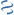MultiGeneBlast hits

Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP015378 : Fictibacillus phosphorivorans strain G25-29 chromosome Total score: 2.5 Cumulative Blast bit score: 379
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
ABE65_003780
1,2-phenylacetyl-CoA epoxidase subunit B
Accession: ANC75977
Location: 720570-720917
NCBI BlastP on this gene
Accession: ANC75977
Location: 720570-720917
NCBI BlastP on this gene
ABE65_003775
1,2-phenylacetyl-CoA epoxidase subunit A
Accession: ANC75976
Location: 719581-720546
NCBI BlastP on this gene
Accession: ANC75976
Location: 719581-720546
NCBI BlastP on this gene
paaA
ABE65_003765
ABE65_003760
ABE65_003755
LysR family transcriptional regulator
Accession: ANC75972
Location: 715168-716046
NCBI BlastP on this gene
Accession: ANC75972
Location: 715168-716046
NCBI BlastP on this gene
ABE65_003750
ABE65_003745
zinc-binding alcohol dehydrogenase
Accession: ANC75970
Location: 713811-714791
NCBI BlastP on this gene
Accession: ANC75970
Location: 713811-714791
NCBI BlastP on this gene
ABE65_003740
ABE65_003735
sugar ABC transporter permease
Accession: ANC75968
Location: 712039-712866
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 100 %
E-value: 1e-57
NCBI BlastP on this gene
Accession: ANC75968
Location: 712039-712866
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 100 %
E-value: 1e-57
NCBI BlastP on this gene
ABE65_003730
ABC transporter permease
Accession: ANC79261
Location: 710680-712008
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 182
Sequence coverage: 82 %
E-value: 4e-50
NCBI BlastP on this gene
Accession: ANC79261
Location: 710680-712008
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 182
Sequence coverage: 82 %
E-value: 4e-50
NCBI BlastP on this gene
ABE65_003725
ABC transporter substrate-binding protein
Accession: ANC75967
Location: 709204-710511
NCBI BlastP on this gene
Accession: ANC75967
Location: 709204-710511
NCBI BlastP on this gene
ABE65_003720
LacI family transcriptional regulator
Accession: ANC75966
Location: 708163-709149
NCBI BlastP on this gene
Accession: ANC75966
Location: 708163-709149
NCBI BlastP on this gene
ABE65_003715
peptide ABC transporter ATP-binding protein
Accession: ANC75965
Location: 707229-707951
NCBI BlastP on this gene
Accession: ANC75965
Location: 707229-707951
NCBI BlastP on this gene
ABE65_003710
glycine/betaine ABC transporter
Accession: ANC79260
Location: 705438-706970
NCBI BlastP on this gene
Accession: ANC79260
Location: 705438-706970
NCBI BlastP on this gene
ABE65_003705
glycine/betaine ABC transporter ATP-binding protein
Accession: ANC75964
Location: 704270-705424
NCBI BlastP on this gene
Accession: ANC75964
Location: 704270-705424
NCBI BlastP on this gene
ABE65_003700
ABE65_003695
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP008820 : Ochrobactrum anthropi strain OAB chromosome 1 Total score: 2.5 Cumulative Blast bit score: 377
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
NMT1/THI5 like family protein
Accession: AIK42988
Location: 2672342-2673340
NCBI BlastP on this gene
Accession: AIK42988
Location: 2672342-2673340
NCBI BlastP on this gene
DR92_2597
binding--dependent transport system inner membrane component family protein
Accession: AIK44254
Location: 2671371-2672228
NCBI BlastP on this gene
Accession: AIK44254
Location: 2671371-2672228
NCBI BlastP on this gene
DR92_2596
binding--dependent transport system inner membrane component family protein
Accession: AIK44308
Location: 2670478-2671371
NCBI BlastP on this gene
Accession: AIK44308
Location: 2670478-2671371
NCBI BlastP on this gene
DR92_2595
ABC transporter family protein
Accession: AIK43610
Location: 2669674-2670441
NCBI BlastP on this gene
Accession: AIK43610
Location: 2669674-2670441
NCBI BlastP on this gene
DR92_2594
hydA
amidase, hydantoinase/carbamoylase family protein
Accession: AIK43518
Location: 2666823-2668070
NCBI BlastP on this gene
Accession: AIK43518
Location: 2666823-2668070
NCBI BlastP on this gene
DR92_2592
HTH-type transcriptional regulator RutR
Accession: AIK45142
Location: 2665905-2666603
NCBI BlastP on this gene
Accession: AIK45142
Location: 2665905-2666603
NCBI BlastP on this gene
rutR
DR92_2589
ABC transporter family protein
Accession: AIK44324
Location: 2664440-2665513
NCBI BlastP on this gene
Accession: AIK44324
Location: 2664440-2665513
NCBI BlastP on this gene
DR92_2590
binding--dependent transport system inner membrane component family protein
Accession: AIK42969
Location: 2663592-2664425
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: AIK42969
Location: 2663592-2664425
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
DR92_2588
binding--dependent transport system inner membrane component family protein
Accession: AIK42897
Location: 2662634-2663587
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Accession: AIK42897
Location: 2662634-2663587
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
DR92_2587
bacterial extracellular solute-binding family protein
Accession: AIK44223
Location: 2661300-2662559
NCBI BlastP on this gene
Accession: AIK44223
Location: 2661300-2662559
NCBI BlastP on this gene
DR92_2586
cys/Met metabolism PLP-dependent enzyme family protein
Accession: AIK44980
Location: 2660095-2661264
NCBI BlastP on this gene
Accession: AIK44980
Location: 2660095-2661264
NCBI BlastP on this gene
DR92_2585
bacterial regulatory s, gntR family protein
Accession: AIK44892
Location: 2659382-2660098
NCBI BlastP on this gene
Accession: AIK44892
Location: 2659382-2660098
NCBI BlastP on this gene
DR92_2584
pld1
DR92_2582
dapA
bacterial regulatory s, gntR family protein
Accession: AIK43481
