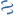MultiGeneBlast hits

Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP040017 : Massilia umbonata strain DSMZ 26121 chromosome Total score: 1.0 Cumulative Blast bit score: 496
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
mannanase
Accession: QCP11084
Location: 2987151-2988455
BlastP hit with man5A
Percentage identity: 55 %
BlastP bit score: 497
Sequence coverage: 89 %
E-value: 6e-170
NCBI BlastP on this gene
Accession: QCP11084
Location: 2987151-2988455
BlastP hit with man5A
Percentage identity: 55 %
BlastP bit score: 497
Sequence coverage: 89 %
E-value: 6e-170
NCBI BlastP on this gene
FCL38_12185
FCL38_12180
FCL38_12175
FCL38_12170
FCL38_12165
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
FP929059 : Eubacterium siraeum V10Sc8a draft genome. Total score: 1.0 Cumulative Blast bit score: 496
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
ES1_21250
ES1_21260
ES1_21270
large conductance mechanosensitive channel protein
Accession: CBL34988
Location: 2293372-2293935
NCBI BlastP on this gene
Accession: CBL34988
Location: 2293372-2293935
NCBI BlastP on this gene
ES1_21300
Predicted glycosylase
Accession: CBL34989
Location: 2294698-2295873
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 496
Sequence coverage: 99 %
E-value: 2e-171
NCBI BlastP on this gene
Accession: CBL34989
Location: 2294698-2295873
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 496
Sequence coverage: 99 %
E-value: 2e-171
NCBI BlastP on this gene
ES1_21320
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003557 : Melioribacter roseus P3M Total score: 1.0 Cumulative Blast bit score: 495
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: AFN75841
Location: 3036326-3037546
BlastP hit with aga27A
Percentage identity: 57 %
BlastP bit score: 495
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
Accession: AFN75841
Location: 3036326-3037546
BlastP hit with aga27A
Percentage identity: 57 %
BlastP bit score: 495
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
MROS_2611
MROS_2610
MROS_2609
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
LT906662 : Thermoanaerobacterium sp. RBIITD genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 494
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
4-O-beta-D-mannosyl-D-glucose phosphorylase
Accession: SNX52632
Location: 39235-40416
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 494
Sequence coverage: 98 %
E-value: 2e-170
NCBI BlastP on this gene
Accession: SNX52632
Location: 39235-40416
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 494
Sequence coverage: 98 %
E-value: 2e-170
NCBI BlastP on this gene
SAMN05660242_0044
Aryl-phospho-beta-D-glucosidase BglC, GH1 family
Accession: SNX52631
Location: 37475-38467
NCBI BlastP on this gene
Accession: SNX52631
Location: 37475-38467
NCBI BlastP on this gene
SAMN05660242_0042
SAMN05660242_0040
SAMN05660242_0039
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
FP929044 : Eubacterium siraeum 70/3 draft genome. Total score: 1.0 Cumulative Blast bit score: 493
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
EUS_08970
large conductance mechanosensitive channel protein
Accession: CBK96124
Location: 898651-899214
NCBI BlastP on this gene
Accession: CBK96124
Location: 898651-899214
NCBI BlastP on this gene
EUS_08940
Predicted glycosylase
Accession: CBK96123
Location: 896713-897888
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 493
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
Accession: CBK96123
Location: 896713-897888
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 493
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
EUS_08920
EUS_08910
EUS_08900
transcription antitermination protein nusG
Accession: CBK96120
Location: 894344-894865
NCBI BlastP on this gene
Accession: CBK96120
Location: 894344-894865
NCBI BlastP on this gene
EUS_08890
preprotein translocase, SecE subunit, bacterial
Accession: CBK96119
Location: 894023-894316
NCBI BlastP on this gene
Accession: CBK96119
Location: 894023-894316
NCBI BlastP on this gene
EUS_08880
EUS_08860
methenyltetrahydrofolate cyclohydrolase
Accession: CBK96117
Location: 892619-893458
NCBI BlastP on this gene
Accession: CBK96117
Location: 892619-893458
NCBI BlastP on this gene
EUS_08850
prolipoprotein diacylglyceryl transferase
Accession: CBK96116
Location: 891480-892619
NCBI BlastP on this gene
Accession: CBK96116
Location: 891480-892619
NCBI BlastP on this gene
EUS_08840
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP013020 : Bacteroides vulgatus strain mpk genome. Total score: 1.0 Cumulative Blast bit score: 481
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
BvMPK_0026
RagB/SusD Domain-Containing Protein
Accession: ALK82667
Location: 34112-35887
NCBI BlastP on this gene
Accession: ALK82667
Location: 34112-35887
NCBI BlastP on this gene
BvMPK_0025
Putative arylsulfate sulfotransferase
Accession: ALK82666
Location: 32111-33910
NCBI BlastP on this gene
Accession: ALK82666
Location: 32111-33910
NCBI BlastP on this gene
BvMPK_0024
Glycosidase-like Protein
Accession: ALK82665
Location: 30819-31997
BlastP hit with unk1
Percentage identity: 63 %
BlastP bit score: 481
Sequence coverage: 97 %
E-value: 2e-165
NCBI BlastP on this gene
Accession: ALK82665
Location: 30819-31997
BlastP hit with unk1
Percentage identity: 63 %
BlastP bit score: 481
Sequence coverage: 97 %
E-value: 2e-165
NCBI BlastP on this gene
BvMPK_0023
Alkaline phosphodiesterase I / Nucleotide pyrophosphatase
Accession: ALK82664
Location: 29759-30661
NCBI BlastP on this gene
Accession: ALK82664
Location: 29759-30661
NCBI BlastP on this gene
BvMPK_0022
putative sodium-dependent mannose transporter
Accession: ALK82663
Location: 27858-29741
NCBI BlastP on this gene
Accession: ALK82663
Location: 27858-29741
NCBI BlastP on this gene
BvMPK_0021
BvMPK_0020
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP042431 : Pseudobacter ginsenosidimutans strain Gsoil 221 chromosome Total score: 1.0 Cumulative Blast bit score: 464
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
glycosidase
Accession: QEC44537
Location: 6020039-6021220
BlastP hit with unk1
Percentage identity: 59 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 6e-159
NCBI BlastP on this gene
Accession: QEC44537
Location: 6020039-6021220
BlastP hit with unk1
Percentage identity: 59 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 6e-159
NCBI BlastP on this gene
FSB84_23725
FSB84_23720
FSB84_23715
FSB84_23710
30S ribosomal protein S12 methylthiotransferase RimO
Accession: QEC44533
Location: 6016715-6018025
NCBI BlastP on this gene
Accession: QEC44533
Location: 6016715-6018025
NCBI BlastP on this gene
rimO
aldehyde dehydrogenase (NADP(+))
Accession: QEC44532
Location: 6014900-6016387
NCBI BlastP on this gene
Accession: QEC44532
Location: 6014900-6016387
NCBI BlastP on this gene
FSB84_23700
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
AP019736 : Alistipes dispar 5CPEGH6 DNA Total score: 1.0 Cumulative Blast bit score: 464
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
hypothetical protein
Accession: BBL06472
Location: 1311959-1313182
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 2e-158
NCBI BlastP on this gene
Accession: BBL06472
Location: 1311959-1313182
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 2e-158
NCBI BlastP on this gene
A5CPEGH6_11100
A5CPEGH6_11090
magnesium transport protein CorA
Accession: BBL06470
Location: 1310153-1311136
NCBI BlastP on this gene
Accession: BBL06470
Location: 1310153-1311136
NCBI BlastP on this gene
corA-2
polysaccharide biosynthesis protein
Accession: BBL06469
Location: 1308334-1309806
NCBI BlastP on this gene
Accession: BBL06469
Location: 1308334-1309806
NCBI BlastP on this gene
A5CPEGH6_11070
DNA polymerase III subunit delta
Accession: BBL06468
Location: 1307168-1308313
NCBI BlastP on this gene
Accession: BBL06468
Location: 1307168-1308313
NCBI BlastP on this gene
A5CPEGH6_11060
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP026604 : Catenovulum sp. CCB-QB4 chromosome Total score: 1.0 Cumulative Blast bit score: 449
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
C2869_05755
dnaJ
C2869_05765
alpha-galactosidase
Accession: AWB65978
Location: 1632344-1633594
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 450
Sequence coverage: 101 %
E-value: 1e-152
NCBI BlastP on this gene
Accession: AWB65978
Location: 1632344-1633594
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 450
Sequence coverage: 101 %
E-value: 1e-152
NCBI BlastP on this gene
C2869_05770
MBL fold metallo-hydrolase
Accession: C2869_05775
Location: 1633700-1633858
NCBI BlastP on this gene
Accession: C2869_05775
Location: 1633700-1633858
NCBI BlastP on this gene
C2869_05775
C2869_05780
C2869_05785
C2869_05790
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP041242 : Lysobacter alkalisoli strain SJ-36 chromosome Total score: 1.0 Cumulative Blast bit score: 447
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
FKV23_01350
FKV23_01345
TRAP transporter substrate-binding protein
Accession: QDH68895
Location: 312857-313888
NCBI BlastP on this gene
Accession: QDH68895
Location: 312857-313888
NCBI BlastP on this gene
FKV23_01340
TRAP transporter small permease
Accession: QDH68894
Location: 312306-312848
NCBI BlastP on this gene
Accession: QDH68894
Location: 312306-312848
NCBI BlastP on this gene
FKV23_01335
TRAP transporter large permease
Accession: QDH68893
Location: 311020-312303
NCBI BlastP on this gene
Accession: QDH68893
Location: 311020-312303
NCBI BlastP on this gene
FKV23_01330
mannanase
Accession: QDH68892
Location: 309522-310880
BlastP hit with man5A
Percentage identity: 51 %
BlastP bit score: 448
Sequence coverage: 91 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: QDH68892
Location: 309522-310880
BlastP hit with man5A
Percentage identity: 51 %
BlastP bit score: 448
Sequence coverage: 91 %
E-value: 2e-150
NCBI BlastP on this gene
FKV23_01325
sugar porter family MFS transporter
Accession: QDH68891
Location: 307717-309159
NCBI BlastP on this gene
Accession: QDH68891
Location: 307717-309159
NCBI BlastP on this gene
FKV23_01320
FKV23_01315
LacI family transcriptional regulator
Accession: QDH68889
Location: 304681-305751
NCBI BlastP on this gene
Accession: QDH68889
Location: 304681-305751
NCBI BlastP on this gene
FKV23_01310
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
AP018694 : Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 446
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
AQPE_1317
AQPE_1316
AQPE_1315
alpha-galactosidase precursor
Accession: BBE17165
Location: 1475360-1476559
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 446
Sequence coverage: 98 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: BBE17165
Location: 1475360-1476559
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 446
Sequence coverage: 98 %
E-value: 2e-151
NCBI BlastP on this gene
AQPE_1314
AQPE_1313
dipeptidyl peptidase IV in 4-hydroxyproline catabolic gene cluster
Accession: BBE17163
Location: 1469815-1472067
NCBI BlastP on this gene
Accession: BBE17163
Location: 1469815-1472067
NCBI BlastP on this gene
AQPE_1312
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP002403 : Ruminococcus albus 7 Total score: 1.0 Cumulative Blast bit score: 440
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
glycosidase related protein
Accession: ADU21379
Location: 950842-952002
BlastP hit with unk1
Percentage identity: 57 %
BlastP bit score: 440
Sequence coverage: 97 %
E-value: 2e-149
NCBI BlastP on this gene
Accession: ADU21379
Location: 950842-952002
BlastP hit with unk1
Percentage identity: 57 %
BlastP bit score: 440
Sequence coverage: 97 %
E-value: 2e-149
