: 80225-80707
NCBI BlastP on this gene
Accession: SNV63641
Location: 80225-80707
NCBI BlastP on this gene
ispF
yabJ_1
lytD
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
LT906477 : Clostridium cochlearium strain NCTC13027 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 120
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase
Accession: SNV74675
Location: 966480-967232
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 121
Sequence coverage: 82 %
E-value: 3e-29
NCBI BlastP on this gene
Accession: SNV74675
Location: 966480-967232
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 121
Sequence coverage: 82 %
E-value: 3e-29
NCBI BlastP on this gene
pgaB
phosphatase-associated protein papQ
Accession: SNV74662
Location: 964484-965506
NCBI BlastP on this gene
Accession: SNV74662
Location: 964484-965506
NCBI BlastP on this gene
papQ
ngr
spxA
spore peptidoglycan hydrolase (N-acetylglucosaminidase)
Accession: SNV74630
Location: 961980-963257
NCBI BlastP on this gene
Accession: SNV74630
Location: 961980-963257
NCBI BlastP on this gene
ydhD
yeeE
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP034842 : Fusobacterium necrophorum subsp. necrophorum strain ATCC 25286 chromosome Total score: 1.0 Cumulative Blast bit score: 120
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
lipopolysaccharide heptosyltransferase family protein
Accession: AZW09987
Location: 2303138-2304211
NCBI BlastP on this gene
Accession: AZW09987
Location: 2303138-2304211
NCBI BlastP on this gene
EO219_10670
EO219_10675
EO219_10680
polysaccharide deacetylase family protein
Accession: AZW09990
Location: 2306319-2307077
NCBI BlastP on this gene
Accession: AZW09990
Location: 2306319-2307077
NCBI BlastP on this gene
EO219_10685
lipopolysaccharide biosynthesis protein
Accession: AZW09991
Location: 2307053-2307811
NCBI BlastP on this gene
Accession: AZW09991
Location: 2307053-2307811
NCBI BlastP on this gene
EO219_10690
galE
polysaccharide deacetylase family protein
Accession: AZW09993
Location: 2308816-2309565
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
Accession: AZW09993
Location: 2308816-2309565
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
EO219_10700
EO219_10705
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP033837 : Fusobacterium necrophorum strain FDAARGOS_565 chromosome Total score: 1.0 Cumulative Blast bit score: 120
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
polysaccharide deacetylase family protein
Accession: AYZ74127
Location: 1928804-1929553
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
Accession: AYZ74127
Location: 1928804-1929553
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
EGX98_08860
EGX98_08855
EGX98_08850
EGX98_08845
O-antigen ligase domain-containing protein
Accession: AYZ74123
Location: 1923301-1924860
NCBI BlastP on this gene
Accession: AYZ74123
Location: 1923301-1924860
NCBI BlastP on this gene
EGX98_08840
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP036171 : Acinetobacter nosocomialis strain KAN02 chromosome Total score: 1.0 Cumulative Blast bit score: 112
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
glucose-6-phosphate isomerase
Accession: QBF79880
Location: 3866789-3868459
NCBI BlastP on this gene
Accession: QBF79880
Location: 3866789-3868459
NCBI BlastP on this gene
KAN02_18540
UDP-glucose/GDP-mannose dehydrogenase family protein
Accession: QBF79881
Location: 3868456-3869718
NCBI BlastP on this gene
Accession: QBF79881
Location: 3868456-3869718
NCBI BlastP on this gene
KAN02_18545
UTP--glucose-1-phosphate uridylyltransferase GalU
Accession: QBF79882
Location: 3869833-3870708
NCBI BlastP on this gene
Accession: QBF79882
Location: 3869833-3870708
NCBI BlastP on this gene
galU
KAN02_18555
KAN02_18560
glycosyltransferase family 2 protein
Accession: QBF79885
Location: 3872196-3873098
NCBI BlastP on this gene
Accession: QBF79885
Location: 3872196-3873098
NCBI BlastP on this gene
KAN02_18565
hypothetical protein
Accession: QBF79886
Location: 3873091-3874032
BlastP hit with GL636894_1983_2612_-
Percentage identity: 39 %
BlastP bit score: 113
Sequence coverage: 67 %
E-value: 3e-26
NCBI BlastP on this gene
Accession: QBF79886
Location: 3873091-3874032
BlastP hit with GL636894_1983_2612_-
Percentage identity: 39 %
BlastP bit score: 113
Sequence coverage: 67 %
E-value: 3e-26
NCBI BlastP on this gene
KAN02_18570
KAN02_18575
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP036267 : Planctomycetes bacterium Mal48 chromosome. Total score: 1.0 Cumulative Blast bit score: 103
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
Mal48_11160
Mal48_11150
Enoyl-[acyl-carrier-protein] reductase [NADPH] FabL
Accession: QDT31878
Location: 1381960-1382706
NCBI BlastP on this gene
Accession: QDT31878
Location: 1381960-1382706
NCBI BlastP on this gene
fabL
Mal48_11130
Mal48_11120
Mal48_11110
Alpha-D-kanosaminyltransferase
Accession: QDT31874
Location: 1378495-1379649
BlastP hit with GL636894_3749_4732_-
Percentage identity: 31 %
BlastP bit score: 104
Sequence coverage: 70 %
E-value: 5e-22
NCBI BlastP on this gene
Accession: QDT31874
Location: 1378495-1379649
BlastP hit with GL636894_3749_4732_-
Percentage identity: 31 %
BlastP bit score: 104
Sequence coverage: 70 %
E-value: 5e-22
NCBI BlastP on this gene
kanE_1
3-oxoacyl-[acyl-carrier-protein] reductase FabG
Accession: QDT31873
Location: 1377675-1378406
NCBI BlastP on this gene
Accession: QDT31873
Location: 1377675-1378406
NCBI BlastP on this gene
fabG_2
D-erythro-7,8-dihydroneopterin triphosphate epimerase
Accession: QDT31872
Location: 1377160-1377528
NCBI BlastP on this gene
Accession: QDT31872
Location: 1377160-1377528
NCBI BlastP on this gene
folX
2-amino-4-hydroxy-6- hydroxymethyldihydropteridine pyrophosphokinase
Accession: QDT31871
Location: 1376489-1377055
NCBI BlastP on this gene
Accession: QDT31871
Location: 1376489-1377055
NCBI BlastP on this gene
folK
Mal48_11060
50S ribosomal protein L31 type B
Accession: QDT31869
Location: 1375278-1375592
NCBI BlastP on this gene
Accession: QDT31869
Location: 1375278-1375592
NCBI BlastP on this gene
rpmE2
Peptide chain release factor RF1
Accession: QDT31868
Location: 1373978-1375048
NCBI BlastP on this gene
Accession: QDT31868
Location: 1373978-1375048
NCBI BlastP on this gene
prfA
Release factor glutamine methyltransferase
Accession: QDT31867
Location: 1372989-1373888
NCBI BlastP on this gene
Accession: QDT31867
Location: 1372989-1373888
NCBI BlastP on this gene
prmC
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP011427 : Pantoea vagans strain ND02 chromosome Total score: 1.0 Cumulative Blast bit score: 103
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
LK04_00315
LK04_00320
universal stress global response regulator UspA
Accession: ALV90686
Location: 72655-73092
NCBI BlastP on this gene
Accession: ALV90686
Location: 72655-73092
NCBI BlastP on this gene
LK04_00325
LK04_00330
LK04_00335
LK04_00340
mannosyltransferase
Accession: ALV90689
Location: 76989-77981
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 104
Sequence coverage: 72 %
E-value: 5e-23
NCBI BlastP on this gene
Accession: ALV90689
Location: 76989-77981
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 104
Sequence coverage: 72 %
E-value: 5e-23
NCBI BlastP on this gene
LK04_00345
LK04_00350
LK04_00355
AraC family transcriptional regulator
Accession: ALV90692
Location: 79399-80400
NCBI BlastP on this gene
Accession: ALV90692
Location: 79399-80400
NCBI BlastP on this gene
LK04_00360
AraC family transcriptional regulator
Accession: ALV90693
Location: 80611-81102
NCBI BlastP on this gene
Accession: ALV90693
Location: 80611-81102
NCBI BlastP on this gene
LK04_00365
LK04_00370
LK04_00375
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP009454 : Pantoea rwandensis strain ND04 Total score: 1.0 Cumulative Blast bit score: 99
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
LH22_03565
universal stress global response regulator UspA
Accession: AIR84583
Location: 784438-784875
NCBI BlastP on this gene
Accession: AIR84583
Location: 784438-784875
NCBI BlastP on this gene
LH22_03570
LH22_03575
LH22_03580
LH22_03585
mannosyltransferase
Accession: AIR84587
Location: 788853-789845
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 99
Sequence coverage: 65 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AIR84587
Location: 788853-789845
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 99
Sequence coverage: 65 %
E-value: 2e-21
NCBI BlastP on this gene
LH22_03590
LH22_03595
LH22_03600
AraC family transcriptional regulator
Accession: AIR84590
Location: 791249-792238
NCBI BlastP on this gene
Accession: AIR84590
Location: 791249-792238
NCBI BlastP on this gene
LH22_03605
AraC family transcriptional regulator
Accession: AIR84591
Location: 792436-792924
NCBI BlastP on this gene
Accession: AIR84591
Location: 792436-792924
NCBI BlastP on this gene
LH22_03610
LH22_03615
LH22_03620
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP035934 : Acinetobacter cumulans strain WCHAc060092 chromosome Total score: 1.0 Cumulative Blast bit score: 92
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
bfr
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: QCO20461
Location: 403353-403808
NCBI BlastP on this gene
Accession: QCO20461
Location: 403353-403808
NCBI BlastP on this gene
C9E88_002450
C9E88_002455
C9E88_002460
glf
C9E88_002470