Location: 2655231-2655863
NCBI BlastP on this gene
Accession: AIK43481
Location: 2655231-2655863
NCBI BlastP on this gene
DR92_2580
Tat (twin-arginine translocation) pathway signal sequence domain protein
Accession: AIK43872
Location: 2653820-2655154
NCBI BlastP on this gene
Accession: AIK43872
Location: 2653820-2655154
NCBI BlastP on this gene
DR92_2579
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP000758 : Ochrobactrum anthropi ATCC 49188 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 377
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Oant_0343
binding-protein-dependent transport systems inner membrane component
Accession: ABS13075
Location: 361121-361981
NCBI BlastP on this gene
Accession: ABS13075
Location: 361121-361981
NCBI BlastP on this gene
Oant_0344
binding-protein-dependent transport systems inner membrane component
Accession: ABS13076
Location: 361978-362871
NCBI BlastP on this gene
Accession: ABS13076
Location: 361978-362871
NCBI BlastP on this gene
Oant_0345
Oant_0346
Oant_0347
amidase, hydantoinase/carbamoylase family
Accession: ABS13079
Location: 365294-366541
NCBI BlastP on this gene
Accession: ABS13079
Location: 365294-366541
NCBI BlastP on this gene
Oant_0348
transcriptional regulator, TetR family
Accession: ABS13080
Location: 366762-367460
NCBI BlastP on this gene
Accession: ABS13080
Location: 366762-367460
NCBI BlastP on this gene
Oant_0349
Oant_0350
Oant_0351
binding-protein-dependent transport systems inner membrane component
Accession: ABS13083
Location: 368940-369773
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: ABS13083
Location: 368940-369773
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Oant_0352
binding-protein-dependent transport systems inner membrane component
Accession: ABS13084
Location: 369778-370731
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Accession: ABS13084
Location: 369778-370731
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Oant_0353
extracellular solute-binding protein family 1
Accession: ABS13085
Location: 370806-372065
NCBI BlastP on this gene
Accession: ABS13085
Location: 370806-372065
NCBI BlastP on this gene
Oant_0354
Oant_0355
Oant_0356
Oant_0357
Oant_0358
Oant_0359
transcriptional regulator, GntR family
Accession: ABS13091
Location: 377472-378134
NCBI BlastP on this gene
Accession: ABS13091
Location: 377472-378134
NCBI BlastP on this gene
Oant_0360
extracellular solute-binding protein family 1
Accession: ABS13092
Location: 378211-379545
NCBI BlastP on this gene
Accession: ABS13092
Location: 378211-379545
NCBI BlastP on this gene
Oant_0361
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP047140 : Streptomyces sp. Tu 2975 chromosome Total score: 2.5 Cumulative Blast bit score: 376
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
GLX30_08700
LacI family transcriptional regulator
Accession: QIP84106
Location: 1985389-1986447
NCBI BlastP on this gene
Accession: QIP84106
Location: 1985389-1986447
NCBI BlastP on this gene
GLX30_08705
glycoside hydrolase family 13 protein
Accession: QIP84107
Location: 1986435-1988084
NCBI BlastP on this gene
Accession: QIP84107
Location: 1986435-1988084
NCBI BlastP on this gene
GLX30_08710
carbohydrate ABC transporter permease
Accession: QIP88438
Location: 1988168-1989022
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 197
Sequence coverage: 99 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: QIP88438
Location: 1988168-1989022
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 197
Sequence coverage: 99 %
E-value: 2e-57
NCBI BlastP on this gene
GLX30_08715
sugar ABC transporter permease
Accession: QIP84108
Location: 1989016-1990026
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
Accession: QIP84108
Location: 1989016-1990026
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
GLX30_08720
extracellular solute-binding protein
Accession: QIP84109
Location: 1990046-1991320
NCBI BlastP on this gene
Accession: QIP84109
Location: 1990046-1991320
NCBI BlastP on this gene
GLX30_08725
LacI family transcriptional regulator
Accession: QIP84110
Location: 1991715-1992761
NCBI BlastP on this gene
Accession: QIP84110
Location: 1991715-1992761
NCBI BlastP on this gene
GLX30_08730
carbohydrate kinase family protein
Accession: QIP84111
Location: 1992758-1993678
NCBI BlastP on this gene
Accession: QIP84111
Location: 1992758-1993678
NCBI BlastP on this gene
GLX30_08735
carbohydrate ABC transporter permease
Accession: QIP88439
Location: 1993773-1994576
NCBI BlastP on this gene
Accession: QIP88439
Location: 1993773-1994576
NCBI BlastP on this gene
GLX30_08740
sugar ABC transporter permease
Accession: QIP84112
Location: 1994680-1995690
NCBI BlastP on this gene
Accession: QIP84112
Location: 1994680-1995690
NCBI BlastP on this gene
GLX30_08745
carbohydrate ABC transporter substrate-binding protein
Accession: QIP84113
Location: 1995797-1997146
NCBI BlastP on this gene
Accession: QIP84113
Location: 1995797-1997146
NCBI BlastP on this gene
GLX30_08750
LacI family transcriptional regulator
Accession: QIP84114
Location: 1997603-1998628
NCBI BlastP on this gene
Accession: QIP84114
Location: 1997603-1998628
NCBI BlastP on this gene
GLX30_08755
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP022604 : Ochrobactrum quorumnocens strain A44 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 376
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
CES85_0286
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV85292
Location: 286302-287159
NCBI BlastP on this gene
Accession: ASV85292
Location: 286302-287159
NCBI BlastP on this gene
CES85_0287
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV85345
Location: 287156-288049