NCBI BlastP on this gene
Rumal_0852
Rumal_0851
Rumal_0850
Rumal_0849
Rumal_0848
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP013020 : Bacteroides vulgatus strain mpk genome. Total score: 1.0 Cumulative Blast bit score: 432
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
TonB family protein / TonB-dependent receptor
Accession: ALK83489
Location: 1053455-1056250
NCBI BlastP on this gene
Accession: ALK83489
Location: 1053455-1056250
NCBI BlastP on this gene
BvMPK_0871
putative outer membrane protein
Accession: ALK83490
Location: 1056272-1057951
NCBI BlastP on this gene
Accession: ALK83490
Location: 1056272-1057951
NCBI BlastP on this gene
BvMPK_0872
Alpha-galactosidase
Accession: ALK83491
Location: 1058161-1059411
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 433
Sequence coverage: 95 %
E-value: 6e-146
NCBI BlastP on this gene
Accession: ALK83491
Location: 1058161-1059411
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 433
Sequence coverage: 95 %
E-value: 6e-146
NCBI BlastP on this gene
BvMPK_0873
BvMPK_0874
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP026604 : Catenovulum sp. CCB-QB4 chromosome Total score: 1.0 Cumulative Blast bit score: 429
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
C2869_03430
C2869_03425
C2869_03420
alpha-galactosidase
Accession: AWB65540
Location: 921837-923051
BlastP hit with aga27A
Percentage identity: 53 %
BlastP bit score: 429
Sequence coverage: 98 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: AWB65540
Location: 921837-923051
BlastP hit with aga27A
Percentage identity: 53 %
BlastP bit score: 429
Sequence coverage: 98 %
E-value: 2e-144
NCBI BlastP on this gene
C2869_03415
C2869_03410
C2869_03405
GNAT family N-acetyltransferase
Accession: AWB65537
Location: 918802-919299
NCBI BlastP on this gene
Accession: AWB65537
Location: 918802-919299
NCBI BlastP on this gene
C2869_03400
C2869_03395
C2869_03390
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003349 : Solitalea canadensis DSM 3403 Total score: 1.0 Cumulative Blast bit score: 428
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
TonB-linked outer membrane protein, SusC/RagA family
Accession: AFD06586
Location: 1819121-1822195
NCBI BlastP on this gene
Accession: AFD06586
Location: 1819121-1822195
NCBI BlastP on this gene
Solca_1511
Solca_1512
Solca_1513
alpha-galactosidase
Accession: AFD06589
Location: 1825111-1826301
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 428
Sequence coverage: 98 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AFD06589
Location: 1825111-1826301
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 428
Sequence coverage: 98 %
E-value: 3e-144
NCBI BlastP on this gene
Solca_1514
Solca_1515
HD superfamily phosphohydrolase
Accession: AFD06591
Location: 1828500-1829381
NCBI BlastP on this gene
Accession: AFD06591
Location: 1828500-1829381
NCBI BlastP on this gene
Solca_1516
Solca_1517
Solca_1518
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP036491 : Bacteroides sp. A1C1 chromosome Total score: 1.0 Cumulative Blast bit score: 422
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
EYA81_15140
PHP domain-containing protein
Accession: QBJ19565
Location: 3692076-3693155
NCBI BlastP on this gene
Accession: QBJ19565
Location: 3692076-3693155
NCBI BlastP on this gene
EYA81_15145
EYA81_15150
glycoside hydrolase family 27 protein
Accession: QBJ19567
Location: 3694674-3695882
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 422
Sequence coverage: 98 %
E-value: 1e-141
NCBI BlastP on this gene
Accession: QBJ19567
Location: 3694674-3695882
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 422
Sequence coverage: 98 %
E-value: 1e-141
NCBI BlastP on this gene
EYA81_15155
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP026100 : Caulobacter flavus strain RHGG3 chromosome Total score: 1.0 Cumulative Blast bit score: 415
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
C1707_15780
C1707_15785
alpha-galactosidase
Accession: AYV47601
Location: 3416557-3417747
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 415
Sequence coverage: 94 %
E-value: 6e-139
NCBI BlastP on this gene
Accession: AYV47601
Location: 3416557-3417747
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 415
Sequence coverage: 94 %
E-value: 6e-139
NCBI BlastP on this gene
C1707_15790
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP000927 : Caulobacter sp. K31 chromosome Total score: 1.0 Cumulative Blast bit score: 413
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
Caul_2125
Caul_2126
putative transposase of insertion sequence
Accession: ABZ71255
Location: 2295162-2295509
NCBI BlastP on this gene
Accession: ABZ71255
Location: 2295162-2295509
NCBI BlastP on this gene
Caul_2127
Caul_2128
Alpha-galactosidase
Accession: ABZ71257
Location: 2296299-2297522
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 413
Sequence coverage: 93 %
E-value: 4e-138
NCBI BlastP on this gene
Accession: ABZ71257
Location: 2296299-2297522
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 413
Sequence coverage: 93 %
E-value: 4e-138
NCBI BlastP on this gene
Caul_2129
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP035501 : Sphingosinicella sp. BN140058 chromosome Total score: 1.0 Cumulative Blast bit score: 410
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
ETR14_01430
ETR14_01435
ETR14_01440
glycoside hydrolase family 27 protein
Accession: QAY75340
Location: 341428-342663
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 410
Sequence coverage: 95 %
E-value: 6e-137
NCBI BlastP on this gene
Accession: QAY75340
Location: 341428-342663
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 410
Sequence coverage: 95 %
E-value: 6e-137
NCBI BlastP on this gene
ETR14_01445
ankyrin repeat domain-containing protein
Accession: QAY75341
Location: 342669-343787
NCBI BlastP on this gene
Accession: QAY75341
Location: 342669-343787
NCBI BlastP on this gene
ETR14_01450
ETR14_01455
DUF1801 domain-containing protein
Accession: QAY75343
Location: 344134-344538
NCBI BlastP on this gene
Accession: QAY75343
Location: 344134-344538
NCBI BlastP on this gene
ETR14_01460
DUF1801 domain-containing protein
Accession: QAY79656
Location: 344557-344988
NCBI BlastP on this gene
Accession: QAY79656
Location: 344557-344988
NCBI BlastP on this gene
ETR14_01465
ETR14_01470
SRPBCC domain-containing protein
Accession: QAY75345
Location: 345307-345717
NCBI BlastP on this gene
Accession: QAY75345
Location: 345307-345717
NCBI BlastP on this gene
ETR14_01475
ETR14_01480
aminoglycoside 3-N-acetyltransferase
Accession: QAY75347
Location: 346085-346933
NCBI BlastP on this gene
Accession: QAY75347
Location: 346085-346933
NCBI BlastP on this gene
aac(3)
FadR family transcriptional regulator
Accession: QAY75348
Location: 346950-347660
NCBI BlastP on this gene
Accession: QAY75348
Location: 346950-347660
NCBI BlastP on this gene
ETR14_01490
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP026100 : Caulobacter flavus strain RHGG3 chromosome Total score: 1.0 Cumulative Blast bit score: 406
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
bifunctional proline dehydrogenase/L-glutamate gamma-semialdehyde dehydrogenase
Accession: AYV46730
Location: 2336845-2339934
NCBI BlastP on this gene
Accession: AYV46730
Location: 2336845-2339934
NCBI BlastP on this gene
C1707_10880
C1707_10875
type II toxin-antitoxin system RelE/ParE family toxin
Accession: AYV46728
Location: 2336263-2336544
NCBI BlastP on this gene
Accession: AYV46728
Location: 2336263-2336544
NCBI BlastP on this gene
C1707_10870
C1707_10865
mannanase
Accession: AYV46726
Location: 2333399-2334730
BlastP hit with man5A
Percentage identity: 47 %
BlastP bit score: 406
Sequence coverage: 89 %
E-value: 2e-134
NCBI BlastP on this gene
Accession: AYV46726
Location: 2333399-2334730
BlastP hit with man5A
Percentage identity: 47 %
BlastP bit score: 406
Sequence coverage: 89 %
E-value: 2e-134
NCBI BlastP on this gene
C1707_10860
C1707_10855
cytochrome bd-I oxidase subunit CydX
Accession: AYV46724
Location: 2332787-2332906
NCBI BlastP on this gene
Accession: AYV46724
Location: 2332787-2332906
NCBI BlastP on this gene
C1707_10850
cytochrome d ubiquinol oxidase subunit II
Accession: AYV46723
Location: 2331624-2332766
NCBI BlastP on this gene
Accession: AYV46723
Location: 2331624-2332766
NCBI BlastP on this gene
cydB
cytochrome d terminal oxidase subunit 1
Accession: AYV46722
Location: 2330030-2331610
NCBI BlastP on this gene
Accession: AYV46722
Location: 2330030-2331610
NCBI BlastP on this gene
C1707_10840
thiol reductant ABC exporter subunit CydD
Accession: AYV46721
Location: 2328262-2329905
NCBI BlastP on this gene
Accession: AYV46721
Location: 2328262-2329905
NCBI BlastP on this gene
cydD
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
AP018694 : Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 406
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
AQPE_4505
AQPE_4506
ferredoxin--NADP(+) reductase
Accession: BBE20316
Location: 5028599-5029249
NCBI BlastP on this gene
Accession: BBE20316
Location: 5028599-5029249
NCBI BlastP on this gene
AQPE_4507
AQPE_4508
acyltransferase family protein
Accession: BBE20318
Location: 5030103-5030594
NCBI BlastP on this gene
Accession: BBE20318
Location: 5030103-5030594
NCBI BlastP on this gene
AQPE_4509
AQPE_4510
alpha-galactosidase precursor
Accession: BBE20320
Location: 5031607-5032782
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 406
Sequence coverage: 95 %
E-value: 8e-136
NCBI BlastP on this gene
Accession: BBE20320
Location: 5031607-5032782
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 406
Sequence coverage: 95 %
E-value: 8e-136
NCBI BlastP on this gene
AQPE_4511
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP016094 : Lacunisphaera limnophila strain IG16b chromosome Total score: 1.0 Cumulative Blast bit score: 391
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
N-carbamoyl-D-amino acid hydrolase
Accession: AOS45569
Location: 3121659-3122534
NCBI BlastP on this gene
Accession: AOS45569
Location: 3121659-3122534
NCBI BlastP on this gene
Verru16b_02652
aguA
Verru16b_02654
rpsA_2
Alpha-galactosidase A precursor
Accession: AOS45573
Location: 3126130-3127332
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 391
Sequence coverage: 100 %
E-value: 8e-130
NCBI BlastP on this gene
Accession: AOS45573
Location: 3126130-3127332
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 391
Sequence coverage: 100 %
E-value: 8e-130
NCBI BlastP on this gene
agaA
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP001614 : Teredinibacter turnerae T7901 Total score: 1.0 Cumulative Blast bit score: 389
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
glycoside hydrolase family 5 domain protein
Accession: ACR14510
Location: 4889231-4890664
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 389
Sequence coverage: 98 %
E-value: 4e-127
NCBI BlastP on this gene
Accession: ACR14510
Location: 4889231-4890664
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 389
Sequence coverage: 98 %
E-value: 4e-127
NCBI BlastP on this gene
TERTU_4415
efflux transporter, RND family, MFP subunit
Accession: ACR13330
Location: 4887601-4888842
NCBI BlastP on this gene
Accession: ACR13330
Location: 4887601-4888842
NCBI BlastP on this gene
TERTU_4413
RND transporter, HAE1/HME family, permease protein
Accession: ACR12116
Location: 4884463-4887582
NCBI BlastP on this gene
Accession: ACR12116
Location: 4884463-4887582
NCBI BlastP on this gene
TERTU_4412
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP000927 : Caulobacter sp. K31 chromosome Total score: 1.0 Cumulative Blast bit score: 385
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