hypothetical protein
Accession: QCO20466
Location: 397956-398933
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 92
Sequence coverage: 81 %
E-value: 6e-19
NCBI BlastP on this gene
Accession: QCO20466
Location: 397956-398933
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 92
Sequence coverage: 81 %
E-value: 6e-19
NCBI BlastP on this gene
C9E88_002475
C9E88_002480
class I SAM-dependent methyltransferase
Accession: QCO22789
Location: 396508-397134
NCBI BlastP on this gene
Accession: QCO22789
Location: 396508-397134
NCBI BlastP on this gene
C9E88_002485
C9E88_002490
ATP-binding cassette domain-containing protein
Accession: QCO20469
Location: 393390-395099
NCBI BlastP on this gene
Accession: QCO20469
Location: 393390-395099
NCBI BlastP on this gene
C9E88_002495
C9E88_002500
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP025618 : Acinetobacter schindleri strain SGAir0122 chromosome Total score: 1.0 Cumulative Blast bit score: 88
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
glycosyltransferase family 1 protein
Accession: C0119_09995
Location: 2538982-2539659
NCBI BlastP on this gene
Accession: C0119_09995
Location: 2538982-2539659
NCBI BlastP on this gene
C0119_09995
C0119_09990
C0119_09985
glycosyltransferase family 25 protein
Accession: AWD70549
Location: 2541258-2542067
NCBI BlastP on this gene
Accession: AWD70549
Location: 2541258-2542067
NCBI BlastP on this gene
C0119_09980
C0119_09975
lipopolysaccharide biosynthesis protein
Accession: AWD70547
Location: 2543723-2544652
NCBI BlastP on this gene
Accession: AWD70547
Location: 2543723-2544652
NCBI BlastP on this gene
C0119_09970
hypothetical protein
Accession: AWD70546
Location: 2544690-2545613
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 88
Sequence coverage: 92 %
E-value: 2e-17
NCBI BlastP on this gene
Accession: AWD70546
Location: 2544690-2545613
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 88
Sequence coverage: 92 %
E-value: 2e-17
NCBI BlastP on this gene
C0119_09965
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP037934 : Marinobacter sp. JH2 chromosome Total score: 1.0 Cumulative Blast bit score: 85
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
MARI_13070
MARI_13080
MARI_13090
MARI_13100
MARI_13110
MARI_13120
hypothetical protein
Accession: QBM17207
Location: 1419559-1420686
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 85
Sequence coverage: 67 %
E-value: 2e-15
NCBI BlastP on this gene
Accession: QBM17207
Location: 1419559-1420686
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 85
Sequence coverage: 67 %
E-value: 2e-15
NCBI BlastP on this gene
MARI_13130
MARI_13140
MARI_13150
MARI_13160
MARI_13170
MARI_13180
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP011427 : Pantoea vagans strain ND02 chromosome Total score: 1.0 Cumulative Blast bit score: 82
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
LysR family transcriptional regulator
Accession: ALV93564
Location: 3454259-3455182
NCBI BlastP on this gene
Accession: ALV93564
Location: 3454259-3455182
NCBI BlastP on this gene
LK04_16040
mannosyltransferase OCH1-related enzyme
Accession: ALV93563
Location: 3453272-3454285
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 82
Sequence coverage: 70 %
E-value: 3e-15
NCBI BlastP on this gene
Accession: ALV93563
Location: 3453272-3454285
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 82
Sequence coverage: 70 %
E-value: 3e-15
NCBI BlastP on this gene
LK04_16035
LK04_16030
LK04_16025
LK04_16020
LK04_16015
LK04_16010
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
CP009454 : Pantoea rwandensis strain ND04 Total score: 1.0 Cumulative Blast bit score: 81
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
LysR family transcriptional regulator
Accession: AIR87520
Location: 4112479-4113402
NCBI BlastP on this gene
Accession: AIR87520
Location: 4112479-4113402
NCBI BlastP on this gene
LH22_19390
mannosyltransferase OCH1-related enzyme
Accession: AIR87519
Location: 4111492-4112505
BlastP hit with GL636894_1983_2612_-
Percentage identity: 34 %
BlastP bit score: 81
Sequence coverage: 70 %
E-value: 6e-15
NCBI BlastP on this gene
Accession: AIR87519
Location: 4111492-4112505
BlastP hit with GL636894_1983_2612_-
Percentage identity: 34 %
BlastP bit score: 81
Sequence coverage: 70 %
E-value: 6e-15
NCBI BlastP on this gene
LH22_19385
LH22_19380
LH22_19375
LH22_19370
LH22_19365
LH22_19360
Query: Acinetobacter baumannii A118 genomic scaffold scaffold1213, whole
301. : CP028287 Acidovorax avenae subsp. avenae strain AA78_5 chromosome. Total score: 1.0 Cumulative Blast bit score: 195
GL636894_155_883_
GL636894_880_1674_-
GL636894_1705_2013_-
GL636894_1983_2612_-
GL636894_2696_3736_-
GL636894_3749_4732_-
GL636894_4735_5544_-
GL636894_5558_6448_-
C8241_01345
C8241_01350
branched-chain amino acid transaminase
Accession: AVS60532
Location: 354758-355696
NCBI BlastP on this gene
Accession: AVS60532
Location: 354758-355696
NCBI BlastP on this gene
C8241_01355
zinc-finger domain-containing protein
Accession: AVS60533
Location: 355738-355944
NCBI BlastP on this gene
Accession: AVS60533
Location: 355738-355944
NCBI BlastP on this gene
C8241_01360
capsular biosynthesis protein
Accession: AVS63922
Location: 355983-356870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 97
Sequence coverage: 67 %
E-value: 9e-21
NCBI BlastP on this gene
Accession: AVS63922
Location: 355983-356870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 97
Sequence coverage: 67 %
E-value: 9e-21
NCBI BlastP on this gene
C8241_01365
C8241_01370
C8241_01375
C8241_01380
C8241_01385
mannosyltransferase
Accession: AVS60536
Location: 360670-361701
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVS60536
Location: 360670-361701
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8241_01390
C8241_01395
LysR family transcriptional regulator
Accession: AVS60538
Location: 362816-363700
NCBI BlastP on this gene
Accession: AVS60538
Location: 362816-363700
NCBI BlastP on this gene
C8241_01400
C8241_01405
302. : CP028290 Acidovorax cattleyae strain CAT98_1 chromosome. Total score: 1.0 Cumulative Blast bit score: 194
C8240_01065
C8240_01070
branched-chain amino acid transaminase
Accession: AVS72831
Location: 348314-349252
NCBI BlastP on this gene
Accession: AVS72831
Location: 348314-349252
NCBI BlastP on this gene
C8240_01075
zinc-finger domain-containing protein
Accession: AVS72832
Location: 349290-349496
NCBI BlastP on this gene
Accession: AVS72832
Location: 349290-349496
NCBI BlastP on this gene
C8240_01080
capsular biosynthesis protein
Accession: AVS76217
Location: 349537-350427
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS76217
Location: 349537-350427
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8240_01085
C8240_01090
C8240_01095
C8240_01100
C8240_01105
C8240_01110
mannosyltransferase
Accession: AVS72838
Location: 354376-355401
BlastP hit with GL636894_1983_2612_-
Percentage identity: 36 %
BlastP bit score: 100
Sequence coverage: 70 %
E-value: 9e-22
NCBI BlastP on this gene
Accession: AVS72838
Location: 354376-355401
BlastP hit with GL636894_1983_2612_-
Percentage identity: 36 %
BlastP bit score: 100
Sequence coverage: 70 %
E-value: 9e-22
NCBI BlastP on this gene
C8240_01115
LysR family transcriptional regulator
Accession: AVS72839
Location: 357086-357406
NCBI BlastP on this gene
Accession: AVS72839
Location: 357086-357406
NCBI BlastP on this gene
C8240_01120
C8240_01125
303. : CP028302 Acidovorax avenae subsp. avenae isolate SF12 chromosome. Total score: 1.0 Cumulative Blast bit score: 193
C8244_01120
C8244_01125
branched-chain amino acid transaminase
Accession: AVT14961
Location: 360944-361882
NCBI BlastP on this gene
Accession: AVT14961
Location: 360944-361882
NCBI BlastP on this gene
C8244_01130
zinc-finger domain-containing protein
Accession: AVT14962
Location: 361924-362130
NCBI BlastP on this gene
Accession: AVT14962
Location: 361924-362130
NCBI BlastP on this gene
C8244_01135
capsular biosynthesis protein
Accession: AVT18179
Location: 362168-363058
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
Accession: AVT18179
Location: 362168-363058
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
C8244_01140
C8244_01145
C8244_01150
C8244_01155
mannosyltransferase
Accession: AVT14966
Location: 366862-367893
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVT14966
Location: 366862-367893
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8244_01160
C8244_01165
LysR family transcriptional regulator
Accession: AVT14968
Location: 369008-369892
NCBI BlastP on this gene
Accession: AVT14968
Location: 369008-369892
NCBI BlastP on this gene
C8244_01170
C8244_01175
304. : CP028297 Acidovorax avenae subsp. avenae strain MOR chromosome. Total score: 1.0 Cumulative Blast bit score: 193
C8236_01480
C8236_01485
branched chain amino acid aminotransferase
Accession: AVS97630
Location: 365975-366913
NCBI BlastP on this gene
Accession: AVS97630
Location: 365975-366913
NCBI BlastP on this gene
C8236_01490
zinc-finger domain-containing protein
Accession: AVS97631
Location: 366955-367161
NCBI BlastP on this gene
Accession: AVS97631
Location: 366955-367161
NCBI BlastP on this gene
C8236_01495
capsular biosynthesis protein
Accession: AVT00759
Location: 367200-368087
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 6e-20
NCBI BlastP on this gene