NCBI BlastP on this gene
Accession: ASV85345
Location: 287156-288049
NCBI BlastP on this gene
CES85_0288
CES85_0289
hydA
amidase, hydantoinase/carbamoylase family protein
Accession: ASV86894
Location: 290492-291754
NCBI BlastP on this gene
Accession: ASV86894
Location: 290492-291754
NCBI BlastP on this gene
CES85_0291
bacterial regulatory, tetR family protein
Accession: ASV87540
Location: 291965-292645
NCBI BlastP on this gene
Accession: ASV87540
Location: 291965-292645
NCBI BlastP on this gene
CES85_0292
CES85_0293
CES85_0294
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV87333
Location: 294125-294958
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: ASV87333
Location: 294125-294958
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 2e-56
NCBI BlastP on this gene
CES85_0295
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV87617
Location: 294963-295847
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 182
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Accession: ASV87617
Location: 294963-295847
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 182
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
CES85_0296
bacterial extracellular solute-binding family protein
Accession: ASV86975
Location: 295993-297252
NCBI BlastP on this gene
Accession: ASV86975
Location: 295993-297252
NCBI BlastP on this gene
CES85_0297
cys/Met metabolism PLP-dependent enzyme family protein
Accession: ASV87147
Location: 297290-298459
NCBI BlastP on this gene
Accession: ASV87147
Location: 297290-298459
NCBI BlastP on this gene
CES85_0298
CES85_0299
CES85_0300
pld1
CES85_0302
CES85_0303
bacterial regulatory, gntR family protein
Accession: ASV87433
Location: 302767-303429
NCBI BlastP on this gene
Accession: ASV87433
Location: 302767-303429
NCBI BlastP on this gene
CES85_0304
tat (twin-arginine translocation) pathway signal sequence domain protein
Accession: ASV87356
Location: 303521-304855
NCBI BlastP on this gene
Accession: ASV87356
Location: 303521-304855
NCBI BlastP on this gene
CES85_0305
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP003359 : Halobacteroides halobius DSM 5150 Total score: 2.5 Cumulative Blast bit score: 376
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Protein of unknown function (DUF2012)
Accession: AGB41482
Location: 1543870-1544850
NCBI BlastP on this gene
Accession: AGB41482
Location: 1543870-1544850
NCBI BlastP on this gene
Halha_1543
Halha_1544
Halha_1545
Halha_1546
trehalose/maltose hydrolase or phosphorylase
Accession: AGB41486
Location: 1549302-1551617
NCBI BlastP on this gene
Accession: AGB41486
Location: 1549302-1551617
NCBI BlastP on this gene
Halha_1547
ABC-type sugar transport system, periplasmic component
Accession: AGB41487
Location: 1551714-1553000
NCBI BlastP on this gene
Accession: AGB41487
Location: 1551714-1553000
NCBI BlastP on this gene
Halha_1548
ABC-type sugar transport system, permease component
Accession: AGB41488
Location: 1553098-1553940
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 195
Sequence coverage: 101 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: AGB41488
Location: 1553098-1553940
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 195
Sequence coverage: 101 %
E-value: 1e-56
NCBI BlastP on this gene
Halha_1549
permease component of ABC-type sugar transporter
Accession: AGB41489
Location: 1553955-1554890
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 181
Sequence coverage: 95 %
E-value: 7e-51
NCBI BlastP on this gene
Accession: AGB41489
Location: 1553955-1554890
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 181
Sequence coverage: 95 %
E-value: 7e-51
NCBI BlastP on this gene
Halha_1550
Halha_1551
Halha_1552
dehydrogenase of unknown specificity, short-chain alcohol dehydrogenase like protein
Accession: AGB41492
Location: 1558204-1558965
NCBI BlastP on this gene
Accession: AGB41492
Location: 1558204-1558965
NCBI BlastP on this gene
Halha_1553
trehalose/maltose hydrolase or phosphorylase
Accession: AGB41493
Location: 1558955-1561306
NCBI BlastP on this gene
Accession: AGB41493
Location: 1558955-1561306
NCBI BlastP on this gene
Halha_1554
ABC-type sugar transport system, periplasmic component
Accession: AGB41494
Location: 1561401-1562645
NCBI BlastP on this gene
Accession: AGB41494
Location: 1561401-1562645
NCBI BlastP on this gene
Halha_1555
ABC-type sugar transport system, permease component
Accession: AGB41495
Location: 1562700-1563533
NCBI BlastP on this gene
Accession: AGB41495
Location: 1562700-1563533
NCBI BlastP on this gene
Halha_1556
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP038147 : Streptomyces sp. S501 chromosome Total score: 2.5 Cumulative Blast bit score: 375
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
D7Y56_09320
D7Y56_09325
XRE family transcriptional regulator
Accession: QBR06106
Location: 1757841-1758719
NCBI BlastP on this gene
Accession: QBR06106
Location: 1757841-1758719
NCBI BlastP on this gene
D7Y56_09330
DUF397 domain-containing protein
Accession: QBR06107
Location: 1758716-1758925
NCBI BlastP on this gene
Accession: QBR06107
Location: 1758716-1758925
NCBI BlastP on this gene
D7Y56_09335
MerR family transcriptional regulator
Accession: QBR06108
Location: 1759002-1759913
NCBI BlastP on this gene
Accession: QBR06108
Location: 1759002-1759913
NCBI BlastP on this gene
D7Y56_09340
D7Y56_09345
RNA polymerase sigma-70 factor
Accession: QBR06110
Location: 1760487-1761386
NCBI BlastP on this gene
Accession: QBR06110
Location: 1760487-1761386
NCBI BlastP on this gene
D7Y56_09350
D7Y56_09355
LacI family transcriptional regulator
Accession: QBR06112
Location: 1762163-1763221
NCBI BlastP on this gene