delta-1-pyrroline-5-carboxylate dehydrogenase
Accession: ABZ70174
Location: 1103095-1106187
NCBI BlastP on this gene
Accession: ABZ70174
Location: 1103095-1106187
NCBI BlastP on this gene
Caul_1044
Glutathione S-transferase domain
Accession: ABZ70173
Location: 1102206-1102844
NCBI BlastP on this gene
Accession: ABZ70173
Location: 1102206-1102844
NCBI BlastP on this gene
Caul_1043
Activator of Hsp90 ATPase 1 family protein
Accession: ABZ70172
Location: 1101772-1102209
NCBI BlastP on this gene
Accession: ABZ70172
Location: 1101772-1102209
NCBI BlastP on this gene
Caul_1042
transcriptional regulator, MarR family
Accession: ABZ70171
Location: 1101437-1101775
NCBI BlastP on this gene
Accession: ABZ70171
Location: 1101437-1101775
NCBI BlastP on this gene
Caul_1041
mannanase, putative
Accession: ABZ70170
Location: 1099968-1101290
BlastP hit with man5A
Percentage identity: 45 %
BlastP bit score: 385
Sequence coverage: 89 %
E-value: 5e-126
NCBI BlastP on this gene
Accession: ABZ70170
Location: 1099968-1101290
BlastP hit with man5A
Percentage identity: 45 %
BlastP bit score: 385
Sequence coverage: 89 %
E-value: 5e-126
NCBI BlastP on this gene
Caul_1040
Caul_1039
Caul_1038
carbamoyl-phosphate synthase, small subunit
Accession: ABZ70167
Location: 1094552-1095718
NCBI BlastP on this gene
Accession: ABZ70167
Location: 1094552-1095718
NCBI BlastP on this gene
Caul_1037
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP041242 : Lysobacter alkalisoli strain SJ-36 chromosome Total score: 1.0 Cumulative Blast bit score: 377
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
glycoside hydrolase family 27 protein
Accession: QDH71259
Location: 3391805-3393013
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 377
Sequence coverage: 99 %
E-value: 4e-124
NCBI BlastP on this gene
Accession: QDH71259
Location: 3391805-3393013
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 377
Sequence coverage: 99 %
E-value: 4e-124
NCBI BlastP on this gene
FKV23_15030
FKV23_15025
FKV23_15020
FKV23_15015
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP011131 : Lysobacter gummosus strain 3.2.11 Total score: 1.0 Cumulative Blast bit score: 373
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase A
Accession: ALN91524
Location: 2850960-2852282
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 373
Sequence coverage: 94 %
E-value: 5e-122
NCBI BlastP on this gene
Accession: ALN91524
Location: 2850960-2852282
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 373
Sequence coverage: 94 %
E-value: 5e-122
NCBI BlastP on this gene
agaA
LG3211_2556
type I phosphodiesterase / nucleotide pyrophosphatase family protein
Accession: ALN91522
Location: 2848884-2850563
NCBI BlastP on this gene
Accession: ALN91522
Location: 2848884-2850563
NCBI BlastP on this gene
LG3211_2555
lumazine-binding family protein
Accession: ALN91521
Location: 2848274-2848720
NCBI BlastP on this gene
Accession: ALN91521
Location: 2848274-2848720
NCBI BlastP on this gene
LG3211_2554
bacterial regulatory helix-turn-helix, lysR family protein
Accession: ALN91520
Location: 2847296-2848177
NCBI BlastP on this gene
Accession: ALN91520
Location: 2847296-2848177
NCBI BlastP on this gene
LG3211_2553
glycosyl hydrolase 9 family protein
Accession: ALN91519
Location: 2845411-2847282
NCBI BlastP on this gene
Accession: ALN91519
Location: 2845411-2847282
NCBI BlastP on this gene
LG3211_2552
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP034550 : Saccharothrix syringae strain NRRL B-16468 chromosome Total score: 1.0 Cumulative Blast bit score: 366
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-L-arabinofuranosidase
Accession: EKG83_29055
Location: 6846426-6847013
NCBI BlastP on this gene
Accession: EKG83_29055
Location: 6846426-6847013
NCBI BlastP on this gene
EKG83_29055
EKG83_29060
alpha-galactosidase
Accession: QFZ20898
Location: 6849888-6851486
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 366
Sequence coverage: 92 %
E-value: 3e-118
NCBI BlastP on this gene
Accession: QFZ20898
Location: 6849888-6851486
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 366
Sequence coverage: 92 %
E-value: 3e-118
NCBI BlastP on this gene
EKG83_29065
EKG83_29070
MarR family transcriptional regulator
Accession: QFZ24575
Location: 6853332-6853697
NCBI BlastP on this gene
Accession: QFZ24575
Location: 6853332-6853697
NCBI BlastP on this gene
EKG83_29075
DHA2 family efflux MFS transporter permease subunit
Accession: QFZ24576
Location: 6853764-6855347
NCBI BlastP on this gene
Accession: QFZ24576
Location: 6853764-6855347
NCBI BlastP on this gene
EKG83_29080
LacI family transcriptional regulator
Accession: QFZ20900
Location: 6855453-6856469
NCBI BlastP on this gene
Accession: QFZ20900
Location: 6855453-6856469
NCBI BlastP on this gene
EKG83_29085
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP042807 : Rhodanobacter glycinis strain T01E-68 chromosome Total score: 1.0 Cumulative Blast bit score: 365
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
CS053_15395
SGNH/GDSL hydrolase family protein
Accession: QEE25735
Location: 3460052-3460909
NCBI BlastP on this gene
Accession: QEE25735
Location: 3460052-3460909
NCBI BlastP on this gene
CS053_15400
CS053_15405
glycoside hydrolase family 27 protein
Accession: QEE25737
Location: 3463075-3464301
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 365
Sequence coverage: 98 %
E-value: 2e-119
NCBI BlastP on this gene
Accession: QEE25737
Location: 3463075-3464301
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 365
Sequence coverage: 98 %
E-value: 2e-119
NCBI BlastP on this gene
CS053_15410
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP046397 : Bacteroides ovatus strain FDAARGOS_733 chromosome Total score: 1.0 Cumulative Blast bit score: 363
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
FOC41_01695
serine/threonine protein phosphatase
Accession: QGT69760
Location: 450147-451598
NCBI BlastP on this gene
Accession: QGT69760
Location: 450147-451598
NCBI BlastP on this gene
FOC41_01700
DUF5009 domain-containing protein
Accession: QGT69761
Location: 451604-453004
NCBI BlastP on this gene
Accession: QGT69761
Location: 451604-453004
NCBI BlastP on this gene
FOC41_01705
FOC41_01710
glycoside hydrolase family 27 protein
Accession: QGT69763
Location: 454200-455384
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 363
Sequence coverage: 100 %
E-value: 1e-118
NCBI BlastP on this gene
Accession: QGT69763
Location: 454200-455384
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 363
Sequence coverage: 100 %
E-value: 1e-118
NCBI BlastP on this gene
FOC41_01715
DUF5009 domain-containing protein
Accession: QGT69764
Location: 456054-457466
NCBI BlastP on this gene
Accession: QGT69764
Location: 456054-457466
NCBI BlastP on this gene
FOC41_01720
FOC41_01725
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
LT607752 : Micromonospora rifamycinica strain DSM 44983 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 361
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
GA0070623_1909
Uncharacterized conserved protein YdhG,
Accession: SCG51287
Location: 2359995-2360477
NCBI BlastP on this gene
Accession: SCG51287
Location: 2359995-2360477
NCBI BlastP on this gene
GA0070623_1910
GA0070623_1911
GA0070623_1912
GA0070623_1913
alpha-galactosidase
Accession: SCG51318
Location: 2364118-2365776
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 361
Sequence coverage: 100 %
E-value: 6e-116
NCBI BlastP on this gene
Accession: SCG51318
Location: 2364118-2365776
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 361
Sequence coverage: 100 %
E-value: 6e-116
NCBI BlastP on this gene
GA0070623_1914
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003777 : Amycolatopsis mediterranei RB Total score: 1.0 Cumulative Blast bit score: 361
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: AGT88368
Location: 8615482-8617062
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: AGT88368
Location: 8615482-8617062
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
galA
glucose/sorbosone dehydrogenase
Accession: AGT88367
Location: 8613194-8615278
NCBI BlastP on this gene
Accession: AGT88367
Location: 8613194-8615278
NCBI BlastP on this gene
B737_7706
B737_7705
B737_7704
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003729 : Amycolatopsis mediterranei S699 Total score: 1.0 Cumulative Blast bit score: 361
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: AFO81240
Location: 8615538-8617118
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: AFO81240
Location: 8615538-8617118
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
galA
glucose/sorbosone dehydrogenase
Accession: AFO81239
Location: 8613250-8615334
NCBI BlastP on this gene
Accession: AFO81239
Location: 8613250-8615334
NCBI BlastP on this gene
AMES_7706
AMES_7705
AMES_7704
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP002896 : Amycolatopsis mediterranei S699 Total score: 1.0 Cumulative Blast bit score: 361
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: AEK46509
Location: 8605621-8607201
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: AEK46509
Location: 8605621-8607201
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
RAM_40210
glucose/sorbosone dehydrogenase
Accession: AEK46508
Location: 8603333-8605417
NCBI BlastP on this gene
Accession: AEK46508
Location: 8603333-8605417
NCBI BlastP on this gene
RAM_40205
RAM_40200
RAM_40195
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP002000 : Amycolatopsis mediterranei U32 Total score: 1.0 Cumulative Blast bit score: 361
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: ADJ49531
Location: 8605565-8607145
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: ADJ49531
Location: 8605565-8607145
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
galA
glucose/sorbosone dehydrogenase
Accession: ADJ49530
Location: 8603277-8605361
NCBI BlastP on this gene
Accession: ADJ49530
Location: 8603277-8605361
NCBI BlastP on this gene
AMED_7822
glycoside hydrolase family protein
Accession: ADJ49529
Location: 8601612-8603162
NCBI BlastP on this gene
Accession: ADJ49529
Location: 8601612-8603162
NCBI BlastP on this gene
AMED_7821
conserved hypothetical protein
Accession: ADJ49528
Location: 8600327-8601484
NCBI BlastP on this gene
Accession: ADJ49528
Location: 8600327-8601484
NCBI BlastP on this gene
AMED_7820
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP002396 : Asticcacaulis excentricus CB 48 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 360
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
Astex_2874
Astex_2875
DNA polymerase III, delta prime subunit
Accession: ADU14513
Location: 559550-560560
NCBI BlastP on this gene
Accession: ADU14513
Location: 559550-560560
NCBI BlastP on this gene
Astex_2876
Astex_2877
Astex_2878
protein of unknown function DUF1304
Accession: ADU14516
Location: 562102-562455
NCBI BlastP on this gene
Accession: ADU14516
Location: 562102-562455
NCBI BlastP on this gene
Astex_2879
Mannan endo-1,4-beta-mannosidase
Accession: ADU14517
Location: 562934-564277
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 360
Sequence coverage: 89 %
E-value: 4e-116
NCBI BlastP on this gene
Accession: ADU14517
Location: 562934-564277
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 360
Sequence coverage: 89 %
E-value: 4e-116
NCBI BlastP on this gene
Astex_2880
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP016174 : Amycolatopsis orientalis strain B-37 Total score: 1.0 Cumulative Blast bit score: 357
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
IclR family transcriptional regulator
Accession: ANN15470
Location: 1500676-1501455
NCBI BlastP on this gene
Accession: ANN15470
Location: 1500676-1501455