Accession: AVT00759
Location: 367200-368087
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 6e-20
NCBI BlastP on this gene
C8236_01500
C8236_01505
C8236_01510
C8236_01515
mannosyltransferase
Accession: AVS97635
Location: 371891-372922
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVS97635
Location: 371891-372922
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C8236_01520
C8236_01525
LysR family transcriptional regulator
Accession: AVS97637
Location: 374037-374921
NCBI BlastP on this gene
Accession: AVS97637
Location: 374037-374921
NCBI BlastP on this gene
C8236_01530
C8236_01535
305. : CP028292 Acidovorax avenae subsp. avenae strain INDB2 chromosome. Total score: 1.0 Cumulative Blast bit score: 193
C8237_01140
C8237_01145
branched-chain amino acid transaminase
Accession: AVS79845
Location: 370722-371660
NCBI BlastP on this gene
Accession: AVS79845
Location: 370722-371660
NCBI BlastP on this gene
C8237_01150
zinc-finger domain-containing protein
Accession: AVS79846
Location: 371702-371908
NCBI BlastP on this gene
Accession: AVS79846
Location: 371702-371908
NCBI BlastP on this gene
C8237_01155
capsular biosynthesis protein
Accession: AVS83051
Location: 371946-372836
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
Accession: AVS83051
Location: 371946-372836
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 95
Sequence coverage: 66 %
E-value: 5e-20
NCBI BlastP on this gene
C8237_01160
C8237_01165
C8237_01170
C8237_01175
C8237_01180
mannosyltransferase
Accession: AVS79851
Location: 376884-377915
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVS79851
Location: 376884-377915
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8237_01185
C8237_01190
LysR family transcriptional regulator
Accession: AVS79853
Location: 379030-379914
NCBI BlastP on this gene
Accession: AVS79853
Location: 379030-379914
NCBI BlastP on this gene
C8237_01195
C8237_01200
306. : CP028303 Acidovorax avenae subsp. avenae strain SH7 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
C7Y68_01460
C7Y68_01465
branched-chain amino acid transaminase
Accession: AVT18817
Location: 379755-380693
NCBI BlastP on this gene
Accession: AVT18817
Location: 379755-380693
NCBI BlastP on this gene
C7Y68_01470
zinc-finger domain-containing protein
Accession: AVT18818
Location: 380735-380941
NCBI BlastP on this gene
Accession: AVT18818
Location: 380735-380941
NCBI BlastP on this gene
C7Y68_01475
capsular biosynthesis protein
Accession: AVT22261
Location: 380980-381870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVT22261
Location: 380980-381870
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C7Y68_01480
C7Y68_01485
C7Y68_01490
C7Y68_01495
mannosyltransferase
Accession: AVT18822
Location: 385674-386705
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVT18822
Location: 385674-386705
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C7Y68_01500
C7Y68_01505
LysR family transcriptional regulator
Accession: AVT18824
Location: 387820-388704
NCBI BlastP on this gene
Accession: AVT18824
Location: 387820-388704
NCBI BlastP on this gene
C7Y68_01510
C7Y68_01515
C7Y68_01520
307. : CP028300 Acidovorax avenae subsp. avenae strain QHB1 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
C8242_01295
C8242_01300
branched-chain amino acid transaminase
Accession: AVT08274
Location: 372258-373196
NCBI BlastP on this gene
Accession: AVT08274
Location: 372258-373196
NCBI BlastP on this gene
C8242_01305
zinc-finger domain-containing protein
Accession: AVT08275
Location: 373238-373444
NCBI BlastP on this gene
Accession: AVT08275
Location: 373238-373444
NCBI BlastP on this gene
C8242_01310
capsular biosynthesis protein
Accession: AVT11131
Location: 373483-374373
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVT11131
Location: 373483-374373
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8242_01315
C8242_01320
C8242_01325
C8242_01330
mannosyltransferase
Accession: AVT08279
Location: 378176-379207
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVT08279
Location: 378176-379207
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C8242_01335
C8242_01340
LysR family transcriptional regulator
Accession: AVT08281
Location: 380322-381206
NCBI BlastP on this gene
Accession: AVT08281
Location: 380322-381206
NCBI BlastP on this gene
C8242_01345
C8242_01350
308. : CP028298 Acidovorax avenae subsp. avenae strain NCT3 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
C8243_00950
C8243_00955
branched-chain amino acid transaminase
Accession: AVT01204
Location: 364567-365505
NCBI BlastP on this gene
Accession: AVT01204
Location: 364567-365505
NCBI BlastP on this gene
C8243_00960
zinc-finger domain-containing protein
Accession: AVT01205
Location: 365547-365753
NCBI BlastP on this gene
Accession: AVT01205
Location: 365547-365753
NCBI BlastP on this gene
C8243_00965
capsular biosynthesis protein
Accession: AVT04260
Location: 365792-366682
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVT04260
Location: 365792-366682
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8243_00970
C8243_00975
C8243_00980
C8243_00985
mannosyltransferase
Accession: AVT01209
Location: 370485-371516
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: AVT01209
Location: 370485-371516
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
C8243_00990
C8243_00995
LysR family transcriptional regulator
Accession: AVT01211
Location: 372631-373515
NCBI BlastP on this gene
Accession: AVT01211
Location: 372631-373515
NCBI BlastP on this gene
C8243_01000
C8243_01005
309. : CP028296 Acidovorax avenae subsp. avenae strain MDB1 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
C8232_00180
C8232_00185
branched-chain amino acid transaminase
Accession: AVS94857
Location: 362617-363555
NCBI BlastP on this gene
Accession: AVS94857
Location: 362617-363555
NCBI BlastP on this gene
C8232_00190
zinc-finger domain-containing protein
Accession: AVS94858
Location: 363597-363803
NCBI BlastP on this gene
Accession: AVS94858
Location: 363597-363803
NCBI BlastP on this gene
C8232_00195
capsular biosynthesis protein
Accession: AVS97109
Location: 363842-364732
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS97109
Location: 363842-364732
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8232_00200
C8232_00205
C8232_00210
C8232_00215
mannosyltransferase
Accession: AVS94862
Location: 368535-369566
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
Accession: AVS94862
Location: 368535-369566
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 7e-21
NCBI BlastP on this gene
C8232_00220
C8232_00225
LysR family transcriptional regulator
Accession: AVS94864
Location: 370681-371565
NCBI BlastP on this gene
Accession: AVS94864
Location: 370681-371565
NCBI BlastP on this gene
C8232_00230
C8232_00235
310. : CP028288 Acidovorax avenae subsp. avenae strain AA81_1 chromosome. Total score: 1.0 Cumulative Blast bit score: 192
C8245_04500
LysR family transcriptional regulator
Accession: AVS65061
Location: 998017-998901
NCBI BlastP on this gene
Accession: AVS65061
Location: 998017-998901
NCBI BlastP on this gene
C8245_04505
C8245_04510
mannosyltransferase
Accession: AVS65063
Location: 1000016-1001047
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 99
Sequence coverage: 66 %
E-value: 4e-21
NCBI BlastP on this gene
Accession: AVS65063
Location: 1000016-1001047
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 99
Sequence coverage: 66 %
E-value: 4e-21
NCBI BlastP on this gene
C8245_04515
glycosyl transferase family 2
Accession: AVS65064
Location: 1001044-1002105
NCBI BlastP on this gene
Accession: AVS65064
Location: 1001044-1002105
NCBI BlastP on this gene
C8245_04520
C8245_04525
C8245_04530
capsular biosynthesis protein
Accession: AVS68648
Location: 1004851-1005738
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS68648
Location: 1004851-1005738
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8245_04535
zinc-finger domain-containing protein
Accession: AVS65067
Location: 1005777-1005983
NCBI BlastP on this gene
Accession: AVS65067
Location: 1005777-1005983
NCBI BlastP on this gene
C8245_04540
branched-chain amino acid transaminase
Accession: AVS65068
Location: 1006025-1006963
NCBI BlastP on this gene
Accession: AVS65068
Location: 1006025-1006963
NCBI BlastP on this gene
C8245_04545
C8245_04550
C8245_04555
311. : CP002521 Acidovorax avenae subsp. avenae ATCC 19860 Total score: 1.0 Cumulative Blast bit score: 192
Acav_0319
Acav_0320
branched-chain amino acid aminotransferase
Accession: ADX44244
Location: 360935-361873
NCBI BlastP on this gene
Accession: ADX44244
Location: 360935-361873
NCBI BlastP on this gene
Acav_0321
Acav_0322
Capsular polysaccharide synthesis
Accession: ADX44246
Location: 362160-363149
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: ADX44246
Location: 362160-363149
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Acav_0323
Acav_0324
Acav_0325
Acav_0326
Capsular polysaccharide synthesis
Accession: ADX44250
Location: 366851-367882
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Accession: ADX44250
Location: 366851-367882
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 5e-21
NCBI BlastP on this gene
Acav_0327
Acav_0328
transcriptional regulator, LysR family
Accession: ADX44252
Location: 368997-369881
NCBI BlastP on this gene
Accession: ADX44252
Location: 368997-369881
NCBI BlastP on this gene