Accession: QBR06112
Location: 1762163-1763221
NCBI BlastP on this gene
D7Y56_09360
glycoside hydrolase family 13 protein
Accession: QBR06113
Location: 1763206-1764882
NCBI BlastP on this gene
Accession: QBR06113
Location: 1763206-1764882
NCBI BlastP on this gene
D7Y56_09365
carbohydrate ABC transporter permease
Accession: QBR06114
Location: 1764981-1765859
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: QBR06114
Location: 1764981-1765859
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
D7Y56_09370
sugar ABC transporter permease
Accession: QBR06115
Location: 1765856-1766872
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: QBR06115
Location: 1765856-1766872
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
D7Y56_09375
extracellular solute-binding protein
Accession: QBR06116
Location: 1766894-1768165
NCBI BlastP on this gene
Accession: QBR06116
Location: 1766894-1768165
NCBI BlastP on this gene
D7Y56_09380
LacI family transcriptional regulator
Accession: QBR06117
Location: 1768526-1769569
NCBI BlastP on this gene
Accession: QBR06117
Location: 1768526-1769569
NCBI BlastP on this gene
D7Y56_09385
inositol phosphorylceramide synthase
Accession: QBR06118
Location: 1769646-1770644
NCBI BlastP on this gene
Accession: QBR06118
Location: 1769646-1770644
NCBI BlastP on this gene
D7Y56_09390
bifunctional [glutamine synthetase]
Accession: QBR06119
Location: 1770729-1773725
NCBI BlastP on this gene
Accession: QBR06119
Location: 1770729-1773725
NCBI BlastP on this gene
D7Y56_09395
putative protein N(5)-glutamine methyltransferase
Accession: QBR06120
Location: 1773885-1774673
NCBI BlastP on this gene
Accession: QBR06120
Location: 1773885-1774673
NCBI BlastP on this gene
D7Y56_09400
D7Y56_09405
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP011340 : Streptomyces pristinaespiralis strain HCCB 10218 Total score: 2.5 Cumulative Blast bit score: 375
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
sulfonate ABC transporter ATP-binding protein
Accession: ALC20168
Location: 2175830-2181247
NCBI BlastP on this gene
Accession: ALC20168
Location: 2175830-2181247
NCBI BlastP on this gene
SPRI_1862
SPRI_1863
LacI family transcriptional regulator
Accession: ALC20170
Location: 2183581-2184630
NCBI BlastP on this gene
Accession: ALC20170
Location: 2183581-2184630
NCBI BlastP on this gene
SPRI_1864
SPRI_1865
ABC transporter permease
Accession: ALC20172
Location: 2186359-2187210
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 196
Sequence coverage: 99 %
E-value: 4e-57
NCBI BlastP on this gene
Accession: ALC20172
Location: 2186359-2187210
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 196
Sequence coverage: 99 %
E-value: 4e-57
NCBI BlastP on this gene
SPRI_1866
ABC transporter permease
Accession: ALC20173
Location: 2187207-2188217
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
Accession: ALC20173
Location: 2187207-2188217
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
SPRI_1867
sugar ABC transporter substrate-binding protein
Accession: ALC20174
Location: 2188237-2189511
NCBI BlastP on this gene
Accession: ALC20174
Location: 2188237-2189511
NCBI BlastP on this gene
SPRI_1868
LacI family transcriptional regulator
Accession: ALC20175
Location: 2189906-2190940
NCBI BlastP on this gene
Accession: ALC20175
Location: 2189906-2190940
NCBI BlastP on this gene
SPRI_1869
SPRI_1870
SPRI_1871
sugar ABC transporter permease
Accession: ALC20178
Location: 2193875-2194786
NCBI BlastP on this gene
Accession: ALC20178
Location: 2193875-2194786
NCBI BlastP on this gene
SPRI_1872
SPRI_1873
SPRI_1874
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP003990 : Streptomyces sp. PAMC26508 Total score: 2.5 Cumulative Blast bit score: 375
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
F750_2078
F750_2079
F750_2080
F750_2081
MerR-family transcriptional regulator
Accession: AGJ54577
Location: 2154094-2155005
NCBI BlastP on this gene
Accession: AGJ54577
Location: 2154094-2155005
NCBI BlastP on this gene
F750_2082
putative integral membrane protein
Accession: AGJ54578
Location: 2155138-2155476
NCBI BlastP on this gene
Accession: AGJ54578
Location: 2155138-2155476
NCBI BlastP on this gene
F750_2083
F750_2084
F750_2085
maltose operon transcriptional repressor MalR
Accession: AGJ54581
Location: 2157258-2158304
NCBI BlastP on this gene
Accession: AGJ54581
Location: 2157258-2158304
NCBI BlastP on this gene
malR
F750_2087
maltose/maltodextrin ABC transpor permease protein MalG
Accession: AGJ54583
Location: 2160076-2160954
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: AGJ54583
Location: 2160076-2160954
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
malG
maltose/maltodextrin ABC transpor permease protein MalF
Accession: AGJ54584
Location: 2160951-2161967
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: AGJ54584
Location: 2160951-2161967
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
malF
maltose/maltodextrin ABC transpor substrate binding periplasmic protein MalE
Accession: AGJ54585
Location: 2161989-2163260
NCBI BlastP on this gene
Accession: AGJ54585
Location: 2161989-2163260
NCBI BlastP on this gene
malE
F750_2091
maltose operon transcriptional repressor MalR
Accession: AGJ54587
Location: 2163519-2164664
NCBI BlastP on this gene
Accession: AGJ54587
Location: 2163519-2164664
NCBI BlastP on this gene
malR
putative integral membrane protein
Accession: AGJ54588
Location: 2164666-2165739
NCBI BlastP on this gene
Accession: AGJ54588
Location: 2164666-2165739
NCBI BlastP on this gene
F750_2093
glutamate-ammonia-ligase adenylyltransferase
Accession: AGJ54589
Location: 2165824-2168820
NCBI BlastP on this gene
Accession: AGJ54589