NCBI BlastP on this gene
SD37_07225
SD37_07220
AraC family transcriptional regulator
Accession: ANN15468
Location: 1499265-1500107
NCBI BlastP on this gene
Accession: ANN15468
Location: 1499265-1500107
NCBI BlastP on this gene
SD37_07215
SD37_07210
SD37_07205
adenosylmethionine-8-amino-7-oxononanoate aminotransferase
Accession: ANN15465
Location: 1496846-1498117
NCBI BlastP on this gene
Accession: ANN15465
Location: 1496846-1498117
NCBI BlastP on this gene
SD37_07200
alpha-galactosidase
Accession: ANN15464
Location: 1494675-1496627
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 3e-113
NCBI BlastP on this gene
Accession: ANN15464
Location: 1494675-1496627
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 3e-113
NCBI BlastP on this gene
SD37_07195
GntR family transcriptional regulator
Accession: ANN15463
Location: 1493938-1494633
NCBI BlastP on this gene
Accession: ANN15463
Location: 1493938-1494633
NCBI BlastP on this gene
SD37_07190
SD37_07185
SD37_07180
SD37_07175
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP010836 : Sphingomonas hengshuiensis strain WHSC-8 Total score: 1.0 Cumulative Blast bit score: 357
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
TS85_09205
succinylglutamate-semialdehyde dehydrogenase
Accession: AJP71926
Location: 2100262-2101677
NCBI BlastP on this gene
Accession: AJP71926
Location: 2100262-2101677
NCBI BlastP on this gene
astD
TS85_09215
TS85_09220
TS85_09225
mannanase
Accession: AJP74397
Location: 2103504-2104844
BlastP hit with man5A
Percentage identity: 43 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 5e-115
NCBI BlastP on this gene
Accession: AJP74397
Location: 2103504-2104844
BlastP hit with man5A
Percentage identity: 43 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 5e-115
NCBI BlastP on this gene
TS85_09230
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP017717 : Nonomuraea sp. ATCC 55076 chromosome Total score: 1.0 Cumulative Blast bit score: 356
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
BKM31_10535
BKM31_10540
BKM31_10545
alpha-galactosidase
Accession: AQZ61851
Location: 2358336-2359913
BlastP hit with aga27A
Percentage identity: 55 %
BlastP bit score: 356
Sequence coverage: 77 %
E-value: 2e-114
NCBI BlastP on this gene
Accession: AQZ61851
Location: 2358336-2359913
BlastP hit with aga27A
Percentage identity: 55 %
BlastP bit score: 356
Sequence coverage: 77 %
E-value: 2e-114
NCBI BlastP on this gene
BKM31_10550
BKM31_10555
BKM31_10560
sugar ABC transporter permease
Accession: AQZ61854
Location: 2362469-2363389
NCBI BlastP on this gene
Accession: AQZ61854
Location: 2362469-2363389
NCBI BlastP on this gene
BKM31_10565
sugar ABC transporter substrate-binding protein
Accession: AQZ61855
Location: 2363391-2364704
NCBI BlastP on this gene
Accession: AQZ61855
Location: 2363391-2364704
NCBI BlastP on this gene
BKM31_10570
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP041061 : Micromonospora sp. HM134 chromosome Total score: 1.0 Cumulative Blast bit score: 353
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
FJK98_09230
FJK98_09225
FJK98_09220
FJK98_09215
alpha-galactosidase
Accession: QDY07330
Location: 2007685-2009343
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 353
Sequence coverage: 85 %
E-value: 7e-113
NCBI BlastP on this gene
Accession: QDY07330
Location: 2007685-2009343
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 353
Sequence coverage: 85 %
E-value: 7e-113
NCBI BlastP on this gene
FJK98_09210
NAD-dependent epimerase/dehydratase family protein
Accession: QDY07329
Location: 2006578-2007492
NCBI BlastP on this gene
Accession: QDY07329
Location: 2006578-2007492
NCBI BlastP on this gene
FJK98_09205
FJK98_09200
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP035501 : Sphingosinicella sp. BN140058 chromosome Total score: 1.0 Cumulative Blast bit score: 353
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
mannanase
Accession: QAY77330
Location: 2871608-2872951
BlastP hit with man5A
Percentage identity: 42 %
BlastP bit score: 353
Sequence coverage: 89 %
E-value: 2e-113
NCBI BlastP on this gene
Accession: QAY77330
Location: 2871608-2872951
BlastP hit with man5A
Percentage identity: 42 %
BlastP bit score: 353
Sequence coverage: 89 %
E-value: 2e-113
NCBI BlastP on this gene
ETR14_13065
ETR14_13060
GNAT family N-acetyltransferase
Accession: QAY77328
Location: 2869194-2869652
NCBI BlastP on this gene
Accession: QAY77328
Location: 2869194-2869652
NCBI BlastP on this gene
ETR14_13055
fumC
ETR14_13045
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP032052 : Streptomyces clavuligerus strain F1D-5 chromosome Total score: 1.0 Cumulative Blast bit score: 353
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
DUF3472 domain-containing protein
Accession: AXU11838
Location: 822688-823896
NCBI BlastP on this gene
Accession: AXU11838
Location: 822688-823896
NCBI BlastP on this gene
D1794_03325
D1794_03320
D1794_03315
D1794_03310
alpha-galactosidase
Accession: AXU11835
Location: 817449-819599
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
Accession: AXU11835
Location: 817449-819599
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
D1794_03305
LacI family transcriptional regulator
Accession: AXU11834
Location: 816173-817315
NCBI BlastP on this gene
Accession: AXU11834
Location: 816173-817315
NCBI BlastP on this gene
D1794_03300
D1794_03295
glycoside hydrolase family 28 protein
Accession: AXU11832
Location: 814203-815642
NCBI BlastP on this gene
Accession: AXU11832
Location: 814203-815642
NCBI BlastP on this gene
D1794_03290
D1794_03285
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP027858 : Streptomyces clavuligerus strain ATCC 27064 chromosome Total score: 1.0 Cumulative Blast bit score: 353
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
DUF3472 domain-containing protein
Accession: QCS04616
Location: 684052-685260
NCBI BlastP on this gene
Accession: QCS04616
Location: 684052-685260
NCBI BlastP on this gene
CRV15_02755
CRV15_02750
CRV15_02745
CRV15_02740
alpha-galactosidase
Accession: QCS04613
Location: 678813-680963
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
Accession: QCS04613
Location: 678813-680963
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
CRV15_02735
LacI family transcriptional regulator
Accession: QCS04612
Location: 677537-678538
NCBI BlastP on this gene
Accession: QCS04612
Location: 677537-678538
NCBI BlastP on this gene
CRV15_02730
CRV15_02725
CRV15_02720
CRV15_02715
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP016559 : Streptomyces clavuligerus strain F613-1 chromosome Total score: 1.0 Cumulative Blast bit score: 353
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
BB341_03170
BB341_03165
BB341_03160
alpha-galactosidase
Accession: ANW17289
Location: 815740-817890
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
Accession: ANW17289
Location: 815740-817890
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
BB341_03155
BB341_03150
BB341_03145
BB341_03140
BB341_03135
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
LT827010 : Actinoplanes sp. SE50/110 isolate ACP50 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 352
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: SLM02731
Location: 6556199-6557776
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
Accession: SLM02731
Location: 6556199-6557776
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
ACSP50_6016
ACSP50_6015
ACSP50_6014
aminoglycoside 2'-N-acetyltransferase
Accession: SLM02728
Location: 6552253-6552792
NCBI BlastP on this gene
Accession: SLM02728
Location: 6552253-6552792
NCBI BlastP on this gene
ACSP50_6013
ACSP50_6012
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP023298 : Actinoplanes sp. SE50 chromosome Total score: 1.0 Cumulative Blast bit score: 352
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
alpha-galactosidase
Accession: ATO85320
Location: 6555329-6556906
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
Accession: ATO85320
Location: 6555329-6556906
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
ACWT_5905
ACWT_5904
Polycystic kidney disease protein 1-like 3
Accession: ATO85318
Location: 6551948-6552502
NCBI BlastP on this gene
Accession: ATO85318
Location: 6551948-6552502
NCBI BlastP on this gene
ACWT_5903
aminoglycoside 2'-N-acetyltransferase
Accession: ATO85317
Location: 6551383-6551922
NCBI BlastP on this gene
Accession: ATO85317
Location: 6551383-6551922
NCBI BlastP on this gene
ACWT_5902
ACWT_5901
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP010836 : Sphingomonas hengshuiensis strain WHSC-8 Total score: 1.0 Cumulative Blast bit score: 352
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
TS85_01410
TS85_01400
mannanase
Accession: AJP70767
Location: 300884-302230
BlastP hit with man5A
Percentage identity: 41 %
BlastP bit score: 352
Sequence coverage: 94 %
E-value: 4e-113
NCBI BlastP on this gene
Accession: AJP70767
Location: 300884-302230
BlastP hit with man5A
Percentage identity: 41 %
BlastP bit score: 352
Sequence coverage: 94 %
E-value: 4e-113
NCBI BlastP on this gene
TS85_01390
TS85_01380
TS85_01375
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003170 : Actinoplanes sp. SE50/110 Total score: 1.0 Cumulative Blast bit score: 352
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
ricin B lectin
Accession: AEV86924
Location: 6556178-6557755
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
Accession: AEV86924
Location: 6556178-6557755
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
ACPL_6037
zinc metalloprotease (elastase)-like protein
Accession: AEV86923
Location: 6553709-6555703
NCBI BlastP on this gene
Accession: AEV86923
Location: 6553709-6555703
NCBI BlastP on this gene
ACPL_6036
Polycystic kidney disease protein 1-like 3
Accession: AEV86922
Location: 6552797-6553351
NCBI BlastP on this gene
Accession: AEV86922
Location: 6552797-6553351
NCBI BlastP on this gene
ACPL_6035
Aminoglycoside 2'-N-acetyltransferase
Accession: AEV86921
Location: 6552232-6552771
NCBI BlastP on this gene
Accession: AEV86921
Location: 6552232-6552771
NCBI BlastP on this gene
aac
putative amino-acid metabolite efflux pump
Accession: AEV86920
Location: 6551223-6552113
NCBI BlastP on this gene
Accession: AEV86920
Location: 6551223-6552113
NCBI BlastP on this gene
ACPL_6033
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
AM420293 : Saccharopolyspora erythraea NRRL2338 complete genome. Total score: 1.0 Cumulative Blast bit score: 350
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
SACE_7060
SACE_7061
two component system sensor kinase
Accession: CAM06223
Location: 7869967-7871508
NCBI BlastP on this gene
Accession: CAM06223
Location: 7869967-7871508
NCBI BlastP on this gene
SACE_7062
two component system response regulator
Accession: CAM06224
Location: 7871517-7872170
NCBI BlastP on this gene
Accession: CAM06224
Location: 7871517-7872170
NCBI BlastP on this gene
SACE_7063
SACE_7064
putative alpha-galactosidase
Accession: CAM06226
Location: 7872998-7874176
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 350
Sequence coverage: 93 %
E-value: 7e-114
NCBI BlastP on this gene
Accession: CAM06226
Location: 7872998-7874176
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 350
Sequence coverage: 93 %
E-value: 7e-114
NCBI BlastP on this gene
agaB2
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003777 : Amycolatopsis mediterranei RB Total score: 1.0 Cumulative Blast bit score: 347
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
B737_3607
B737_3606
B737_3605
alpha-galactosidase
Accession: AGT84268
Location: 3962936-3964891
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
Accession: AGT84268
Location: 3962936-3964891