Acav_0329
protein of unknown function DUF1228
Accession: ADX44253
Location: 369951-371225
NCBI BlastP on this gene
Accession: ADX44253
Location: 369951-371225
NCBI BlastP on this gene
Acav_0330
312. : CP028295 Acidovorax avenae subsp. avenae strain MD5 chromosome. Total score: 1.0 Cumulative Blast bit score: 191
C8246_13775
LysR family transcriptional regulator
Accession: AVS92686
Location: 3063597-3064481
NCBI BlastP on this gene
Accession: AVS92686
Location: 3063597-3064481
NCBI BlastP on this gene
C8246_13780
C8246_13785
mannosyltransferase
Accession: AVS92688
Location: 3065596-3066633
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVS92688
Location: 3065596-3066633
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
C8246_13790
glycosyl transferase family 2
Accession: AVS92689
Location: 3066630-3067691
NCBI BlastP on this gene
Accession: AVS92689
Location: 3066630-3067691
NCBI BlastP on this gene
C8246_13795
C8246_13800
C8246_13805
capsular biosynthesis protein
Accession: AVS94633
Location: 3070437-3071327
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS94633
Location: 3070437-3071327
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8246_13810
zinc-finger domain-containing protein
Accession: AVS92692
Location: 3071366-3071572
NCBI BlastP on this gene
Accession: AVS92692
Location: 3071366-3071572
NCBI BlastP on this gene
C8246_13815
branched-chain amino acid transaminase
Accession: AVS92693
Location: 3071614-3072552
NCBI BlastP on this gene
Accession: AVS92693
Location: 3071614-3072552
NCBI BlastP on this gene
C8246_13820
C8246_13825
C8246_13830
313. : CP028294 Acidovorax avenae subsp. avenae strain KL3 chromosome. Total score: 1.0 Cumulative Blast bit score: 191
C8238_01105
C8238_01110
branched-chain amino acid transaminase
Accession: AVS87029
Location: 353399-354337
NCBI BlastP on this gene
Accession: AVS87029
Location: 353399-354337
NCBI BlastP on this gene
C8238_01115
zinc-finger domain-containing protein
Accession: AVS87030
Location: 354379-354585
NCBI BlastP on this gene
Accession: AVS87030
Location: 354379-354585
NCBI BlastP on this gene
C8238_01120
capsular biosynthesis protein
Accession: AVS90195
Location: 354624-355514
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS90195
Location: 354624-355514
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8238_01125
C8238_01130
C8238_01135
C8238_01140
mannosyltransferase
Accession: AVS87034
Location: 359317-360354
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVS87034
Location: 359317-360354
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
C8238_01145
C8238_01150
LysR family transcriptional regulator
Accession: AVS87036
Location: 361469-362353
NCBI BlastP on this gene
Accession: AVS87036
Location: 361469-362353
NCBI BlastP on this gene
C8238_01155
C8238_01160
314. : CP028293 Acidovorax avenae subsp. avenae strain INV chromosome. Total score: 1.0 Cumulative Blast bit score: 191
C8239_01575
C8239_01580
branched-chain amino acid transaminase
Accession: AVS83605
Location: 354178-355116
NCBI BlastP on this gene
Accession: AVS83605
Location: 354178-355116
NCBI BlastP on this gene
C8239_01585
zinc-finger domain-containing protein
Accession: AVS83606
Location: 355158-355364
NCBI BlastP on this gene
Accession: AVS83606
Location: 355158-355364
NCBI BlastP on this gene
C8239_01590
capsular biosynthesis protein
Accession: AVS86546
Location: 355403-356293
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: AVS86546
Location: 355403-356293
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 66 %
E-value: 2e-19
NCBI BlastP on this gene
C8239_01595
C8239_01600
C8239_01605
C8239_01610
mannosyltransferase
Accession: AVS83610
Location: 360097-361134
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVS83610
Location: 360097-361134
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 97
Sequence coverage: 66 %
E-value: 1e-20
NCBI BlastP on this gene
C8239_01615
C8239_01620
LysR family transcriptional regulator
Accession: AVS83612
Location: 362249-363133
NCBI BlastP on this gene
Accession: AVS83612
Location: 362249-363133
NCBI BlastP on this gene
C8239_01625
C8239_01630
315. : CP028289 Acidovorax avenae subsp. avenae strain AA99_2 chromosome. Total score: 1.0 Cumulative Blast bit score: 191
C8247_01030
C8247_01035
branched-chain amino acid transaminase
Accession: AVS69168
Location: 354803-355741
NCBI BlastP on this gene
Accession: AVS69168
Location: 354803-355741
NCBI BlastP on this gene
C8247_01040
zinc-finger domain-containing protein
Accession: AVS69169
Location: 355783-355989
NCBI BlastP on this gene
Accession: AVS69169
Location: 355783-355989
NCBI BlastP on this gene
C8247_01045
capsular biosynthesis protein
Accession: AVS72347
Location: 356028-356918
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 3e-19
NCBI BlastP on this gene
Accession: AVS72347
Location: 356028-356918
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 93
Sequence coverage: 66 %
E-value: 3e-19
NCBI BlastP on this gene
C8247_01050
C8247_01055
C8247_01060
C8247_01065
mannosyltransferase
Accession: AVS69173
Location: 360722-361753
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 6e-21
NCBI BlastP on this gene
Accession: AVS69173
Location: 360722-361753
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 98
Sequence coverage: 66 %
E-value: 6e-21
NCBI BlastP on this gene
C8247_01070
C8247_01075
LysR family transcriptional regulator
Accession: AVS69175
Location: 362868-363752
NCBI BlastP on this gene
Accession: AVS69175
Location: 362868-363752
NCBI BlastP on this gene
C8247_01080
C8247_01085
316. : CP044016 Arachidicoccus sp. B3-10 chromosome Total score: 1.0 Cumulative Blast bit score: 174
glycerol-3-phosphate dehydrogenase
Accession: QES89184
Location: 2575778-2576725
NCBI BlastP on this gene
Accession: QES89184
Location: 2575778-2576725
NCBI BlastP on this gene
E0W69_011090
NAD-dependent epimerase/dehydratase family protein
Accession: E0W69_011095
Location: 2576739-2577721
NCBI BlastP on this gene
Accession: E0W69_011095
Location: 2576739-2577721
NCBI BlastP on this gene
E0W69_011095
E0W69_011100
capsular biosynthesis protein
Accession: QES89186
Location: 2579014-2579943
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 69
Sequence coverage: 48 %
E-value: 6e-11
NCBI BlastP on this gene
Accession: QES89186
Location: 2579014-2579943
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 69
Sequence coverage: 48 %
E-value: 6e-11
NCBI BlastP on this gene
E0W69_011105
glycosyltransferase family 4 protein
Accession: QES89187
Location: 2579991-2581187
NCBI BlastP on this gene
Accession: QES89187
Location: 2579991-2581187
NCBI BlastP on this gene
E0W69_011110
glycosyltransferase family 4 protein
Accession: QES89188
Location: 2581205-2582293
NCBI BlastP on this gene
Accession: QES89188
Location: 2581205-2582293
NCBI BlastP on this gene
E0W69_011115
E0W69_011120
hypothetical protein
Accession: QES89190
Location: 2583330-2584277
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 105
Sequence coverage: 67 %
E-value: 2e-23
NCBI BlastP on this gene
Accession: QES89190
Location: 2583330-2584277
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 105
Sequence coverage: 67 %
E-value: 2e-23
NCBI BlastP on this gene
E0W69_011125
E0W69_011130
glycosyltransferase family 2 protein
Accession: QES89192
Location: 2585610-2586629
NCBI BlastP on this gene
Accession: QES89192
Location: 2585610-2586629
NCBI BlastP on this gene
E0W69_011135
beta-1,6-N-acetylglucosaminyltransferase
Accession: QES89193
Location: 2586640-2587530
NCBI BlastP on this gene
Accession: QES89193
Location: 2586640-2587530
NCBI BlastP on this gene
E0W69_011140
317. : CP049868 Pedobacter sp. HDW13 chromosome Total score: 1.0 Cumulative Blast bit score: 172
G7074_10860
G7074_10865
glycosyltransferase family 4 protein
Accession: QIL39724
Location: 2563276-2564181
NCBI BlastP on this gene
Accession: QIL39724
Location: 2563276-2564181
NCBI BlastP on this gene
G7074_10870
G7074_10875
G7074_10880
G7074_10885
hypothetical protein
Accession: QIL39728
Location: 2567273-2567776
BlastP hit with GL636894_880_1674_-
Percentage identity: 36 %
BlastP bit score: 83
Sequence coverage: 44 %
E-value: 4e-16
NCBI BlastP on this gene
Accession: QIL39728
Location: 2567273-2567776
BlastP hit with GL636894_880_1674_-
Percentage identity: 36 %
BlastP bit score: 83
Sequence coverage: 44 %
E-value: 4e-16
NCBI BlastP on this gene
G7074_10890
hypothetical protein
Accession: QIL39729
Location: 2567746-2568003
BlastP hit with GL636894_880_1674_-
Percentage identity: 50 %
BlastP bit score: 89
Sequence coverage: 31 %
E-value: 4e-19
NCBI BlastP on this gene
Accession: QIL39729
Location: 2567746-2568003
BlastP hit with GL636894_880_1674_-
Percentage identity: 50 %
BlastP bit score: 89
Sequence coverage: 31 %
E-value: 4e-19
NCBI BlastP on this gene
G7074_10895
ABC transporter ATP-binding protein
Accession: G7074_10900
Location: 2568109-2569943
NCBI BlastP on this gene
Accession: G7074_10900
Location: 2568109-2569943
NCBI BlastP on this gene
G7074_10900
glycosyltransferase family 2 protein
Accession: QIL39730
Location: 2570072-2571058
NCBI BlastP on this gene
Accession: QIL39730
Location: 2570072-2571058
NCBI BlastP on this gene
G7074_10905
glycosyltransferase family 2 protein
Accession: G7074_10910
Location: 2571059-2571933
NCBI BlastP on this gene
Accession: G7074_10910