Location: 2165824-2168820
NCBI BlastP on this gene
F750_2094
methylase of polypeptide chain release factors
Accession: AGJ54590
Location: 2168980-2169768
NCBI BlastP on this gene
Accession: AGJ54590
Location: 2168980-2169768
NCBI BlastP on this gene
F750_2095
3',5'-cyclic-nucleotide phosphodiesterase
Accession: AGJ54591
Location: 2169983-2170726
NCBI BlastP on this gene
Accession: AGJ54591
Location: 2169983-2170726
NCBI BlastP on this gene
F750_2096
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP002475 : Streptomyces pratensis ATCC 33331 Total score: 2.5 Cumulative Blast bit score: 375
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
alpha/beta hydrolase fold protein
Accession: ADW06012
Location: 5315785-5316660
NCBI BlastP on this gene
Accession: ADW06012
Location: 5315785-5316660
NCBI BlastP on this gene
Sfla_4611
Sfla_4609
ATP-binding region ATPase domain protein
Accession: ADW06010
Location: 5313884-5314321
NCBI BlastP on this gene
Accession: ADW06010
Location: 5313884-5314321
NCBI BlastP on this gene
Sfla_4608
helix-turn-helix domain protein
Accession: ADW06009
Location: 5312841-5313719
NCBI BlastP on this gene
Accession: ADW06009
Location: 5312841-5313719
NCBI BlastP on this gene
Sfla_4607
protein of unknown function DUF397
Accession: ADW06008
Location: 5312635-5312844
NCBI BlastP on this gene
Accession: ADW06008
Location: 5312635-5312844
NCBI BlastP on this gene
Sfla_4606
transcriptional regulator, MerR family
Accession: ADW06007
Location: 5311647-5312558
NCBI BlastP on this gene
Accession: ADW06007
Location: 5311647-5312558
NCBI BlastP on this gene
Sfla_4605
Sfla_4604
GCN5-related N-acetyltransferase
Accession: ADW06005
Location: 5310619-5311176
NCBI BlastP on this gene
Accession: ADW06005
Location: 5310619-5311176
NCBI BlastP on this gene
Sfla_4603
transcriptional regulator, LacI family
Accession: ADW06004
Location: 5309450-5310496
NCBI BlastP on this gene
Accession: ADW06004
Location: 5309450-5310496
NCBI BlastP on this gene
Sfla_4602
alpha amylase catalytic region
Accession: ADW06003
Location: 5307777-5309453
NCBI BlastP on this gene
Accession: ADW06003
Location: 5307777-5309453
NCBI BlastP on this gene
Sfla_4601
binding-protein-dependent transport systems inner membrane component
Accession: ADW06002
Location: 5306800-5307678
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: ADW06002
Location: 5306800-5307678
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Sfla_4600
binding-protein-dependent transport systems inner membrane component
Accession: ADW06001
Location: 5305787-5306803
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: ADW06001
Location: 5305787-5306803
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Sfla_4599
extracellular solute-binding protein family 1
Accession: ADW06000
Location: 5304494-5305765
NCBI BlastP on this gene
Accession: ADW06000
Location: 5304494-5305765
NCBI BlastP on this gene
Sfla_4598
transcriptional regulator, LacI family
Accession: ADW05999
Location: 5303090-5304235
NCBI BlastP on this gene
Accession: ADW05999
Location: 5303090-5304235
NCBI BlastP on this gene
Sfla_4597
Sfla_4596
(Glutamate--ammonia-ligase) adenylyltransferase
Accession: ADW05997
Location: 5298934-5301930
NCBI BlastP on this gene
Accession: ADW05997
Location: 5298934-5301930
NCBI BlastP on this gene
Sfla_4595
modification methylase, HemK family
Accession: ADW05996
Location: 5297986-5298774
NCBI BlastP on this gene
Accession: ADW05996
Location: 5297986-5298774
NCBI BlastP on this gene
Sfla_4594
Sfla_4593
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP035945 : Blautia producta strain PMF1 chromosome Total score: 2.5 Cumulative Blast bit score: 373
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
gutB_1
Bifunctional purine biosynthesis protein PurH
Accession: QBE96980
Location: 2872205-2873383
NCBI BlastP on this gene
Accession: QBE96980
Location: 2872205-2873383
NCBI BlastP on this gene
purH
PMF13cell1_02528
Ribosomal RNA large subunit methyltransferase I
Accession: QBE96978
Location: 2869806-2871005
NCBI BlastP on this gene
Accession: QBE96978
Location: 2869806-2871005
NCBI BlastP on this gene
rlmI
PMF13cell1_02526
PMF13cell1_02525
map_2
PMF13cell1_02523
PMF13cell1_02522
PMF13cell1_02521
Inner membrane ABC transporter permease protein YcjP
Accession: QBE96971
Location: 2864487-2865332
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 2e-54
NCBI BlastP on this gene
Accession: QBE96971
Location: 2864487-2865332
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 2e-54
NCBI BlastP on this gene
ycjP_6
Inner membrane ABC transporter permease protein YcjO
Accession: QBE96970
Location: 2863557-2864474
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 184
Sequence coverage: 100 %
E-value: 3e-52
NCBI BlastP on this gene
Accession: QBE96970
Location: 2863557-2864474
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 184
Sequence coverage: 100 %
E-value: 3e-52
NCBI BlastP on this gene
ycjO_3
PMF13cell1_02518
Multiple sugar-binding protein
Accession: QBE96968
Location: 2860640-2861938
NCBI BlastP on this gene
Accession: QBE96968
Location: 2860640-2861938
NCBI BlastP on this gene
msmE_19
putative sensor-like histidine kinase
Accession: QBE96967
Location: 2858814-2860628
NCBI BlastP on this gene
Accession: QBE96967
Location: 2858814-2860628
NCBI BlastP on this gene
PMF13cell1_02516
putative response regulatory protein
Accession: QBE96966
Location: 2857235-2858809
NCBI BlastP on this gene
Accession: QBE96966
Location: 2857235-2858809
NCBI BlastP on this gene
PMF13cell1_02515
rbr1_2
cmk_4
Gamma-glutamylputrescine oxidoreductase
Accession: QBE96963
Location: 2853801-2855261
NCBI BlastP on this gene
Accession: QBE96963