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
galA
TetR family transcriptional regulator
Accession: AGT84267
Location: 3961989-3962633
NCBI BlastP on this gene
Accession: AGT84267
Location: 3961989-3962633
NCBI BlastP on this gene
B737_3603
carbon-monoxide dehydrogenase small subunit
Accession: AGT84266
Location: 3961397-3961885
NCBI BlastP on this gene
Accession: AGT84266
Location: 3961397-3961885
NCBI BlastP on this gene
B737_3602
carbon-monoxide dehydrogenase medium subunit
Accession: AGT84265
Location: 3960405-3961400
NCBI BlastP on this gene
Accession: AGT84265
Location: 3960405-3961400
NCBI BlastP on this gene
B737_3601
carbon-monoxide dehydrogenase large subunit
Accession: AGT84264
Location: 3958192-3960408
NCBI BlastP on this gene
Accession: AGT84264
Location: 3958192-3960408
NCBI BlastP on this gene
B737_3600
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
CP003729 : Amycolatopsis mediterranei S699 Total score: 1.0 Cumulative Blast bit score: 347
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
AMES_3607
AMES_3606
AMES_3605
alpha-galactosidase
Accession: AFO77140
Location: 3962999-3964954
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
Accession: AFO77140
Location: 3962999-3964954
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
galA
TetR family transcriptional regulator
Accession: AFO77139
Location: 3962052-3962696
NCBI BlastP on this gene
Accession: AFO77139
Location: 3962052-3962696
NCBI BlastP on this gene
AMES_3603
carbon-monoxide dehydrogenase small subunit
Accession: AFO77138
Location: 3961460-3961948
NCBI BlastP on this gene
Accession: AFO77138
Location: 3961460-3961948
NCBI BlastP on this gene
AMES_3602
carbon-monoxide dehydrogenase medium subunit
Accession: AFO77137
Location: 3960468-3961463
NCBI BlastP on this gene
Accession: AFO77137
Location: 3960468-3961463
NCBI BlastP on this gene
AMES_3601
carbon-monoxide dehydrogenase large subunit
Accession: AFO77136
Location: 3958255-3960471
NCBI BlastP on this gene
Accession: AFO77136
Location: 3958255-3960471
NCBI BlastP on this gene
AMES_3600
Query: Cellvibrio mixtus Unk1 (unk1), Unk2 (unk2), Man5A (man5A), and
301. : CP040017 Massilia umbonata strain DSMZ 26121 chromosome Total score: 1.0 Cumulative Blast bit score: 496
GH130
Location: 112-1293
Location: 112-1293
unk1
Unk2
Location: 1297-2529
Location: 1297-2529
unk2
GH5 7|GH5
Location: 2542-3912
Location: 2542-3912
man5A
GH27
Location: 4040-5257
Location: 4040-5257
aga27A
mannanase
Accession: QCP11084
Location: 2987151-2988455
BlastP hit with man5A
Percentage identity: 55 %
BlastP bit score: 497
Sequence coverage: 89 %
E-value: 6e-170
NCBI BlastP on this gene
Accession: QCP11084
Location: 2987151-2988455
BlastP hit with man5A
Percentage identity: 55 %
BlastP bit score: 497
Sequence coverage: 89 %
E-value: 6e-170
NCBI BlastP on this gene
FCL38_12185
FCL38_12180
FCL38_12175
FCL38_12170
FCL38_12165
302. : FP929059 Eubacterium siraeum V10Sc8a draft genome. Total score: 1.0 Cumulative Blast bit score: 496
ES1_21250
ES1_21260
ES1_21270
large conductance mechanosensitive channel protein
Accession: CBL34988
Location: 2293372-2293935
NCBI BlastP on this gene
Accession: CBL34988
Location: 2293372-2293935
NCBI BlastP on this gene
ES1_21300
Predicted glycosylase
Accession: CBL34989
Location: 2294698-2295873
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 496
Sequence coverage: 99 %
E-value: 2e-171
NCBI BlastP on this gene
Accession: CBL34989
Location: 2294698-2295873
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 496
Sequence coverage: 99 %
E-value: 2e-171
NCBI BlastP on this gene
ES1_21320
303. : CP003557 Melioribacter roseus P3M Total score: 1.0 Cumulative Blast bit score: 495
alpha-galactosidase
Accession: AFN75841
Location: 3036326-3037546
BlastP hit with aga27A
Percentage identity: 57 %
BlastP bit score: 495
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
Accession: AFN75841
Location: 3036326-3037546
BlastP hit with aga27A
Percentage identity: 57 %
BlastP bit score: 495
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
MROS_2611
MROS_2610
MROS_2609
304. : LT906662 Thermoanaerobacterium sp. RBIITD genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 494
4-O-beta-D-mannosyl-D-glucose phosphorylase
Accession: SNX52632
Location: 39235-40416
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 494
Sequence coverage: 98 %
E-value: 2e-170
NCBI BlastP on this gene
Accession: SNX52632
Location: 39235-40416
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 494
Sequence coverage: 98 %
E-value: 2e-170
NCBI BlastP on this gene
SAMN05660242_0044
Aryl-phospho-beta-D-glucosidase BglC, GH1 family
Accession: SNX52631
Location: 37475-38467
NCBI BlastP on this gene
Accession: SNX52631
Location: 37475-38467
NCBI BlastP on this gene
SAMN05660242_0042
SAMN05660242_0040
SAMN05660242_0039
305. : FP929044 Eubacterium siraeum 70/3 draft genome. Total score: 1.0 Cumulative Blast bit score: 493
EUS_08970
large conductance mechanosensitive channel protein
Accession: CBK96124
Location: 898651-899214
NCBI BlastP on this gene
Accession: CBK96124
Location: 898651-899214
NCBI BlastP on this gene
EUS_08940
Predicted glycosylase
Accession: CBK96123
Location: 896713-897888
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 493
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
Accession: CBK96123
Location: 896713-897888
BlastP hit with unk1
Percentage identity: 60 %
BlastP bit score: 493
Sequence coverage: 99 %
E-value: 3e-170
NCBI BlastP on this gene
EUS_08920
EUS_08910
EUS_08900
transcription antitermination protein nusG
Accession: CBK96120
Location: 894344-894865
NCBI BlastP on this gene
Accession: CBK96120
Location: 894344-894865
NCBI BlastP on this gene
EUS_08890
preprotein translocase, SecE subunit, bacterial
Accession: CBK96119
Location: 894023-894316
NCBI BlastP on this gene
Accession: CBK96119
Location: 894023-894316
NCBI BlastP on this gene
EUS_08880
EUS_08860
methenyltetrahydrofolate cyclohydrolase
Accession: CBK96117
Location: 892619-893458
NCBI BlastP on this gene
Accession: CBK96117
Location: 892619-893458
NCBI BlastP on this gene
EUS_08850
prolipoprotein diacylglyceryl transferase
Accession: CBK96116
Location: 891480-892619
NCBI BlastP on this gene
Accession: CBK96116
Location: 891480-892619
NCBI BlastP on this gene
EUS_08840
306. : CP013020 Bacteroides vulgatus strain mpk genome. Total score: 1.0 Cumulative Blast bit score: 481
BvMPK_0026
RagB/SusD Domain-Containing Protein
Accession: ALK82667
Location: 34112-35887
NCBI BlastP on this gene
Accession: ALK82667
Location: 34112-35887
NCBI BlastP on this gene
BvMPK_0025
Putative arylsulfate sulfotransferase
Accession: ALK82666
Location: 32111-33910
NCBI BlastP on this gene
Accession: ALK82666
Location: 32111-33910
NCBI BlastP on this gene
BvMPK_0024
Glycosidase-like Protein
Accession: ALK82665
Location: 30819-31997
BlastP hit with unk1
Percentage identity: 63 %
BlastP bit score: 481
Sequence coverage: 97 %
E-value: 2e-165
NCBI BlastP on this gene
Accession: ALK82665
Location: 30819-31997
BlastP hit with unk1
Percentage identity: 63 %
BlastP bit score: 481
Sequence coverage: 97 %
E-value: 2e-165
NCBI BlastP on this gene
BvMPK_0023
Alkaline phosphodiesterase I / Nucleotide pyrophosphatase
Accession: ALK82664
Location: 29759-30661
NCBI BlastP on this gene
Accession: ALK82664
Location: 29759-30661
NCBI BlastP on this gene
BvMPK_0022
putative sodium-dependent mannose transporter
Accession: ALK82663
Location: 27858-29741
NCBI BlastP on this gene
Accession: ALK82663
Location: 27858-29741
NCBI BlastP on this gene
BvMPK_0021
BvMPK_0020
307. : CP042431 Pseudobacter ginsenosidimutans strain Gsoil 221 chromosome Total score: 1.0 Cumulative Blast bit score: 464
glycosidase
Accession: QEC44537
Location: 6020039-6021220
BlastP hit with unk1
Percentage identity: 59 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 6e-159
NCBI BlastP on this gene
Accession: QEC44537
Location: 6020039-6021220
BlastP hit with unk1
Percentage identity: 59 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 6e-159
NCBI BlastP on this gene
FSB84_23725
FSB84_23720
FSB84_23715
FSB84_23710
30S ribosomal protein S12 methylthiotransferase RimO
Accession: QEC44533
Location: 6016715-6018025
NCBI BlastP on this gene
Accession: QEC44533
Location: 6016715-6018025
NCBI BlastP on this gene
rimO
aldehyde dehydrogenase (NADP(+))
Accession: QEC44532
Location: 6014900-6016387
NCBI BlastP on this gene
Accession: QEC44532
Location: 6014900-6016387
NCBI BlastP on this gene
FSB84_23700
308. : AP019736 Alistipes dispar 5CPEGH6 DNA Total score: 1.0 Cumulative Blast bit score: 464
hypothetical protein
Accession: BBL06472
Location: 1311959-1313182
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 2e-158
NCBI BlastP on this gene
Accession: BBL06472
Location: 1311959-1313182
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 2e-158
NCBI BlastP on this gene
A5CPEGH6_11100
A5CPEGH6_11090
magnesium transport protein CorA
Accession: BBL06470
Location: 1310153-1311136
NCBI BlastP on this gene
Accession: BBL06470
Location: 1310153-1311136
NCBI BlastP on this gene
corA-2
polysaccharide biosynthesis protein
Accession: BBL06469
Location: 1308334-1309806
NCBI BlastP on this gene
Accession: BBL06469
Location: 1308334-1309806
NCBI BlastP on this gene
A5CPEGH6_11070
DNA polymerase III subunit delta
Accession: BBL06468
Location: 1307168-1308313
NCBI BlastP on this gene
Accession: BBL06468
Location: 1307168-1308313
NCBI BlastP on this gene
A5CPEGH6_11060
309. : CP026604 Catenovulum sp. CCB-QB4 chromosome Total score: 1.0 Cumulative Blast bit score: 449
C2869_05755
dnaJ
C2869_05765
alpha-galactosidase
Accession: AWB65978
Location: 1632344-1633594
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 450
Sequence coverage: 101 %
E-value: 1e-152
NCBI BlastP on this gene
Accession: AWB65978
Location: 1632344-1633594
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 450
Sequence coverage: 101 %
E-value: 1e-152
NCBI BlastP on this gene
C2869_05770
MBL fold metallo-hydrolase
Accession: C2869_05775
Location: 1633700-1633858
NCBI BlastP on this gene
Accession: C2869_05775
Location: 1633700-1633858
NCBI BlastP on this gene
C2869_05775
C2869_05780
C2869_05785
C2869_05790
310. : CP041242 Lysobacter alkalisoli strain SJ-36 chromosome Total score: 1.0 Cumulative Blast bit score: 447
FKV23_01350
FKV23_01345
TRAP transporter substrate-binding protein
Accession: QDH68895
Location: 312857-313888
NCBI BlastP on this gene
Accession: QDH68895
Location: 312857-313888
NCBI BlastP on this gene
FKV23_01340
TRAP transporter small permease
Accession: QDH68894
Location: 312306-312848
NCBI BlastP on this gene
Accession: QDH68894
Location: 312306-312848
NCBI BlastP on this gene
FKV23_01335
TRAP transporter large permease
Accession: QDH68893
Location: 311020-312303
NCBI BlastP on this gene
Accession: QDH68893
Location: 311020-312303
NCBI BlastP on this gene
FKV23_01330
mannanase
Accession: QDH68892
Location: 309522-310880
BlastP hit with man5A
Percentage identity: 51 %
BlastP bit score: 448
Sequence coverage: 91 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: QDH68892
Location: 309522-310880
BlastP hit with man5A
Percentage identity: 51 %
BlastP bit score: 448
Sequence coverage: 91 %
E-value: 2e-150
NCBI BlastP on this gene
FKV23_01325
sugar porter family MFS transporter
Accession: QDH68891
Location: 307717-309159
NCBI BlastP on this gene
Accession: QDH68891
Location: 307717-309159
NCBI BlastP on this gene
FKV23_01320
FKV23_01315
LacI family transcriptional regulator
Accession: QDH68889
Location: 304681-305751
NCBI BlastP on this gene
Accession: QDH68889
Location: 304681-305751
NCBI BlastP on this gene
FKV23_01310
311. : AP018694 Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 446