Location: 2571059-2571933
NCBI BlastP on this gene
G7074_10910
methylmalonyl Co-A mutase-associated GTPase MeaB
Accession: QIL39731
Location: 2571993-2572889
NCBI BlastP on this gene
Accession: QIL39731
Location: 2571993-2572889
NCBI BlastP on this gene
meaB
aminopeptidase P family protein
Accession: QIL39732
Location: 2572941-2574230
NCBI BlastP on this gene
Accession: QIL39732
Location: 2572941-2574230
NCBI BlastP on this gene
G7074_10920
318. : CP018259 Acinetobacter bereziniae strain XH901 Total score: 1.0 Cumulative Blast bit score: 167
BSR55_03865
BSR55_03860
lipid A biosynthesis acyltransferase
Accession: ATZ62537
Location: 828310-829176
NCBI BlastP on this gene
Accession: ATZ62537
Location: 828310-829176
NCBI BlastP on this gene
BSR55_03855
capsular biosynthesis protein
Accession: ATZ62536
Location: 827288-828295
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 73
Sequence coverage: 61 %
E-value: 2e-12
NCBI BlastP on this gene
Accession: ATZ62536
Location: 827288-828295
BlastP hit with GL636894_1983_2612_-
Percentage identity: 37 %
BlastP bit score: 73
Sequence coverage: 61 %
E-value: 2e-12
NCBI BlastP on this gene
BSR55_03850
BSR55_03845
BSR55_03840
BSR55_03835
BSR55_03830
hypothetical protein
Accession: ATZ62531
Location: 822890-823804
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 94
Sequence coverage: 67 %
E-value: 2e-19
NCBI BlastP on this gene
Accession: ATZ62531
Location: 822890-823804
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 94
Sequence coverage: 67 %
E-value: 2e-19
NCBI BlastP on this gene
BSR55_03825
BSR55_03820
capsular polysaccharide biosynthesis protein
Accession: ATZ62529
Location: 821027-822034
NCBI BlastP on this gene
Accession: ATZ62529
Location: 821027-822034
NCBI BlastP on this gene
BSR55_03815
BSR55_03810
BSR55_03805
319. : CP037934 Marinobacter sp. JH2 chromosome Total score: 1.0 Cumulative Blast bit score: 144
hypothetical protein
Accession: QBM18131
Location: 2476210-2477229
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 102 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QBM18131
Location: 2476210-2477229
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 102 %
E-value: 2e-36
NCBI BlastP on this gene
MARI_22600
MARI_22590
MARI_22580
lipid A biosynthesis lauroyltransferase
Accession: QBM18128
Location: 2473173-2474132
NCBI BlastP on this gene
Accession: QBM18128
Location: 2473173-2474132
NCBI BlastP on this gene
lpxL_1
3-deoxy-D-manno-octulosonic acid transferase
Accession: QBM18127
Location: 2471817-2473070
NCBI BlastP on this gene
Accession: QBM18127
Location: 2471817-2473070
NCBI BlastP on this gene
waaA
tolC
320. : CP002637 Selenomonas sputigena ATCC 35185 Total score: 1.0 Cumulative Blast bit score: 140
Selsp_1174
polysaccharide pyruvyl transferase CsaB
Accession: AEC00133
Location: 1314550-1315668
NCBI BlastP on this gene
Accession: AEC00133
Location: 1314550-1315668
NCBI BlastP on this gene
Selsp_1173
UspA domain-containing protein
Accession: AEC00132
Location: 1313992-1314372
NCBI BlastP on this gene
Accession: AEC00132
Location: 1313992-1314372
NCBI BlastP on this gene
Selsp_1172
Selsp_1171
prolipoprotein diacylglyceryl transferase
Accession: AEC00130
Location: 1312190-1313005
NCBI BlastP on this gene
Accession: AEC00130
Location: 1312190-1313005
NCBI BlastP on this gene
Selsp_1170
polysaccharide deacetylase
Accession: AEC00129
Location: 1311206-1312054
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 140
Sequence coverage: 99 %
E-value: 6e-36
NCBI BlastP on this gene
Accession: AEC00129
Location: 1311206-1312054
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 140
Sequence coverage: 99 %
E-value: 6e-36
NCBI BlastP on this gene
Selsp_1169
Selsp_1168
Selsp_1167
biotin/lipoyl attachment domain-containing protein
Accession: AEC00126
Location: 1305941-1306927
NCBI BlastP on this gene
Accession: AEC00126
Location: 1305941-1306927
NCBI BlastP on this gene
Selsp_1166
Selsp_1165
321. : CP010086 Clostridium beijerinckii strain NCIMB 14988 Total score: 1.0 Cumulative Blast bit score: 136
LF65_00314
LF65_00317
LF65_00318
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AJG96988
Location: 318116-318595
NCBI BlastP on this gene
Accession: AJG96988
Location: 318116-318595
NCBI BlastP on this gene
LF65_00319
LF65_00321
polysaccharide deacetylase
Accession: AJG96991
Location: 320024-320983
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 136
Sequence coverage: 82 %
E-value: 5e-34
NCBI BlastP on this gene
Accession: AJG96991
Location: 320024-320983
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 136
Sequence coverage: 82 %
E-value: 5e-34
NCBI BlastP on this gene
LF65_00322
LF65_00323
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: AJG96993
Location: 322387-323433
NCBI BlastP on this gene
Accession: AJG96993
Location: 322387-323433
NCBI BlastP on this gene
LF65_00324
MarR family transcriptional regulator
Accession: AJG96994
Location: 323646-324080
NCBI BlastP on this gene
Accession: AJG96994
Location: 323646-324080
NCBI BlastP on this gene
LF65_00325
LF65_00326
LF65_00327
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: AJG96997
Location: 325224-327806
NCBI BlastP on this gene
Accession: AJG96997
Location: 325224-327806
NCBI BlastP on this gene
LF65_00328
322. : CP016090 Clostridium beijerinckii strain BAS/B3/I/124 Total score: 1.0 Cumulative Blast bit score: 135
methyl-accepting chemotaxis protein McpB
Accession: AQS02926
Location: 312480-314957
NCBI BlastP on this gene
Accession: AQS02926
Location: 312480-314957
NCBI BlastP on this gene
mcpB_1
CLBIJ_03080
CLBIJ_03090
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AQS02929
Location: 316870-317349
NCBI BlastP on this gene
Accession: AQS02929
Location: 316870-317349
NCBI BlastP on this gene
ispF
CLBIJ_03110
RNA polymerase sigma factor SigI
Accession: AQS02931
Location: 317731-318762
NCBI BlastP on this gene
Accession: AQS02931
Location: 317731-318762
NCBI BlastP on this gene
sigI_1
poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase precursor
Accession: AQS02932
Location: 318778-319737
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 135
Sequence coverage: 82 %
E-value: 8e-34
NCBI BlastP on this gene
Accession: AQS02932
Location: 318778-319737
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 135
Sequence coverage: 82 %
E-value: 8e-34
NCBI BlastP on this gene
icaB
rsiV
bioB_1
CLBIJ_03160
CLBIJ_03170
quaternary ammonium compound-resistance protein SugE
Accession: AQS02937
Location: 323352-323678
NCBI BlastP on this gene
Accession: AQS02937
Location: 323352-323678
NCBI BlastP on this gene
sugE_1
adhE_1
323. : LN908213 Clostridium beijerinckii isolate C. beijerinckii DSM 6423 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 134
Methyl-accepting chemotaxis protein
Accession: CUU45382
Location: 313057-315534
NCBI BlastP on this gene
Accession: CUU45382
Location: 313057-315534
NCBI BlastP on this gene
CIBE_0297
conserved protein of unknown function
Accession: CUU45383
Location: 315909-316313
NCBI BlastP on this gene
Accession: CUU45383
Location: 315909-316313
NCBI BlastP on this gene
CIBE_0298
conserved protein of unknown function
Accession: CUU45384
Location: 316584-317339
NCBI BlastP on this gene
Accession: CUU45384
Location: 316584-317339
NCBI BlastP on this gene
CIBE_0299
2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase
Accession: CUU45385
Location: 317443-317919
NCBI BlastP on this gene
Accession: CUU45385
Location: 317443-317919
NCBI BlastP on this gene
ispF
conserved protein of unknown function
Accession: CUU45386
Location: 317993-318163
NCBI BlastP on this gene
Accession: CUU45386
Location: 317993-318163
NCBI BlastP on this gene
CIBE_0301
conserved protein of unknown function
Accession: CUU45387
Location: 318300-319331
NCBI BlastP on this gene
Accession: CUU45387
Location: 318300-319331
NCBI BlastP on this gene
CIBE_0302
Polysaccharide deacetylase
Accession: CUU45388
Location: 319347-320405
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 4e-33
NCBI BlastP on this gene
Accession: CUU45388
Location: 319347-320405
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 4e-33
NCBI BlastP on this gene
CIBE_0303
conserved protein of unknown function
Accession: CUU45389
Location: 320709-321464
NCBI BlastP on this gene
Accession: CUU45389
Location: 320709-321464
NCBI BlastP on this gene
CIBE_0304
CIBE_0305
fused acetaldehyde-CoA dehydrogenase;
Accession: CUU45391
Location: 323285-325867
NCBI BlastP on this gene
Accession: CUU45391
Location: 323285-325867
NCBI BlastP on this gene
adhE
324. : CP043998 Clostridium diolis strain DSM 15410 chromosome Total score: 1.0 Cumulative Blast bit score: 134
methyl-accepting chemotaxis protein
Accession: QES71561
Location: 296925-299402
NCBI BlastP on this gene
Accession: QES71561
Location: 296925-299402
NCBI BlastP on this gene
F3K33_01475
F3K33_01490
F3K33_01495
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: QES71564
Location: 301311-301787
NCBI BlastP on this gene
Accession: QES71564
Location: 301311-301787
NCBI BlastP on this gene
F3K33_01500
anti-sigma factor domain-containing protein
Accession: QES71565
Location: 302169-303200
NCBI BlastP on this gene
Accession: QES71565
Location: 302169-303200
NCBI BlastP on this gene
F3K33_01505
polysaccharide deacetylase family protein
Accession: QES71566
Location: 303216-304175