Location: 2853801-2855261
NCBI BlastP on this gene
puuB
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP030862 : Streptomyces globosus strain LZH-48 chromosome Total score: 2.5 Cumulative Blast bit score: 373
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
DUF3372 domain-containing protein
Accession: AXE26134
Location: 5434344-5439752
NCBI BlastP on this gene
Accession: AXE26134
Location: 5434344-5439752
NCBI BlastP on this gene
C0216_24115
C0216_24120
LacI family transcriptional regulator
Accession: AXE26136
Location: 5441840-5442871
NCBI BlastP on this gene
Accession: AXE26136
Location: 5441840-5442871
NCBI BlastP on this gene
C0216_24125
C0216_24130
ABC transporter permease
Accession: AXE26138
Location: 5444547-5445455
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 91 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: AXE26138
Location: 5444547-5445455
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 91 %
E-value: 2e-57
NCBI BlastP on this gene
C0216_24135
sugar ABC transporter permease
Accession: AXE26139
Location: 5445458-5446489
BlastP hit with WP_039959892.1
Percentage identity: 32 %
BlastP bit score: 176
Sequence coverage: 102 %
E-value: 1e-48
NCBI BlastP on this gene
Accession: AXE26139
Location: 5445458-5446489
BlastP hit with WP_039959892.1
Percentage identity: 32 %
BlastP bit score: 176
Sequence coverage: 102 %
E-value: 1e-48
NCBI BlastP on this gene
C0216_24140
sugar ABC transporter substrate-binding protein
Accession: AXE26140
Location: 5446498-5447772
NCBI BlastP on this gene
Accession: AXE26140
Location: 5446498-5447772
NCBI BlastP on this gene
C0216_24145
C0216_24150
LacI family transcriptional regulator
Accession: AXE27717
Location: 5449649-5450695
NCBI BlastP on this gene
Accession: AXE27717
Location: 5449649-5450695
NCBI BlastP on this gene
C0216_24155
C0216_24160
GNAT family N-acetyltransferase
Accession: AXE26143
Location: 5451674-5452108
NCBI BlastP on this gene
Accession: AXE26143
Location: 5451674-5452108
NCBI BlastP on this gene
C0216_24165
C0216_24170
DUF397 domain-containing protein
Accession: AXE26145
Location: 5453350-5453607
NCBI BlastP on this gene
Accession: AXE26145
Location: 5453350-5453607
NCBI BlastP on this gene
C0216_24175
C0216_24180
C0216_24185
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP015775 : Ochrobactrum pseudogrignonense strain K8 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 372
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
nitrate ABC transporter substrate-binding protein
Accession: ANG95191
Location: 223392-224390
NCBI BlastP on this gene
Accession: ANG95191
Location: 223392-224390
NCBI BlastP on this gene
A8A54_01035
A8A54_01040
A8A54_01045
sulfonate ABC transporter ATP-binding protein
Accession: ANG95194
Location: 226309-227130
NCBI BlastP on this gene
Accession: ANG95194
Location: 226309-227130
NCBI BlastP on this gene
A8A54_01050
A8A54_01055
A8A54_01060
TetR family transcriptional regulator
Accession: ANG95197
Location: 230167-230847
NCBI BlastP on this gene
Accession: ANG95197
Location: 230167-230847
NCBI BlastP on this gene
A8A54_01065
DNA mismatch repair protein MutT
Accession: ANG95198
Location: 230844-231242
NCBI BlastP on this gene
Accession: ANG95198
Location: 230844-231242
NCBI BlastP on this gene
A8A54_01070
glycerol-3-phosphate ABC transporter ATP-binding protein
Accession: ANG95199
Location: 231239-232312
NCBI BlastP on this gene
Accession: ANG95199
Location: 231239-232312
NCBI BlastP on this gene
A8A54_01075
ABC transporter permease
Accession: ANG95200
Location: 232327-233160
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: ANG95200
Location: 232327-233160
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
A8A54_01080
ABC transporter permease
Accession: ANG95201
Location: 233165-234118
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 178
Sequence coverage: 90 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: ANG95201
Location: 233165-234118
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 178
Sequence coverage: 90 %
E-value: 1e-49
NCBI BlastP on this gene
A8A54_01085
ABC transporter substrate-binding protein
Accession: ANG95202
Location: 234194-235453
NCBI BlastP on this gene
Accession: ANG95202
Location: 234194-235453
NCBI BlastP on this gene
A8A54_01090
A8A54_01095
GntR family transcriptional regulator
Accession: ANG95204
Location: 236655-237371
NCBI BlastP on this gene
Accession: ANG95204
Location: 236655-237371
NCBI BlastP on this gene
A8A54_01100
A8A54_01105
A8A54_01110
dihydrodipicolinate synthase family protein
Accession: ANG95207
Location: 240058-240960
NCBI BlastP on this gene
Accession: ANG95207
Location: 240058-240960
NCBI BlastP on this gene
A8A54_01115
GntR family transcriptional regulator
Accession: ANG98310
Location: 241002-241661
NCBI BlastP on this gene
Accession: ANG98310
Location: 241002-241661
NCBI BlastP on this gene
A8A54_01120
ABC transporter substrate-binding protein
Accession: ANG95208
Location: 241739-243073
NCBI BlastP on this gene
Accession: ANG95208
Location: 241739-243073
NCBI BlastP on this gene
A8A54_01125
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP044970 : Ochrobactrum anthropi strain T16R-87 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 371
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
ABC transporter substrate-binding protein
Accession: QFP62524
Location: 1044343-1045341
NCBI BlastP on this gene
Accession: QFP62524
Location: 1044343-1045341
NCBI BlastP on this gene
FT787_05130
FT787_05125
FT787_05120
ABC transporter ATP-binding protein
Accession: QFP62521
Location: 1041621-1042442
NCBI BlastP on this gene
Accession: QFP62521
Location: 1041621-1042442