AQPE_1317
AQPE_1316
AQPE_1315
alpha-galactosidase precursor
Accession: BBE17165
Location: 1475360-1476559
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 446
Sequence coverage: 98 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: BBE17165
Location: 1475360-1476559
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 446
Sequence coverage: 98 %
E-value: 2e-151
NCBI BlastP on this gene
AQPE_1314
AQPE_1313
dipeptidyl peptidase IV in 4-hydroxyproline catabolic gene cluster
Accession: BBE17163
Location: 1469815-1472067
NCBI BlastP on this gene
Accession: BBE17163
Location: 1469815-1472067
NCBI BlastP on this gene
AQPE_1312
312. : CP002403 Ruminococcus albus 7 Total score: 1.0 Cumulative Blast bit score: 440
glycosidase related protein
Accession: ADU21379
Location: 950842-952002
BlastP hit with unk1
Percentage identity: 57 %
BlastP bit score: 440
Sequence coverage: 97 %
E-value: 2e-149
NCBI BlastP on this gene
Accession: ADU21379
Location: 950842-952002
BlastP hit with unk1
Percentage identity: 57 %
BlastP bit score: 440
Sequence coverage: 97 %
E-value: 2e-149
NCBI BlastP on this gene
Rumal_0852
Rumal_0851
Rumal_0850
Rumal_0849
Rumal_0848
313. : CP013020 Bacteroides vulgatus strain mpk genome. Total score: 1.0 Cumulative Blast bit score: 432
TonB family protein / TonB-dependent receptor
Accession: ALK83489
Location: 1053455-1056250
NCBI BlastP on this gene
Accession: ALK83489
Location: 1053455-1056250
NCBI BlastP on this gene
BvMPK_0871
putative outer membrane protein
Accession: ALK83490
Location: 1056272-1057951
NCBI BlastP on this gene
Accession: ALK83490
Location: 1056272-1057951
NCBI BlastP on this gene
BvMPK_0872
Alpha-galactosidase
Accession: ALK83491
Location: 1058161-1059411
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 433
Sequence coverage: 95 %
E-value: 6e-146
NCBI BlastP on this gene
Accession: ALK83491
Location: 1058161-1059411
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 433
Sequence coverage: 95 %
E-value: 6e-146
NCBI BlastP on this gene
BvMPK_0873
BvMPK_0874
314. : CP026604 Catenovulum sp. CCB-QB4 chromosome Total score: 1.0 Cumulative Blast bit score: 429
C2869_03430
C2869_03425
C2869_03420
alpha-galactosidase
Accession: AWB65540
Location: 921837-923051
BlastP hit with aga27A
Percentage identity: 53 %
BlastP bit score: 429
Sequence coverage: 98 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: AWB65540
Location: 921837-923051
BlastP hit with aga27A
Percentage identity: 53 %
BlastP bit score: 429
Sequence coverage: 98 %
E-value: 2e-144
NCBI BlastP on this gene
C2869_03415
C2869_03410
C2869_03405
GNAT family N-acetyltransferase
Accession: AWB65537
Location: 918802-919299
NCBI BlastP on this gene
Accession: AWB65537
Location: 918802-919299
NCBI BlastP on this gene
C2869_03400
C2869_03395
C2869_03390
315. : CP003349 Solitalea canadensis DSM 3403 Total score: 1.0 Cumulative Blast bit score: 428
TonB-linked outer membrane protein, SusC/RagA family
Accession: AFD06586
Location: 1819121-1822195
NCBI BlastP on this gene
Accession: AFD06586
Location: 1819121-1822195
NCBI BlastP on this gene
Solca_1511
Solca_1512
Solca_1513
alpha-galactosidase
Accession: AFD06589
Location: 1825111-1826301
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 428
Sequence coverage: 98 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AFD06589
Location: 1825111-1826301
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 428
Sequence coverage: 98 %
E-value: 3e-144
NCBI BlastP on this gene
Solca_1514
Solca_1515
HD superfamily phosphohydrolase
Accession: AFD06591
Location: 1828500-1829381
NCBI BlastP on this gene
Accession: AFD06591
Location: 1828500-1829381
NCBI BlastP on this gene
Solca_1516
Solca_1517
Solca_1518
316. : CP036491 Bacteroides sp. A1C1 chromosome Total score: 1.0 Cumulative Blast bit score: 422
EYA81_15140
PHP domain-containing protein
Accession: QBJ19565
Location: 3692076-3693155
NCBI BlastP on this gene
Accession: QBJ19565
Location: 3692076-3693155
NCBI BlastP on this gene
EYA81_15145
EYA81_15150
glycoside hydrolase family 27 protein
Accession: QBJ19567
Location: 3694674-3695882
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 422
Sequence coverage: 98 %
E-value: 1e-141
NCBI BlastP on this gene
Accession: QBJ19567
Location: 3694674-3695882
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 422
Sequence coverage: 98 %
E-value: 1e-141
NCBI BlastP on this gene
EYA81_15155
317. : CP026100 Caulobacter flavus strain RHGG3 chromosome Total score: 1.0 Cumulative Blast bit score: 415
C1707_15780
C1707_15785
alpha-galactosidase
Accession: AYV47601
Location: 3416557-3417747
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 415
Sequence coverage: 94 %
E-value: 6e-139
NCBI BlastP on this gene
Accession: AYV47601
Location: 3416557-3417747
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 415
Sequence coverage: 94 %
E-value: 6e-139
NCBI BlastP on this gene
C1707_15790
318. : CP000927 Caulobacter sp. K31 chromosome Total score: 1.0 Cumulative Blast bit score: 413
Caul_2125
Caul_2126
putative transposase of insertion sequence
Accession: ABZ71255
Location: 2295162-2295509
NCBI BlastP on this gene
Accession: ABZ71255
Location: 2295162-2295509
NCBI BlastP on this gene
Caul_2127
Caul_2128
Alpha-galactosidase
Accession: ABZ71257
Location: 2296299-2297522
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 413
Sequence coverage: 93 %
E-value: 4e-138
NCBI BlastP on this gene
Accession: ABZ71257
Location: 2296299-2297522
BlastP hit with aga27A
Percentage identity: 54 %
BlastP bit score: 413
Sequence coverage: 93 %
E-value: 4e-138
NCBI BlastP on this gene
Caul_2129
319. : CP035501 Sphingosinicella sp. BN140058 chromosome Total score: 1.0 Cumulative Blast bit score: 410
ETR14_01430
ETR14_01435
ETR14_01440
glycoside hydrolase family 27 protein
Accession: QAY75340
Location: 341428-342663
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 410
Sequence coverage: 95 %
E-value: 6e-137
NCBI BlastP on this gene
Accession: QAY75340
Location: 341428-342663
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 410
Sequence coverage: 95 %
E-value: 6e-137
NCBI BlastP on this gene
ETR14_01445
ankyrin repeat domain-containing protein
Accession: QAY75341
Location: 342669-343787
NCBI BlastP on this gene
Accession: QAY75341
Location: 342669-343787
NCBI BlastP on this gene
ETR14_01450
ETR14_01455
DUF1801 domain-containing protein
Accession: QAY75343
Location: 344134-344538
NCBI BlastP on this gene
Accession: QAY75343
Location: 344134-344538
NCBI BlastP on this gene
ETR14_01460
DUF1801 domain-containing protein
Accession: QAY79656
Location: 344557-344988
NCBI BlastP on this gene
Accession: QAY79656
Location: 344557-344988
NCBI BlastP on this gene
ETR14_01465
ETR14_01470
SRPBCC domain-containing protein
Accession: QAY75345
Location: 345307-345717
NCBI BlastP on this gene
Accession: QAY75345
Location: 345307-345717
NCBI BlastP on this gene
ETR14_01475
ETR14_01480
aminoglycoside 3-N-acetyltransferase
Accession: QAY75347
Location: 346085-346933
NCBI BlastP on this gene
Accession: QAY75347
Location: 346085-346933
NCBI BlastP on this gene
aac(3)
FadR family transcriptional regulator
Accession: QAY75348
Location: 346950-347660
NCBI BlastP on this gene
Accession: QAY75348
Location: 346950-347660
NCBI BlastP on this gene
ETR14_01490
320. : CP026100 Caulobacter flavus strain RHGG3 chromosome Total score: 1.0 Cumulative Blast bit score: 406
bifunctional proline dehydrogenase/L-glutamate gamma-semialdehyde dehydrogenase
Accession: AYV46730
Location: 2336845-2339934
NCBI BlastP on this gene
Accession: AYV46730
Location: 2336845-2339934
NCBI BlastP on this gene
C1707_10880
C1707_10875
type II toxin-antitoxin system RelE/ParE family toxin
Accession: AYV46728
Location: 2336263-2336544
NCBI BlastP on this gene
Accession: AYV46728
Location: 2336263-2336544
NCBI BlastP on this gene
C1707_10870
C1707_10865
mannanase
Accession: AYV46726
Location: 2333399-2334730
BlastP hit with man5A
Percentage identity: 47 %
BlastP bit score: 406
Sequence coverage: 89 %
E-value: 2e-134
NCBI BlastP on this gene
Accession: AYV46726
Location: 2333399-2334730
BlastP hit with man5A
Percentage identity: 47 %
BlastP bit score: 406
Sequence coverage: 89 %
E-value: 2e-134
NCBI BlastP on this gene
C1707_10860
C1707_10855
cytochrome bd-I oxidase subunit CydX
Accession: AYV46724
Location: 2332787-2332906
NCBI BlastP on this gene
Accession: AYV46724
Location: 2332787-2332906
NCBI BlastP on this gene
C1707_10850
cytochrome d ubiquinol oxidase subunit II
Accession: AYV46723
Location: 2331624-2332766
NCBI BlastP on this gene
Accession: AYV46723
Location: 2331624-2332766
NCBI BlastP on this gene
cydB
cytochrome d terminal oxidase subunit 1
Accession: AYV46722
Location: 2330030-2331610
NCBI BlastP on this gene
Accession: AYV46722
Location: 2330030-2331610
NCBI BlastP on this gene
C1707_10840
thiol reductant ABC exporter subunit CydD
Accession: AYV46721
Location: 2328262-2329905
NCBI BlastP on this gene
Accession: AYV46721
Location: 2328262-2329905
NCBI BlastP on this gene
cydD
321. : AP018694 Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 406
AQPE_4505
AQPE_4506
ferredoxin--NADP(+) reductase
Accession: BBE20316
Location: 5028599-5029249
NCBI BlastP on this gene
Accession: BBE20316
Location: 5028599-5029249
NCBI BlastP on this gene
AQPE_4507
AQPE_4508
acyltransferase family protein
Accession: BBE20318
Location: 5030103-5030594
NCBI BlastP on this gene
Accession: BBE20318
Location: 5030103-5030594
NCBI BlastP on this gene
AQPE_4509
AQPE_4510
alpha-galactosidase precursor
Accession: BBE20320
Location: 5031607-5032782
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 406
Sequence coverage: 95 %
E-value: 8e-136
NCBI BlastP on this gene
Accession: BBE20320
Location: 5031607-5032782
BlastP hit with aga27A
Percentage identity: 52 %
BlastP bit score: 406
Sequence coverage: 95 %
E-value: 8e-136
NCBI BlastP on this gene
AQPE_4511
322. : CP016094 Lacunisphaera limnophila strain IG16b chromosome Total score: 1.0 Cumulative Blast bit score: 391
N-carbamoyl-D-amino acid hydrolase
Accession: AOS45569
Location: 3121659-3122534
NCBI BlastP on this gene
Accession: AOS45569
Location: 3121659-3122534
NCBI BlastP on this gene
Verru16b_02652
aguA
Verru16b_02654
rpsA_2
Alpha-galactosidase A precursor
Accession: AOS45573
Location: 3126130-3127332
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 391
Sequence coverage: 100 %
E-value: 8e-130
NCBI BlastP on this gene
Accession: AOS45573
Location: 3126130-3127332
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 391
Sequence coverage: 100 %
E-value: 8e-130
NCBI BlastP on this gene
agaA
323. : CP001614 Teredinibacter turnerae T7901 Total score: 1.0 Cumulative Blast bit score: 389
glycoside hydrolase family 5 domain protein
Accession: ACR14510
Location: 4889231-4890664
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 389
Sequence coverage: 98 %
E-value: 4e-127
NCBI BlastP on this gene
Accession: ACR14510
Location: 4889231-4890664
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 389
Sequence coverage: 98 %
E-value: 4e-127
NCBI BlastP on this gene
TERTU_4415
efflux transporter, RND family, MFP subunit
Accession: ACR13330
Location: 4887601-4888842
NCBI BlastP on this gene
Accession: ACR13330
Location: 4887601-4888842
NCBI BlastP on this gene
TERTU_4413
RND transporter, HAE1/HME family, permease protein
Accession: ACR12116
Location: 4884463-4887582
NCBI BlastP on this gene
Accession: ACR12116
Location: 4884463-4887582
NCBI BlastP on this gene
TERTU_4412
324. : CP000927 Caulobacter sp. K31 chromosome Total score: 1.0 Cumulative Blast bit score: 385
delta-1-pyrroline-5-carboxylate dehydrogenase
Accession: ABZ70174
Location: 1103095-1106187