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
Accession: QES71566
Location: 303216-304175
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
F3K33_01510
DUF3298 and DUF4163 domain-containing protein
Accession: QES71567
Location: 304582-305337
NCBI BlastP on this gene
Accession: QES71567
Location: 304582-305337
NCBI BlastP on this gene
F3K33_01515
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: QES71568
Location: 305579-306625
NCBI BlastP on this gene
Accession: QES71568
Location: 305579-306625
NCBI BlastP on this gene
hydE
MarR family transcriptional regulator
Accession: QES71569
Location: 306838-307272
NCBI BlastP on this gene
Accession: QES71569
Location: 306838-307272
NCBI BlastP on this gene
F3K33_01525
F3K33_01530
multidrug efflux SMR transporter
Accession: QES71571
Location: 307791-308117
NCBI BlastP on this gene
Accession: QES71571
Location: 307791-308117
NCBI BlastP on this gene
F3K33_01535
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: QES71572
Location: 308416-311010
NCBI BlastP on this gene
Accession: QES71572
Location: 308416-311010
NCBI BlastP on this gene
adhE
325. : CP014331 Clostridium sp. MF28, genome. Total score: 1.0 Cumulative Blast bit score: 134
AXY43_15660
AXY43_15675
AXY43_15680
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AVK49323
Location: 3657982-3658458
NCBI BlastP on this gene
Accession: AVK49323
Location: 3657982-3658458
NCBI BlastP on this gene
AXY43_15685
AXY43_15690
polysaccharide deacetylase
Accession: AVK49325
Location: 3659887-3660846
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
Accession: AVK49325
Location: 3659887-3660846
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
AXY43_15695
AXY43_15700
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: AVK49327
Location: 3662250-3663296
NCBI BlastP on this gene
Accession: AVK49327
Location: 3662250-3663296
NCBI BlastP on this gene
AXY43_15705
MarR family transcriptional regulator
Accession: AVK49328
Location: 3663509-3663943
NCBI BlastP on this gene
Accession: AVK49328
Location: 3663509-3663943
NCBI BlastP on this gene
AXY43_15710
AXY43_15715
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: AVK49329
Location: 3665086-3667680
NCBI BlastP on this gene
Accession: AVK49329
Location: 3665086-3667680
NCBI BlastP on this gene
AXY43_15720
326. : CP011966 Clostridium beijerinckii NRRL B-598 chromosome Total score: 1.0 Cumulative Blast bit score: 134
methyl-accepting chemotaxis protein
Accession: ALB48351
Location: 301218-303695
NCBI BlastP on this gene
Accession: ALB48351
Location: 301218-303695
NCBI BlastP on this gene
X276_25350
X276_25335
X276_25330
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: ALB48349
Location: 305604-306080
NCBI BlastP on this gene
Accession: ALB48349
Location: 305604-306080
NCBI BlastP on this gene
X276_25325
anti-sigma factor domain-containing protein
Accession: ALB48348
Location: 306462-307493
NCBI BlastP on this gene
Accession: ALB48348
Location: 306462-307493
NCBI BlastP on this gene
X276_25320
polysaccharide deacetylase family protein
Accession: ALB48347
Location: 307509-308468
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
Accession: ALB48347
Location: 307509-308468
BlastP hit with GL636894_4735_5544_-
Percentage identity: 37 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 3e-33
NCBI BlastP on this gene
X276_25315
DUF3298/DUF4163 domain-containing protein
Accession: ALB48346
Location: 308875-309630
NCBI BlastP on this gene
Accession: ALB48346
Location: 308875-309630
NCBI BlastP on this gene
X276_25310
[FeFe] hydrogenase H-cluster radical SAM maturase HydE
Accession: ALB48345
Location: 309872-310918
NCBI BlastP on this gene
Accession: ALB48345
Location: 309872-310918
NCBI BlastP on this gene
hydE
QacE family quaternary ammonium compound efflux SMR transporter
Accession: X276_27430
Location: 311030-311143
NCBI BlastP on this gene
Accession: X276_27430
Location: 311030-311143
NCBI BlastP on this gene
X276_27430
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: ALB48344
Location: 311442-314036
NCBI BlastP on this gene
Accession: ALB48344
Location: 311442-314036
NCBI BlastP on this gene
X276_25300
327. : CP006777 Clostridium beijerinckii ATCC 35702 Total score: 1.0 Cumulative Blast bit score: 134
methyl-accepting chemotaxis sensory transducer
Accession: AIU02043
Location: 352392-354869
NCBI BlastP on this gene
Accession: AIU02043
Location: 352392-354869
NCBI BlastP on this gene
Cbs_0294
Cbs_0295
Cbs_0296
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: AIU00312
Location: 356778-357254
NCBI BlastP on this gene
Accession: AIU00312
Location: 356778-357254
NCBI BlastP on this gene
ispF
Cbs_0298
polysaccharide deacetylase
Accession: AIU03183
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: AIU03183
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Cbs_0299
Cbs_0300
Cbs_0301
MarR family transcriptional regulator
Accession: AIU03625
Location: 362304-362738
NCBI BlastP on this gene
Accession: AIU03625
Location: 362304-362738
NCBI BlastP on this gene
Cbs_0302
Cbs_0303
small multidrug resistance protein
Accession: AIU03858
Location: 363257-363583
NCBI BlastP on this gene
Accession: AIU03858
Location: 363257-363583
NCBI BlastP on this gene
Cbs_0304
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: AIU02688
Location: 363882-366476
NCBI BlastP on this gene
Accession: AIU02688
Location: 363882-366476
NCBI BlastP on this gene
Cbs_0305
328. : CP000721 Clostridium beijerinckii NCIMB 8052 Total score: 1.0 Cumulative Blast bit score: 134
methyl-accepting chemotaxis sensory transducer
Accession: ABR32482
Location: 352392-354869
NCBI BlastP on this gene
Accession: ABR32482
Location: 352392-354869
NCBI BlastP on this gene
Cbei_0294
Cbei_0295
Cbei_0296
2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: ABR32485
Location: 356778-357254
NCBI BlastP on this gene
Accession: ABR32485
Location: 356778-357254
NCBI BlastP on this gene
Cbei_0297
Cbei_0298
polysaccharide deacetylase
Accession: ABR32487
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: ABR32487
Location: 358683-359642
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 134
Sequence coverage: 82 %
E-value: 2e-33
NCBI BlastP on this gene
Cbei_0299
Cbei_0300
Cbei_0301
transcriptional regulator, MarR family
Accession: ABR32490
Location: 362304-362738
NCBI BlastP on this gene
Accession: ABR32490
Location: 362304-362738
NCBI BlastP on this gene
Cbei_0302
Cbei_0303
small multidrug resistance protein
Accession: ABR32492
Location: 363257-363583
NCBI BlastP on this gene
Accession: ABR32492
Location: 363257-363583
NCBI BlastP on this gene
Cbei_0304
iron-containing alcohol dehydrogenase
Accession: ABR32493
Location: 363882-366476
NCBI BlastP on this gene
Accession: ABR32493
Location: 363882-366476
NCBI BlastP on this gene
Cbei_0305
329. : CP002637 Selenomonas sputigena ATCC 35185 Total score: 1.0 Cumulative Blast bit score: 130
phospho-N-acetylmuramoyl-pentapeptide- transferase
Accession: AEC00777
Location: 2004406-2005380
NCBI BlastP on this gene
Accession: AEC00777
Location: 2004406-2005380
NCBI BlastP on this gene
Selsp_1822
UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-
Accession: AEC00778
Location: 2005377-2008277
NCBI BlastP on this gene
Accession: AEC00778
Location: 2005377-2008277
NCBI BlastP on this gene
Selsp_1823
Selsp_1824
S-adenosyl-methyltransferase MraW
Accession: AEC00780
Location: 2008764-2009699
NCBI BlastP on this gene
Accession: AEC00780
Location: 2008764-2009699
NCBI BlastP on this gene
Selsp_1825
Selsp_1826
polysaccharide deacetylase
Accession: AEC00782
Location: 2010387-2011193
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 130
Sequence coverage: 82 %
E-value: 1e-32
NCBI BlastP on this gene
Accession: AEC00782
Location: 2010387-2011193
BlastP hit with GL636894_4735_5544_-
Percentage identity: 38 %
BlastP bit score: 130
Sequence coverage: 82 %
E-value: 1e-32
NCBI BlastP on this gene
Selsp_1827
330. : CP014331 Clostridium sp. MF28, genome. Total score: 1.0 Cumulative Blast bit score: 127
polysaccharide deacetylase
Accession: AVK51319
Location: 6105005-6105919
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 9e-31
NCBI BlastP on this gene
Accession: AVK51319
Location: 6105005-6105919
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 9e-31
NCBI BlastP on this gene
AXY43_26710
AXY43_26705
AXY43_26700
AXY43_26695
AXY43_26690
AXY43_26685
AXY43_26680
AXY43_26675
331. : CP010086 Clostridium beijerinckii strain NCIMB 14988 Total score: 1.0 Cumulative Blast bit score: 127
LF65_03181
YgiQ family radical SAM protein
Accession: AJG99747
Location: 3511379-3513358
NCBI BlastP on this gene
Accession: AJG99747
Location: 3511379-3513358
NCBI BlastP on this gene
LF65_03182
LF65_03184
LF65_03185
polysaccharide deacetylase
Accession: AJG99751
Location: 3515874-3516788
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 1e-30
NCBI BlastP on this gene
Accession: AJG99751
Location: 3515874-3516788
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 127
Sequence coverage: 85 %
E-value: 1e-30
NCBI BlastP on this gene
LF65_03186
332. : CP016090 Clostridium beijerinckii strain BAS/B3/I/124 Total score: 1.0 Cumulative Blast bit score: 126
poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase precursor
Accession: AQS05090
Location: 2750134-2751048
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 126