NCBI BlastP on this gene
FT787_05115
hydA
FT787_05105
HTH-type transcriptional regulator RutR
Accession: QFP62518
Location: 1037905-1038603
NCBI BlastP on this gene
Accession: QFP62518
Location: 1037905-1038603
NCBI BlastP on this gene
rutR
NUDIX domain-containing protein
Accession: QFP62517
Location: 1037510-1037908
NCBI BlastP on this gene
Accession: QFP62517
Location: 1037510-1037908
NCBI BlastP on this gene
FT787_05095
sn-glycerol-3-phosphate ABC transporter ATP-binding protein UgpC
Accession: QFP62516
Location: 1036440-1037513
NCBI BlastP on this gene
Accession: QFP62516
Location: 1036440-1037513
NCBI BlastP on this gene
ugpC
carbohydrate ABC transporter permease
Accession: QFP62515
Location: 1035488-1036321
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: QFP62515
Location: 1035488-1036321
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
FT787_05085
sugar ABC transporter permease
Accession: QFP62514
Location: 1034524-1035483
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 177
Sequence coverage: 90 %
E-value: 2e-49
NCBI BlastP on this gene
Accession: QFP62514
Location: 1034524-1035483
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 177
Sequence coverage: 90 %
E-value: 2e-49
NCBI BlastP on this gene
FT787_05080
sugar ABC transporter substrate-binding protein
Accession: QFP62513
Location: 1033189-1034448
NCBI BlastP on this gene
Accession: QFP62513
Location: 1033189-1034448
NCBI BlastP on this gene
FT787_05075
FT787_05070
FadR family transcriptional regulator
Accession: QFP62511
Location: 1031271-1031987
NCBI BlastP on this gene
Accession: QFP62511
Location: 1031271-1031987
NCBI BlastP on this gene
FT787_05065
FT787_05060
FT787_05055
FT787_05050
dihydrodipicolinate synthase family protein
Accession: QFP62509
Location: 1027809-1028711
NCBI BlastP on this gene
Accession: QFP62509
Location: 1027809-1028711
NCBI BlastP on this gene
FT787_05045
GntR family transcriptional regulator
Accession: QFP62508
Location: 1027108-1027770
NCBI BlastP on this gene
Accession: QFP62508
Location: 1027108-1027770
NCBI BlastP on this gene
FT787_05040
sugar ABC transporter substrate-binding protein
Accession: QFP62507
Location: 1025697-1027031
NCBI BlastP on this gene
Accession: QFP62507
Location: 1025697-1027031
NCBI BlastP on this gene
FT787_05035
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP021332 : Maritalea myrionectae strain HL2708#5 plasmid pHL2708Y3 Total score: 2.5 Cumulative Blast bit score: 371
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
MXMO3_03650
ADP-ribosyl-[dinitrogen reductase] hydrolase
Accession: AVX06154
Location: 39690-40097
NCBI BlastP on this gene
Accession: AVX06154
Location: 39690-40097
NCBI BlastP on this gene
MXMO3_03651
HTH-type transcriptional regulator RafR
Accession: AVX06155
Location: 40227-41258
NCBI BlastP on this gene
Accession: AVX06155
Location: 40227-41258
NCBI BlastP on this gene
MXMO3_03652
MXMO3_03653
MXMO3_03654
mannose-1-phosphate guanylyltransferase
Accession: AVX06158
Location: 45326-45682
NCBI BlastP on this gene
Accession: AVX06158
Location: 45326-45682
NCBI BlastP on this gene
MXMO3_03655
MXMO3_03656
inner membrane ABC transporter permease protein
Accession: AVX06160
Location: 46724-47563
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 194
Sequence coverage: 94 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: AVX06160
Location: 46724-47563
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 194
Sequence coverage: 94 %
E-value: 2e-56
NCBI BlastP on this gene
MXMO3_03657
Sn-glycerol-3-phosphate transport system permease protein UgpA
Accession: AVX06161
Location: 47565-48497
BlastP hit with WP_039959892.1
Percentage identity: 37 %
BlastP bit score: 177
Sequence coverage: 82 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: AVX06161
Location: 47565-48497
BlastP hit with WP_039959892.1
Percentage identity: 37 %
BlastP bit score: 177
Sequence coverage: 82 %
E-value: 1e-49
NCBI BlastP on this gene
MXMO3_03658
MXMO3_03659
putative HTH-type transcriptional regulator YcbG
Accession: AVX06163
Location: 49914-50660
NCBI BlastP on this gene
Accession: AVX06163
Location: 49914-50660
NCBI BlastP on this gene
MXMO3_03660
mercuric resistance operon regulatory protein
Accession: AVX06164
Location: 50815-51222
NCBI BlastP on this gene
Accession: AVX06164
Location: 50815-51222
NCBI BlastP on this gene
MXMO3_03661
MXMO3_03662
mercuric transport protein periplasmic component
Accession: AVX06166
Location: 51942-52235
NCBI BlastP on this gene
Accession: AVX06166
Location: 51942-52235
NCBI BlastP on this gene
MXMO3_03663
MXMO3_03664
MXMO3_03665
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP015736 : Shinella sp. HZN7 Total score: 2.5 Cumulative Blast bit score: 371
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
shn_16825
spermidine/putrescine ABC transporter ATP-binding protein
Accession: ANH05539
Location: 3499244-3500242
NCBI BlastP on this gene
Accession: ANH05539
Location: 3499244-3500242
NCBI BlastP on this gene
shn_16830
shn_16835
shn_16840
shn_16845
LysR family transcriptional regulator
Accession: ANH05543
Location: 3503131-3504066
NCBI BlastP on this gene
Accession: ANH05543
Location: 3503131-3504066
NCBI BlastP on this gene
shn_16850
shn_16855
mandelate racemase/muconate lactonizing protein
Accession: ANH05545
Location: 3505480-3506586
NCBI BlastP on this gene
Accession: ANH05545
Location: 3505480-3506586
NCBI BlastP on this gene
shn_16860
hypothetical protein
Accession: ANH06816
Location: 3506607-3507440
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 187
Sequence coverage: 97 %
E-value: 8e-54
NCBI BlastP on this gene
Accession: ANH06816
Location: 3506607-3507440