NCBI BlastP on this gene
Accession: ABZ70174
Location: 1103095-1106187
NCBI BlastP on this gene
Caul_1044
Glutathione S-transferase domain
Accession: ABZ70173
Location: 1102206-1102844
NCBI BlastP on this gene
Accession: ABZ70173
Location: 1102206-1102844
NCBI BlastP on this gene
Caul_1043
Activator of Hsp90 ATPase 1 family protein
Accession: ABZ70172
Location: 1101772-1102209
NCBI BlastP on this gene
Accession: ABZ70172
Location: 1101772-1102209
NCBI BlastP on this gene
Caul_1042
transcriptional regulator, MarR family
Accession: ABZ70171
Location: 1101437-1101775
NCBI BlastP on this gene
Accession: ABZ70171
Location: 1101437-1101775
NCBI BlastP on this gene
Caul_1041
mannanase, putative
Accession: ABZ70170
Location: 1099968-1101290
BlastP hit with man5A
Percentage identity: 45 %
BlastP bit score: 385
Sequence coverage: 89 %
E-value: 5e-126
NCBI BlastP on this gene
Accession: ABZ70170
Location: 1099968-1101290
BlastP hit with man5A
Percentage identity: 45 %
BlastP bit score: 385
Sequence coverage: 89 %
E-value: 5e-126
NCBI BlastP on this gene
Caul_1040
Caul_1039
Caul_1038
carbamoyl-phosphate synthase, small subunit
Accession: ABZ70167
Location: 1094552-1095718
NCBI BlastP on this gene
Accession: ABZ70167
Location: 1094552-1095718
NCBI BlastP on this gene
Caul_1037
325. : CP041242 Lysobacter alkalisoli strain SJ-36 chromosome Total score: 1.0 Cumulative Blast bit score: 377
glycoside hydrolase family 27 protein
Accession: QDH71259
Location: 3391805-3393013
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 377
Sequence coverage: 99 %
E-value: 4e-124
NCBI BlastP on this gene
Accession: QDH71259
Location: 3391805-3393013
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 377
Sequence coverage: 99 %
E-value: 4e-124
NCBI BlastP on this gene
FKV23_15030
FKV23_15025
FKV23_15020
FKV23_15015
326. : CP011131 Lysobacter gummosus strain 3.2.11 Total score: 1.0 Cumulative Blast bit score: 373
alpha-galactosidase A
Accession: ALN91524
Location: 2850960-2852282
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 373
Sequence coverage: 94 %
E-value: 5e-122
NCBI BlastP on this gene
Accession: ALN91524
Location: 2850960-2852282
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 373
Sequence coverage: 94 %
E-value: 5e-122
NCBI BlastP on this gene
agaA
LG3211_2556
type I phosphodiesterase / nucleotide pyrophosphatase family protein
Accession: ALN91522
Location: 2848884-2850563
NCBI BlastP on this gene
Accession: ALN91522
Location: 2848884-2850563
NCBI BlastP on this gene
LG3211_2555
lumazine-binding family protein
Accession: ALN91521
Location: 2848274-2848720
NCBI BlastP on this gene
Accession: ALN91521
Location: 2848274-2848720
NCBI BlastP on this gene
LG3211_2554
bacterial regulatory helix-turn-helix, lysR family protein
Accession: ALN91520
Location: 2847296-2848177
NCBI BlastP on this gene
Accession: ALN91520
Location: 2847296-2848177
NCBI BlastP on this gene
LG3211_2553
glycosyl hydrolase 9 family protein
Accession: ALN91519
Location: 2845411-2847282
NCBI BlastP on this gene
Accession: ALN91519
Location: 2845411-2847282
NCBI BlastP on this gene
LG3211_2552
327. : CP034550 Saccharothrix syringae strain NRRL B-16468 chromosome Total score: 1.0 Cumulative Blast bit score: 366
alpha-L-arabinofuranosidase
Accession: EKG83_29055
Location: 6846426-6847013
NCBI BlastP on this gene
Accession: EKG83_29055
Location: 6846426-6847013
NCBI BlastP on this gene
EKG83_29055
EKG83_29060
alpha-galactosidase
Accession: QFZ20898
Location: 6849888-6851486
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 366
Sequence coverage: 92 %
E-value: 3e-118
NCBI BlastP on this gene
Accession: QFZ20898
Location: 6849888-6851486
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 366
Sequence coverage: 92 %
E-value: 3e-118
NCBI BlastP on this gene
EKG83_29065
EKG83_29070
MarR family transcriptional regulator
Accession: QFZ24575
Location: 6853332-6853697
NCBI BlastP on this gene
Accession: QFZ24575
Location: 6853332-6853697
NCBI BlastP on this gene
EKG83_29075
DHA2 family efflux MFS transporter permease subunit
Accession: QFZ24576
Location: 6853764-6855347
NCBI BlastP on this gene
Accession: QFZ24576
Location: 6853764-6855347
NCBI BlastP on this gene
EKG83_29080
LacI family transcriptional regulator
Accession: QFZ20900
Location: 6855453-6856469
NCBI BlastP on this gene
Accession: QFZ20900
Location: 6855453-6856469
NCBI BlastP on this gene
EKG83_29085
328. : CP042807 Rhodanobacter glycinis strain T01E-68 chromosome Total score: 1.0 Cumulative Blast bit score: 365
CS053_15395
SGNH/GDSL hydrolase family protein
Accession: QEE25735
Location: 3460052-3460909
NCBI BlastP on this gene
Accession: QEE25735
Location: 3460052-3460909
NCBI BlastP on this gene
CS053_15400
CS053_15405
glycoside hydrolase family 27 protein
Accession: QEE25737
Location: 3463075-3464301
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 365
Sequence coverage: 98 %
E-value: 2e-119
NCBI BlastP on this gene
Accession: QEE25737
Location: 3463075-3464301
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 365
Sequence coverage: 98 %
E-value: 2e-119
NCBI BlastP on this gene
CS053_15410
329. : CP046397 Bacteroides ovatus strain FDAARGOS_733 chromosome Total score: 1.0 Cumulative Blast bit score: 363
FOC41_01695
serine/threonine protein phosphatase
Accession: QGT69760
Location: 450147-451598
NCBI BlastP on this gene
Accession: QGT69760
Location: 450147-451598
NCBI BlastP on this gene
FOC41_01700
DUF5009 domain-containing protein
Accession: QGT69761
Location: 451604-453004
NCBI BlastP on this gene
Accession: QGT69761
Location: 451604-453004
NCBI BlastP on this gene
FOC41_01705
FOC41_01710
glycoside hydrolase family 27 protein
Accession: QGT69763
Location: 454200-455384
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 363
Sequence coverage: 100 %
E-value: 1e-118
NCBI BlastP on this gene
Accession: QGT69763
Location: 454200-455384
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 363
Sequence coverage: 100 %
E-value: 1e-118
NCBI BlastP on this gene
FOC41_01715
DUF5009 domain-containing protein
Accession: QGT69764
Location: 456054-457466
NCBI BlastP on this gene
Accession: QGT69764
Location: 456054-457466
NCBI BlastP on this gene
FOC41_01720
FOC41_01725
330. : LT607752 Micromonospora rifamycinica strain DSM 44983 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 361
GA0070623_1909
Uncharacterized conserved protein YdhG,
Accession: SCG51287
Location: 2359995-2360477
NCBI BlastP on this gene
Accession: SCG51287
Location: 2359995-2360477
NCBI BlastP on this gene
GA0070623_1910
GA0070623_1911
GA0070623_1912
GA0070623_1913
alpha-galactosidase
Accession: SCG51318
Location: 2364118-2365776
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 361
Sequence coverage: 100 %
E-value: 6e-116
NCBI BlastP on this gene
Accession: SCG51318
Location: 2364118-2365776
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 361
Sequence coverage: 100 %
E-value: 6e-116
NCBI BlastP on this gene
GA0070623_1914
331. : CP003777 Amycolatopsis mediterranei RB Total score: 1.0 Cumulative Blast bit score: 361
alpha-galactosidase
Accession: AGT88368
Location: 8615482-8617062
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: AGT88368
Location: 8615482-8617062
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
galA
glucose/sorbosone dehydrogenase
Accession: AGT88367
Location: 8613194-8615278
NCBI BlastP on this gene
Accession: AGT88367
Location: 8613194-8615278
NCBI BlastP on this gene
B737_7706
B737_7705
B737_7704
332. : CP003729 Amycolatopsis mediterranei S699 Total score: 1.0 Cumulative Blast bit score: 361
alpha-galactosidase
Accession: AFO81240
Location: 8615538-8617118
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: AFO81240
Location: 8615538-8617118
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
galA
glucose/sorbosone dehydrogenase
Accession: AFO81239
Location: 8613250-8615334
NCBI BlastP on this gene
Accession: AFO81239
Location: 8613250-8615334
NCBI BlastP on this gene
AMES_7706
AMES_7705
AMES_7704
333. : CP002896 Amycolatopsis mediterranei S699 Total score: 1.0 Cumulative Blast bit score: 361
alpha-galactosidase
Accession: AEK46509
Location: 8605621-8607201
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: AEK46509
Location: 8605621-8607201
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
RAM_40210
glucose/sorbosone dehydrogenase
Accession: AEK46508
Location: 8603333-8605417
NCBI BlastP on this gene
Accession: AEK46508
Location: 8603333-8605417
NCBI BlastP on this gene
RAM_40205
RAM_40200
RAM_40195
334. : CP002000 Amycolatopsis mediterranei U32 Total score: 1.0 Cumulative Blast bit score: 361
alpha-galactosidase
Accession: ADJ49531
Location: 8605565-8607145
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
Accession: ADJ49531
Location: 8605565-8607145
BlastP hit with aga27A
Percentage identity: 48 %
BlastP bit score: 361
Sequence coverage: 96 %
E-value: 2e-116
NCBI BlastP on this gene
galA
glucose/sorbosone dehydrogenase
Accession: ADJ49530
Location: 8603277-8605361
NCBI BlastP on this gene
Accession: ADJ49530
Location: 8603277-8605361
NCBI BlastP on this gene
AMED_7822
glycoside hydrolase family protein
Accession: ADJ49529
Location: 8601612-8603162
NCBI BlastP on this gene
Accession: ADJ49529
Location: 8601612-8603162
NCBI BlastP on this gene
AMED_7821
conserved hypothetical protein
Accession: ADJ49528
Location: 8600327-8601484
NCBI BlastP on this gene
Accession: ADJ49528
Location: 8600327-8601484
NCBI BlastP on this gene
AMED_7820
335. : CP002396 Asticcacaulis excentricus CB 48 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 360
Astex_2874
Astex_2875
DNA polymerase III, delta prime subunit
Accession: ADU14513
Location: 559550-560560
NCBI BlastP on this gene
Accession: ADU14513
Location: 559550-560560
NCBI BlastP on this gene
Astex_2876
Astex_2877
Astex_2878
protein of unknown function DUF1304
Accession: ADU14516
Location: 562102-562455
NCBI BlastP on this gene
Accession: ADU14516
Location: 562102-562455
NCBI BlastP on this gene
Astex_2879
Mannan endo-1,4-beta-mannosidase
Accession: ADU14517
Location: 562934-564277
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 360
Sequence coverage: 89 %
E-value: 4e-116
NCBI BlastP on this gene
Accession: ADU14517
Location: 562934-564277
BlastP hit with man5A
Percentage identity: 44 %
BlastP bit score: 360
Sequence coverage: 89 %
E-value: 4e-116
NCBI BlastP on this gene
Astex_2880
336. : CP016174 Amycolatopsis orientalis strain B-37 Total score: 1.0 Cumulative Blast bit score: 357
IclR family transcriptional regulator
Accession: ANN15470
Location: 1500676-1501455
NCBI BlastP on this gene
Accession: ANN15470
Location: 1500676-1501455
NCBI BlastP on this gene
SD37_07225
SD37_07220
AraC family transcriptional regulator
Accession: ANN15468
Location: 1499265-1500107
NCBI BlastP on this gene
Accession: ANN15468
Location: 1499265-1500107
NCBI BlastP on this gene
SD37_07215
SD37_07210
SD37_07205
adenosylmethionine-8-amino-7-oxononanoate aminotransferase
Accession: ANN15465
Location: 1496846-1498117
NCBI BlastP on this gene
Accession: ANN15465
Location: 1496846-1498117
NCBI BlastP on this gene
SD37_07200
alpha-galactosidase
Accession: ANN15464
Location: 1494675-1496627
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 3e-113
NCBI BlastP on this gene
Accession: ANN15464
Location: 1494675-1496627
BlastP hit with aga27A
Percentage identity: 49 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 3e-113
NCBI BlastP on this gene
SD37_07195
GntR family transcriptional regulator
Accession: ANN15463
Location: 1493938-1494633
NCBI BlastP on this gene
Accession: ANN15463
Location: 1493938-1494633
NCBI BlastP on this gene
SD37_07190
SD37_07185
SD37_07180
SD37_07175