Sequence coverage: 85 %
E-value: 2e-30
NCBI BlastP on this gene
Accession: AQS05090
Location: 2750134-2751048
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 126
Sequence coverage: 85 %
E-value: 2e-30
NCBI BlastP on this gene
pgaB
CLBIJ_25200
CLBIJ_25190
CLBIJ_25180
CLBIJ_25170
CLBIJ_25160
small, acid-soluble spore protein beta
Accession: AQS05084
Location: 2746964-2747170
NCBI BlastP on this gene
Accession: AQS05084
Location: 2746964-2747170
NCBI BlastP on this gene
CLBIJ_25150
CLBIJ_25140
CLBIJ_25130
CLBIJ_25120
CLBIJ_25110
333. : CP043998 Clostridium diolis strain DSM 15410 chromosome Total score: 1.0 Cumulative Blast bit score: 125
polysaccharide deacetylase family protein
Accession: QES73502
Location: 2638497-2639411
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: QES73502
Location: 2638497-2639411
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
F3K33_12030
F3K33_12025
F3K33_12020
F3K33_12015
alpha/beta-type small acid-soluble spore protein
Accession: QES73498
Location: 2635135-2635341
NCBI BlastP on this gene
Accession: QES73498
Location: 2635135-2635341
NCBI BlastP on this gene
F3K33_12010
F3K33_12005
F3K33_12000
334. : CP011966 Clostridium beijerinckii NRRL B-598 chromosome Total score: 1.0 Cumulative Blast bit score: 125
polysaccharide deacetylase family protein
Accession: ALB46438
Location: 2662157-2663071
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: ALB46438
Location: 2662157-2663071
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
X276_14905
X276_14910
X276_14915
X276_14920
alpha/beta-type small acid-soluble spore protein
Accession: ALB46442
Location: 2658795-2659001
NCBI BlastP on this gene
Accession: ALB46442
Location: 2658795-2659001
NCBI BlastP on this gene
X276_14925
X276_14930
X276_14935
335. : CP006777 Clostridium beijerinckii ATCC 35702 Total score: 1.0 Cumulative Blast bit score: 125
polysaccharide deacetylase
Accession: AIU03179
Location: 2712562-2713476
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: AIU03179
Location: 2712562-2713476
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Cbs_2350
Cbs_2349
Cbs_2348
Cbs_2347
Cbs_2346
small acid-soluble spore protein, alpha/beta type
Accession: AIU03854
Location: 2709191-2709397
NCBI BlastP on this gene
Accession: AIU03854
Location: 2709191-2709397
NCBI BlastP on this gene
Cbs_2345
Cbs_2344
336. : CP000721 Clostridium beijerinckii NCIMB 8052 Total score: 1.0 Cumulative Blast bit score: 125
polysaccharide deacetylase
Accession: ABR34510
Location: 2712567-2713481
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Accession: ABR34510
Location: 2712567-2713481
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 125
Sequence coverage: 85 %
E-value: 4e-30
NCBI BlastP on this gene
Cbei_2350
Cbei_2349
Cbei_2348
Cbei_2347
Cbei_2346
small acid-soluble spore protein, alpha/beta type
Accession: ABR34505
Location: 2709196-2709402
NCBI BlastP on this gene
Accession: ABR34505
Location: 2709196-2709402
NCBI BlastP on this gene
Cbei_2345
Cbei_2344
337. : LN908213 Clostridium beijerinckii isolate C. beijerinckii DSM 6423 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 123
conserved exported protein of unknown function
Accession: CUU47882
Location: 2805018-2805932
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 123
Sequence coverage: 85 %
E-value: 2e-29
NCBI BlastP on this gene
Accession: CUU47882
Location: 2805018-2805932
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 123
Sequence coverage: 85 %
E-value: 2e-29
NCBI BlastP on this gene
CIBE_2781
conserved protein of unknown function
Accession: CUU47881
Location: 2804007-2804312
NCBI BlastP on this gene
Accession: CUU47881
Location: 2804007-2804312
NCBI BlastP on this gene
CIBE_2780
conserved protein of unknown function
Accession: CUU47880
Location: 2803182-2803688
NCBI BlastP on this gene
Accession: CUU47880
Location: 2803182-2803688
NCBI BlastP on this gene
CIBE_2779
conserved protein of unknown function
Accession: CUU47879
Location: 2801031-2803007
NCBI BlastP on this gene
Accession: CUU47879
Location: 2801031-2803007
NCBI BlastP on this gene
CIBE_2778
conserved protein of unknown function
Accession: CUU47878
Location: 2800724-2800954
NCBI BlastP on this gene
Accession: CUU47878
Location: 2800724-2800954
NCBI BlastP on this gene
CIBE_2777
conserved protein of unknown function
Accession: CUU47877
Location: 2800408-2800569
NCBI BlastP on this gene
Accession: CUU47877
Location: 2800408-2800569
NCBI BlastP on this gene
CIBE_2776
Small, acid-soluble spore protein beta
Accession: CUU47876
Location: 2799941-2800147
NCBI BlastP on this gene
Accession: CUU47876
Location: 2799941-2800147
NCBI BlastP on this gene
CIBE_2775
338. : LT906477 Clostridium cochlearium strain NCTC13027 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 122
tpl_1
metal-binding protein of ferredoxin fold
Accession: SNV63480
Location: 70754-70957
NCBI BlastP on this gene
Accession: SNV63480
Location: 70754-70957
NCBI BlastP on this gene
SAMEA4530647_00079
SAMEA4530647_00080
SAMEA4530647_00081
corrin/porphyrin methyltransferase
Accession: SNV63526
Location: 72087-72935
NCBI BlastP on this gene
Accession: SNV63526
Location: 72087-72935
NCBI BlastP on this gene
rsmI
iap
transition state regulatory protein AbrB
Accession: SNV63556
Location: 74352-74594
NCBI BlastP on this gene
Accession: SNV63556
Location: 74352-74594
NCBI BlastP on this gene
abrB
tal
polysaccharide deacetylase
Accession: SNV63583
Location: 75801-76688
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 122
Sequence coverage: 86 %
E-value: 4e-29
NCBI BlastP on this gene
Accession: SNV63583
Location: 75801-76688
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 122
Sequence coverage: 86 %
E-value: 4e-29
NCBI BlastP on this gene
icaB
glcK
SAMEA4530647_00088
hexA_1
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
Accession: SNV63641
Location: 80225-80707
NCBI BlastP on this gene
Accession: SNV63641
Location: 80225-80707
NCBI BlastP on this gene
ispF
yabJ_1
lytD
339. : LT906477 Clostridium cochlearium strain NCTC13027 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 120
polysaccharide deacetylase
Accession: SNV74675
Location: 966480-967232
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 121
Sequence coverage: 82 %
E-value: 3e-29
NCBI BlastP on this gene
Accession: SNV74675
Location: 966480-967232
BlastP hit with GL636894_4735_5544_-
Percentage identity: 34 %
BlastP bit score: 121
Sequence coverage: 82 %
E-value: 3e-29
NCBI BlastP on this gene
pgaB
phosphatase-associated protein papQ
Accession: SNV74662
Location: 964484-965506
NCBI BlastP on this gene
Accession: SNV74662
Location: 964484-965506
NCBI BlastP on this gene
papQ
ngr
spxA
spore peptidoglycan hydrolase (N-acetylglucosaminidase)
Accession: SNV74630
Location: 961980-963257
NCBI BlastP on this gene
Accession: SNV74630
Location: 961980-963257
NCBI BlastP on this gene
ydhD
yeeE
340. : CP034842 Fusobacterium necrophorum subsp. necrophorum strain ATCC 25286 chromosome Total score: 1.0 Cumulative Blast bit score: 120
lipopolysaccharide heptosyltransferase family protein
Accession: AZW09987
Location: 2303138-2304211
NCBI BlastP on this gene
Accession: AZW09987
Location: 2303138-2304211
NCBI BlastP on this gene
EO219_10670
EO219_10675
EO219_10680
polysaccharide deacetylase family protein
Accession: AZW09990
Location: 2306319-2307077
NCBI BlastP on this gene
Accession: AZW09990
Location: 2306319-2307077
NCBI BlastP on this gene
EO219_10685
lipopolysaccharide biosynthesis protein
Accession: AZW09991
Location: 2307053-2307811
NCBI BlastP on this gene
Accession: AZW09991
Location: 2307053-2307811
NCBI BlastP on this gene
EO219_10690
galE
polysaccharide deacetylase family protein
Accession: AZW09993
Location: 2308816-2309565
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
Accession: AZW09993
Location: 2308816-2309565
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
EO219_10700
EO219_10705
341. : CP033837 Fusobacterium necrophorum strain FDAARGOS_565 chromosome Total score: 1.0 Cumulative Blast bit score: 120
polysaccharide deacetylase family protein
Accession: AYZ74127
Location: 1928804-1929553
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
Accession: AYZ74127
Location: 1928804-1929553
BlastP hit with GL636894_4735_5544_-
Percentage identity: 33 %
BlastP bit score: 121
Sequence coverage: 86 %
E-value: 3e-29
NCBI BlastP on this gene
EGX98_08860
EGX98_08855
EGX98_08850
EGX98_08845
O-antigen ligase domain-containing protein
Accession: AYZ74123
Location: 1923301-1924860
NCBI BlastP on this gene
Accession: AYZ74123
Location: 1923301-1924860
NCBI BlastP on this gene
EGX98_08840
342. : CP036171 Acinetobacter nosocomialis strain KAN02 chromosome Total score: 1.0 Cumulative Blast bit score: 112
glucose-6-phosphate isomerase
Accession: QBF79880
Location: 3866789-3868459
NCBI BlastP on this gene
Accession: QBF79880
Location: 3866789-3868459
NCBI BlastP on this gene
KAN02_18540
UDP-glucose/GDP-mannose dehydrogenase family protein
Accession: QBF79881
Location: 3868456-3869718
NCBI BlastP on this gene
Accession: QBF79881
Location: 3868456-3869718
NCBI BlastP on this gene
KAN02_18545
UTP--glucose-1-phosphate uridylyltransferase GalU
Accession: QBF79882
Location: 3869833-3870708
NCBI BlastP on this gene
Accession: QBF79882
Location: 3869833-3870708
NCBI BlastP on this gene
galU
KAN02_18555
KAN02_18560
glycosyltransferase family 2 protein
Accession: QBF79885
Location: 3872196-3873098
NCBI BlastP on this gene
Accession: QBF79885
Location: 3872196-3873098
NCBI BlastP on this gene
KAN02_18565
hypothetical protein
Accession: QBF79886
Location: 3873091-3874032
BlastP hit with GL636894_1983_2612_-
Percentage identity: 39 %
BlastP bit score: 113
Sequence coverage: 67 %
E-value: 3e-26
NCBI BlastP on this gene
Accession: QBF79886
Location: 3873091-3874032
BlastP hit with GL636894_1983_2612_-
Percentage identity: 39 %
BlastP bit score: 113
Sequence coverage: 67 %
E-value: 3e-26
NCBI BlastP on this gene
KAN02_18570
KAN02_18575
343. : CP036267 Planctomycetes bacterium Mal48 chromosome. Total score: 1.0 Cumulative Blast bit score: 103
Mal48_11160
Mal48_11150
Enoyl-[acyl-carrier-protein] reductase [NADPH] FabL
Accession: QDT31878
Location: 1381960-1382706
NCBI BlastP on this gene
Accession: QDT31878
Location: 1381960-1382706
NCBI BlastP on this gene
fabL
Mal48_11130
Mal48_11120
Mal48_11110
Alpha-D-kanosaminyltransferase
Accession: QDT31874
Location: 1378495-1379649
BlastP hit with GL636894_3749_4732_-
Percentage identity: 31 %
BlastP bit score: 104
Sequence coverage: 70 %
E-value: 5e-22
NCBI BlastP on this gene
Accession: QDT31874
Location: 1378495-1379649
BlastP hit with GL636894_3749_4732_-
Percentage identity: 31 %
BlastP bit score: 104
Sequence coverage: 70 %
E-value: 5e-22
NCBI BlastP on this gene
kanE_1
3-oxoacyl-[acyl-carrier-protein] reductase FabG
Accession: QDT31873
Location: 1377675-1378406
NCBI BlastP on this gene
Accession: QDT31873
Location: 1377675-1378406
NCBI BlastP on this gene
fabG_2
D-erythro-7,8-dihydroneopterin triphosphate epimerase
Accession: QDT31872
Location: 1377160-1377528
NCBI BlastP on this gene
Accession: QDT31872
Location: 1377160-1377528
NCBI BlastP on this gene
folX
2-amino-4-hydroxy-6- hydroxymethyldihydropteridine pyrophosphokinase
Accession: QDT31871
Location: 1376489-1377055
NCBI BlastP on this gene
Accession: QDT31871
Location: 1376489-1377055
NCBI BlastP on this gene
folK
Mal48_11060
50S ribosomal protein L31 type B
Accession: QDT31869
Location: 1375278-1375592
NCBI BlastP on this gene
Accession: QDT31869
Location: 1375278-1375592
NCBI BlastP on this gene
rpmE2
Peptide chain release factor RF1
Accession: QDT31868
Location: 1373978-1375048
NCBI BlastP on this gene
Accession: QDT31868
Location: 1373978-1375048
NCBI BlastP on this gene
prfA
Release factor glutamine methyltransferase
Accession: QDT31867
Location: 1372989-1373888
NCBI BlastP on this gene
Accession: QDT31867
Location: 1372989-1373888
NCBI BlastP on this gene
prmC
344. : CP011427 Pantoea vagans strain ND02 chromosome Total score: 1.0 Cumulative Blast bit score: 103
LK04_00315
LK04_00320
universal stress global response regulator UspA
Accession: ALV90686
Location: 72655-73092
NCBI BlastP on this gene
Accession: ALV90686
Location: 72655-73092
NCBI BlastP on this gene
LK04_00325
LK04_00330
LK04_00335
LK04_00340
mannosyltransferase
Accession: ALV90689
Location: 76989-77981
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 104
Sequence coverage: 72 %
E-value: 5e-23
NCBI BlastP on this gene
Accession: ALV90689
Location: 76989-77981
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 104
Sequence coverage: 72 %
E-value: 5e-23
NCBI BlastP on this gene
LK04_00345
LK04_00350
LK04_00355
AraC family transcriptional regulator
Accession: ALV90692
Location: 79399-80400
NCBI BlastP on this gene
Accession: ALV90692
Location: 79399-80400
NCBI BlastP on this gene
LK04_00360
AraC family transcriptional regulator
Accession: ALV90693
Location: 80611-81102
NCBI BlastP on this gene
Accession: ALV90693
Location: 80611-81102
NCBI BlastP on this gene
LK04_00365
LK04_00370
LK04_00375
345. : CP009454 Pantoea rwandensis strain ND04 Total score: 1.0 Cumulative Blast bit score: 99
LH22_03565
universal stress global response regulator UspA
Accession: AIR84583
Location: 784438-784875
NCBI BlastP on this gene
Accession: AIR84583
Location: 784438-784875
NCBI BlastP on this gene
LH22_03570
LH22_03575
LH22_03580
LH22_03585
mannosyltransferase
Accession: AIR84587
Location: 788853-789845
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 99
Sequence coverage: 65 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AIR84587
Location: 788853-789845
BlastP hit with GL636894_1983_2612_-
Percentage identity: 35 %
BlastP bit score: 99
Sequence coverage: 65 %
E-value: 2e-21
NCBI BlastP on this gene
LH22_03590
LH22_03595
LH22_03600
AraC family transcriptional regulator
Accession: AIR84590
Location: 791249-792238
NCBI BlastP on this gene
Accession: AIR84590
Location: 791249-792238
NCBI BlastP on this gene
LH22_03605
AraC family transcriptional regulator
Accession: AIR84591
Location: 792436-792924
NCBI BlastP on this gene
Accession: AIR84591
Location: 792436-792924
NCBI BlastP on this gene
LH22_03610
LH22_03615
LH22_03620
346. : CP035934 Acinetobacter cumulans strain WCHAc060092 chromosome Total score: 1.0 Cumulative Blast bit score: 92
bfr
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: QCO20461
Location: 403353-403808
NCBI BlastP on this gene
Accession: QCO20461
Location: 403353-403808
NCBI BlastP on this gene
C9E88_002450
C9E88_002455
C9E88_002460
glf
C9E88_002470
hypothetical protein
Accession: QCO20466
Location: 397956-398933
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 92
Sequence coverage: 81 %
E-value: 6e-19
NCBI BlastP on this gene
Accession: QCO20466
Location: 397956-398933
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 92
Sequence coverage: 81 %
E-value: 6e-19
NCBI BlastP on this gene
C9E88_002475
C9E88_002480
class I SAM-dependent methyltransferase
Accession: QCO22789
Location: 396508-397134
NCBI BlastP on this gene
Accession: QCO22789
Location: 396508-397134
NCBI BlastP on this gene
C9E88_002485
C9E88_002490
ATP-binding cassette domain-containing protein
Accession: QCO20469
Location: 393390-395099
NCBI BlastP on this gene
Accession: QCO20469
Location: 393390-395099
NCBI BlastP on this gene
C9E88_002495
C9E88_002500
347. : CP025618 Acinetobacter schindleri strain SGAir0122 chromosome Total score: 1.0 Cumulative Blast bit score: 88
glycosyltransferase family 1 protein
Accession: C0119_09995
Location: 2538982-2539659
NCBI BlastP on this gene
Accession: C0119_09995
Location: 2538982-2539659
NCBI BlastP on this gene
C0119_09995
C0119_09990
C0119_09985
glycosyltransferase family 25 protein
Accession: AWD70549
Location: 2541258-2542067
NCBI BlastP on this gene
Accession: AWD70549
Location: 2541258-2542067
NCBI BlastP on this gene
C0119_09980
C0119_09975
lipopolysaccharide biosynthesis protein
Accession: AWD70547
Location: 2543723-2544652
NCBI BlastP on this gene
Accession: AWD70547
Location: 2543723-2544652
NCBI BlastP on this gene
C0119_09970
hypothetical protein
Accession: AWD70546
Location: 2544690-2545613
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 88
Sequence coverage: 92 %
E-value: 2e-17
NCBI BlastP on this gene
Accession: AWD70546
Location: 2544690-2545613
BlastP hit with GL636894_1983_2612_-
Percentage identity: 32 %
BlastP bit score: 88
Sequence coverage: 92 %
E-value: 2e-17
NCBI BlastP on this gene
C0119_09965
348. : CP037934 Marinobacter sp. JH2 chromosome Total score: 1.0 Cumulative Blast bit score: 85
MARI_13070
MARI_13080
MARI_13090
MARI_13100
MARI_13110
MARI_13120
hypothetical protein
Accession: QBM17207
Location: 1419559-1420686
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 85
Sequence coverage: 67 %
E-value: 2e-15
NCBI BlastP on this gene
Accession: QBM17207
Location: 1419559-1420686
BlastP hit with GL636894_3749_4732_-
Percentage identity: 32 %
BlastP bit score: 85
Sequence coverage: 67 %
E-value: 2e-15
NCBI BlastP on this gene
MARI_13130
MARI_13140
MARI_13150
MARI_13160
MARI_13170
MARI_13180
349. : CP011427 Pantoea vagans strain ND02 chromosome Total score: 1.0 Cumulative Blast bit score: 82
LysR family transcriptional regulator
Accession: ALV93564
Location: 3454259-3455182
NCBI BlastP on this gene
Accession: ALV93564
Location: 3454259-3455182
NCBI BlastP on this gene
LK04_16040
mannosyltransferase OCH1-related enzyme
Accession: ALV93563
Location: 3453272-3454285
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 82
Sequence coverage: 70 %
E-value: 3e-15
NCBI BlastP on this gene
Accession: ALV93563
Location: 3453272-3454285
BlastP hit with GL636894_1983_2612_-
Percentage identity: 33 %
BlastP bit score: 82
Sequence coverage: 70 %
E-value: 3e-15
NCBI BlastP on this gene
LK04_16035
LK04_16030
LK04_16025
LK04_16020
LK04_16015
LK04_16010
350. : CP009454 Pantoea rwandensis strain ND04 Total score: 1.0 Cumulative Blast bit score: 81
LysR family transcriptional regulator
Accession: AIR87520
Location: 4112479-4113402
NCBI BlastP on this gene
Accession: AIR87520
Location: 4112479-4113402
NCBI BlastP on this gene
LH22_19390
mannosyltransferase OCH1-related enzyme
Accession: AIR87519
Location: 4111492-4112505
BlastP hit with GL636894_1983_2612_-
Percentage identity: 34 %
BlastP bit score: 81
Sequence coverage: 70 %
E-value: 6e-15
NCBI BlastP on this gene
Accession: AIR87519
Location: 4111492-4112505
BlastP hit with GL636894_1983_2612_-
Percentage identity: 34 %
BlastP bit score: 81
Sequence coverage: 70 %
E-value: 6e-15
NCBI BlastP on this gene
LH22_19385
LH22_19380
LH22_19375
LH22_19370
LH22_19365
LH22_19360

Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.