337. : CP010836 Sphingomonas hengshuiensis strain WHSC-8 Total score: 1.0 Cumulative Blast bit score: 357
TS85_09205
succinylglutamate-semialdehyde dehydrogenase
Accession: AJP71926
Location: 2100262-2101677
NCBI BlastP on this gene
Accession: AJP71926
Location: 2100262-2101677
NCBI BlastP on this gene
astD
TS85_09215
TS85_09220
TS85_09225
mannanase
Accession: AJP74397
Location: 2103504-2104844
BlastP hit with man5A
Percentage identity: 43 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 5e-115
NCBI BlastP on this gene
Accession: AJP74397
Location: 2103504-2104844
BlastP hit with man5A
Percentage identity: 43 %
BlastP bit score: 357
Sequence coverage: 92 %
E-value: 5e-115
NCBI BlastP on this gene
TS85_09230
338. : CP017717 Nonomuraea sp. ATCC 55076 chromosome Total score: 1.0 Cumulative Blast bit score: 356
BKM31_10535
BKM31_10540
BKM31_10545
alpha-galactosidase
Accession: AQZ61851
Location: 2358336-2359913
BlastP hit with aga27A
Percentage identity: 55 %
BlastP bit score: 356
Sequence coverage: 77 %
E-value: 2e-114
NCBI BlastP on this gene
Accession: AQZ61851
Location: 2358336-2359913
BlastP hit with aga27A
Percentage identity: 55 %
BlastP bit score: 356
Sequence coverage: 77 %
E-value: 2e-114
NCBI BlastP on this gene
BKM31_10550
BKM31_10555
BKM31_10560
sugar ABC transporter permease
Accession: AQZ61854
Location: 2362469-2363389
NCBI BlastP on this gene
Accession: AQZ61854
Location: 2362469-2363389
NCBI BlastP on this gene
BKM31_10565
sugar ABC transporter substrate-binding protein
Accession: AQZ61855
Location: 2363391-2364704
NCBI BlastP on this gene
Accession: AQZ61855
Location: 2363391-2364704
NCBI BlastP on this gene
BKM31_10570
339. : CP041061 Micromonospora sp. HM134 chromosome Total score: 1.0 Cumulative Blast bit score: 353
FJK98_09230
FJK98_09225
FJK98_09220
FJK98_09215
alpha-galactosidase
Accession: QDY07330
Location: 2007685-2009343
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 353
Sequence coverage: 85 %
E-value: 7e-113
NCBI BlastP on this gene
Accession: QDY07330
Location: 2007685-2009343
BlastP hit with aga27A
Percentage identity: 51 %
BlastP bit score: 353
Sequence coverage: 85 %
E-value: 7e-113
NCBI BlastP on this gene
FJK98_09210
NAD-dependent epimerase/dehydratase family protein
Accession: QDY07329
Location: 2006578-2007492
NCBI BlastP on this gene
Accession: QDY07329
Location: 2006578-2007492
NCBI BlastP on this gene
FJK98_09205
FJK98_09200
340. : CP035501 Sphingosinicella sp. BN140058 chromosome Total score: 1.0 Cumulative Blast bit score: 353
mannanase
Accession: QAY77330
Location: 2871608-2872951
BlastP hit with man5A
Percentage identity: 42 %
BlastP bit score: 353
Sequence coverage: 89 %
E-value: 2e-113
NCBI BlastP on this gene
Accession: QAY77330
Location: 2871608-2872951
BlastP hit with man5A
Percentage identity: 42 %
BlastP bit score: 353
Sequence coverage: 89 %
E-value: 2e-113
NCBI BlastP on this gene
ETR14_13065
ETR14_13060
GNAT family N-acetyltransferase
Accession: QAY77328
Location: 2869194-2869652
NCBI BlastP on this gene
Accession: QAY77328
Location: 2869194-2869652
NCBI BlastP on this gene
ETR14_13055
fumC
ETR14_13045
341. : CP032052 Streptomyces clavuligerus strain F1D-5 chromosome Total score: 1.0 Cumulative Blast bit score: 353
DUF3472 domain-containing protein
Accession: AXU11838
Location: 822688-823896
NCBI BlastP on this gene
Accession: AXU11838
Location: 822688-823896
NCBI BlastP on this gene
D1794_03325
D1794_03320
D1794_03315
D1794_03310
alpha-galactosidase
Accession: AXU11835
Location: 817449-819599
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
Accession: AXU11835
Location: 817449-819599
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
D1794_03305
LacI family transcriptional regulator
Accession: AXU11834
Location: 816173-817315
NCBI BlastP on this gene
Accession: AXU11834
Location: 816173-817315
NCBI BlastP on this gene
D1794_03300
D1794_03295
glycoside hydrolase family 28 protein
Accession: AXU11832
Location: 814203-815642
NCBI BlastP on this gene
Accession: AXU11832
Location: 814203-815642
NCBI BlastP on this gene
D1794_03290
D1794_03285
342. : CP027858 Streptomyces clavuligerus strain ATCC 27064 chromosome Total score: 1.0 Cumulative Blast bit score: 353
DUF3472 domain-containing protein
Accession: QCS04616
Location: 684052-685260
NCBI BlastP on this gene
Accession: QCS04616
Location: 684052-685260
NCBI BlastP on this gene
CRV15_02755
CRV15_02750
CRV15_02745
CRV15_02740
alpha-galactosidase
Accession: QCS04613
Location: 678813-680963
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
Accession: QCS04613
Location: 678813-680963
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
CRV15_02735
LacI family transcriptional regulator
Accession: QCS04612
Location: 677537-678538
NCBI BlastP on this gene
Accession: QCS04612
Location: 677537-678538
NCBI BlastP on this gene
CRV15_02730
CRV15_02725
CRV15_02720
CRV15_02715
343. : CP016559 Streptomyces clavuligerus strain F613-1 chromosome Total score: 1.0 Cumulative Blast bit score: 353
BB341_03170
BB341_03165
BB341_03160
alpha-galactosidase
Accession: ANW17289
Location: 815740-817890
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
Accession: ANW17289
Location: 815740-817890
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 353
Sequence coverage: 96 %
E-value: 8e-111
NCBI BlastP on this gene
BB341_03155
BB341_03150
BB341_03145
BB341_03140
BB341_03135
344. : LT827010 Actinoplanes sp. SE50/110 isolate ACP50 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 352
alpha-galactosidase
Accession: SLM02731
Location: 6556199-6557776
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
Accession: SLM02731
Location: 6556199-6557776
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
ACSP50_6016
ACSP50_6015
ACSP50_6014
aminoglycoside 2'-N-acetyltransferase
Accession: SLM02728
Location: 6552253-6552792
NCBI BlastP on this gene
Accession: SLM02728
Location: 6552253-6552792
NCBI BlastP on this gene
ACSP50_6013
ACSP50_6012
345. : CP023298 Actinoplanes sp. SE50 chromosome Total score: 1.0 Cumulative Blast bit score: 352
alpha-galactosidase
Accession: ATO85320
Location: 6555329-6556906
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
Accession: ATO85320
Location: 6555329-6556906
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
ACWT_5905
ACWT_5904
Polycystic kidney disease protein 1-like 3
Accession: ATO85318
Location: 6551948-6552502
NCBI BlastP on this gene
Accession: ATO85318
Location: 6551948-6552502
NCBI BlastP on this gene
ACWT_5903
aminoglycoside 2'-N-acetyltransferase
Accession: ATO85317
Location: 6551383-6551922
NCBI BlastP on this gene
Accession: ATO85317
Location: 6551383-6551922
NCBI BlastP on this gene
ACWT_5902
ACWT_5901
346. : CP010836 Sphingomonas hengshuiensis strain WHSC-8 Total score: 1.0 Cumulative Blast bit score: 352
TS85_01410
TS85_01400
mannanase
Accession: AJP70767
Location: 300884-302230
BlastP hit with man5A
Percentage identity: 41 %
BlastP bit score: 352
Sequence coverage: 94 %
E-value: 4e-113
NCBI BlastP on this gene
Accession: AJP70767
Location: 300884-302230
BlastP hit with man5A
Percentage identity: 41 %
BlastP bit score: 352
Sequence coverage: 94 %
E-value: 4e-113
NCBI BlastP on this gene
TS85_01390
TS85_01380
TS85_01375
347. : CP003170 Actinoplanes sp. SE50/110 Total score: 1.0 Cumulative Blast bit score: 352
ricin B lectin
Accession: AEV86924
Location: 6556178-6557755
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
Accession: AEV86924
Location: 6556178-6557755
BlastP hit with aga27A
Percentage identity: 44 %
BlastP bit score: 352
Sequence coverage: 101 %
E-value: 1e-112
NCBI BlastP on this gene
ACPL_6037
zinc metalloprotease (elastase)-like protein
Accession: AEV86923
Location: 6553709-6555703
NCBI BlastP on this gene
Accession: AEV86923
Location: 6553709-6555703
NCBI BlastP on this gene
ACPL_6036
Polycystic kidney disease protein 1-like 3
Accession: AEV86922
Location: 6552797-6553351
NCBI BlastP on this gene
Accession: AEV86922
Location: 6552797-6553351
NCBI BlastP on this gene
ACPL_6035
Aminoglycoside 2'-N-acetyltransferase
Accession: AEV86921
Location: 6552232-6552771
NCBI BlastP on this gene
Accession: AEV86921
Location: 6552232-6552771
NCBI BlastP on this gene
aac
putative amino-acid metabolite efflux pump
Accession: AEV86920
Location: 6551223-6552113
NCBI BlastP on this gene
Accession: AEV86920
Location: 6551223-6552113
NCBI BlastP on this gene
ACPL_6033
348. : AM420293 Saccharopolyspora erythraea NRRL2338 complete genome. Total score: 1.0 Cumulative Blast bit score: 350
SACE_7060
SACE_7061
two component system sensor kinase
Accession: CAM06223
Location: 7869967-7871508
NCBI BlastP on this gene
Accession: CAM06223
Location: 7869967-7871508
NCBI BlastP on this gene
SACE_7062
two component system response regulator
Accession: CAM06224
Location: 7871517-7872170
NCBI BlastP on this gene
Accession: CAM06224
Location: 7871517-7872170
NCBI BlastP on this gene
SACE_7063
SACE_7064
putative alpha-galactosidase
Accession: CAM06226
Location: 7872998-7874176
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 350
Sequence coverage: 93 %
E-value: 7e-114
NCBI BlastP on this gene
Accession: CAM06226
Location: 7872998-7874176
BlastP hit with aga27A
Percentage identity: 47 %
BlastP bit score: 350
Sequence coverage: 93 %
E-value: 7e-114
NCBI BlastP on this gene
agaB2
349. : CP003777 Amycolatopsis mediterranei RB Total score: 1.0 Cumulative Blast bit score: 347
B737_3607
B737_3606
B737_3605
alpha-galactosidase
Accession: AGT84268
Location: 3962936-3964891
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
Accession: AGT84268
Location: 3962936-3964891
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
galA
TetR family transcriptional regulator
Accession: AGT84267
Location: 3961989-3962633
NCBI BlastP on this gene
Accession: AGT84267
Location: 3961989-3962633
NCBI BlastP on this gene
B737_3603
carbon-monoxide dehydrogenase small subunit
Accession: AGT84266
Location: 3961397-3961885
NCBI BlastP on this gene
Accession: AGT84266
Location: 3961397-3961885
NCBI BlastP on this gene
B737_3602
carbon-monoxide dehydrogenase medium subunit
Accession: AGT84265
Location: 3960405-3961400
NCBI BlastP on this gene
Accession: AGT84265
Location: 3960405-3961400
NCBI BlastP on this gene
B737_3601
carbon-monoxide dehydrogenase large subunit
Accession: AGT84264
Location: 3958192-3960408
NCBI BlastP on this gene
Accession: AGT84264
Location: 3958192-3960408
NCBI BlastP on this gene
B737_3600
350. : CP003729 Amycolatopsis mediterranei S699 Total score: 1.0 Cumulative Blast bit score: 347
AMES_3607
AMES_3606
AMES_3605
alpha-galactosidase
Accession: AFO77140
Location: 3962999-3964954
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
Accession: AFO77140
Location: 3962999-3964954
BlastP hit with aga27A
Percentage identity: 46 %
BlastP bit score: 347
Sequence coverage: 94 %
E-value: 2e-109
NCBI BlastP on this gene
galA
TetR family transcriptional regulator
Accession: AFO77139
Location: 3962052-3962696
NCBI BlastP on this gene
Accession: AFO77139
Location: 3962052-3962696
NCBI BlastP on this gene
AMES_3603
carbon-monoxide dehydrogenase small subunit
Accession: AFO77138
Location: 3961460-3961948
NCBI BlastP on this gene
Accession: AFO77138
Location: 3961460-3961948
NCBI BlastP on this gene
AMES_3602
carbon-monoxide dehydrogenase medium subunit
Accession: AFO77137
Location: 3960468-3961463
NCBI BlastP on this gene
Accession: AFO77137
Location: 3960468-3961463
NCBI BlastP on this gene
AMES_3601
carbon-monoxide dehydrogenase large subunit
Accession: AFO77136
Location: 3958255-3960471
NCBI BlastP on this gene
Accession: AFO77136
Location: 3958255-3960471
NCBI BlastP on this gene
AMES_3600

Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.