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 187
Sequence coverage: 97 %
E-value: 8e-54
NCBI BlastP on this gene
shn_16865
hypothetical protein
Accession: ANH05546
Location: 3507497-3508393
BlastP hit with WP_039959892.1
Percentage identity: 38 %
BlastP bit score: 184
Sequence coverage: 83 %
E-value: 3e-52
NCBI BlastP on this gene
Accession: ANH05546
Location: 3507497-3508393
BlastP hit with WP_039959892.1
Percentage identity: 38 %
BlastP bit score: 184
Sequence coverage: 83 %
E-value: 3e-52
NCBI BlastP on this gene
shn_16870
shn_16875
shn_16880
shn_16885
shn_16890
shn_16895
shn_16900
sugar ABC transporter ATP-binding protein
Accession: ANH05553
Location: 3515516-3516598
NCBI BlastP on this gene
Accession: ANH05553
Location: 3515516-3516598
NCBI BlastP on this gene
shn_16905
shn_16910
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
LT907981 : Streptomyces sp. 2323.1 genome assembly, chromosome: I. Total score: 2.5 Cumulative Blast bit score: 369
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
SAMN06272775_5116
Cupin domain-containing protein
Accession: SOE14153
Location: 5838589-5839062
NCBI BlastP on this gene
Accession: SOE14153
Location: 5838589-5839062
NCBI BlastP on this gene
SAMN06272775_5115
SAMN06272775_5113
SAMN06272775_5112
Transposase DDE domain-containing protein
Accession: SOE14150
Location: 5837176-5837532
NCBI BlastP on this gene
Accession: SOE14150
Location: 5837176-5837532
NCBI BlastP on this gene
SAMN06272775_5111
Histidine kinase-like ATPase domain-containing protein
Accession: SOE14149
Location: 5836135-5836599
NCBI BlastP on this gene
Accession: SOE14149
Location: 5836135-5836599
NCBI BlastP on this gene
SAMN06272775_5110
Helix-turn-helix domain-containing protein
Accession: SOE14148
Location: 5835150-5835980
NCBI BlastP on this gene
Accession: SOE14148
Location: 5835150-5835980
NCBI BlastP on this gene
SAMN06272775_5109
SAMN06272775_5108
SAMN06272775_5107
transcriptional regulator, LacI family
Accession: SOE14145
Location: 5832559-5833617
NCBI BlastP on this gene
Accession: SOE14145
Location: 5832559-5833617
NCBI BlastP on this gene
SAMN06272775_5106
SAMN06272775_5105
arabinogalactan oligomer / maltooligosaccharide transport system permease protein
Accession: SOE14143
Location: 5830084-5830971
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 3e-54
NCBI BlastP on this gene
Accession: SOE14143
Location: 5830084-5830971
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 3e-54
NCBI BlastP on this gene
SAMN06272775_5104
arabinogalactan oligomer / maltooligosaccharide transport system permease protein
Accession: SOE14142
Location: 5829146-5830081
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 180
Sequence coverage: 102 %
E-value: 2e-50
NCBI BlastP on this gene
Accession: SOE14142
Location: 5829146-5830081
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 180
Sequence coverage: 102 %
E-value: 2e-50
NCBI BlastP on this gene
SAMN06272775_5103
carbohydrate ABC transporter substrate-binding protein, CUT1 family
Accession: SOE14140
Location: 5827872-5829149
NCBI BlastP on this gene
Accession: SOE14140
Location: 5827872-5829149
NCBI BlastP on this gene
SAMN06272775_5102
SAMN06272775_5101
transcriptional regulator, LacI family
Accession: SOE14138
Location: 5824905-5825954
NCBI BlastP on this gene
Accession: SOE14138
Location: 5824905-5825954
NCBI BlastP on this gene
SAMN06272775_5100
SAMN06272775_5099
SAMN06272775_5098
SAMN06272775_5097
glutamate-ammonia-ligase adenylyltransferase
Accession: SOE14134
Location: 5819176-5822202
NCBI BlastP on this gene
Accession: SOE14134
Location: 5819176-5822202
NCBI BlastP on this gene
SAMN06272775_5096
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP002131 : Thermosediminibacter oceani DSM 16646 Total score: 2.5 Cumulative Blast bit score: 369
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Toce_1757
conserved hypothetical protein
Accession: ADL08490
Location: 1755530-1755979
NCBI BlastP on this gene
Accession: ADL08490
Location: 1755530-1755979
NCBI BlastP on this gene
Toce_1756
major facilitator superfamily MFS 1
Accession: ADL08489
Location: 1754147-1755400
NCBI BlastP on this gene
Accession: ADL08489
Location: 1754147-1755400
NCBI BlastP on this gene
Toce_1755
conserved hypothetical protein
Accession: ADL08488
Location: 1753494-1754036
NCBI BlastP on this gene
Accession: ADL08488
Location: 1753494-1754036
NCBI BlastP on this gene
Toce_1754
4Fe-4S ferredoxin iron-sulfur binding domain protein
Accession: ADL08487
Location: 1752621-1753376
NCBI BlastP on this gene
Accession: ADL08487
Location: 1752621-1753376
NCBI BlastP on this gene
Toce_1753
conserved hypothetical protein
Accession: ADL08486
Location: 1752164-1752334
NCBI BlastP on this gene
Accession: ADL08486
Location: 1752164-1752334
NCBI BlastP on this gene
Toce_1752
death-on-curing family protein
Accession: ADL08485
Location: 1751781-1752164
NCBI BlastP on this gene
Accession: ADL08485
Location: 1751781-1752164
NCBI BlastP on this gene
Toce_1751
Toce_1750
Peptidoglycan-binding domain 1 protein
Accession: ADL08483
Location: 1749229-1750689
NCBI BlastP on this gene
Accession: ADL08483
Location: 1749229-1750689
NCBI BlastP on this gene
Toce_1749
Toce_1748
RNA polymerase sigma factor, sigma-70 family
Accession: ADL08481
Location: 1748032-1748457
NCBI BlastP on this gene
Accession: ADL08481
Location: 1748032-1748457
NCBI BlastP on this gene
Toce_1747
carbohydrate ABC transporter membrane protein 2, CUT1 family
Accession: ADL08480
Location: 1747168-1748001
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 189
Sequence coverage: 100 %
E-value: 1e-54
NCBI BlastP on this gene
Accession: ADL08480
Location: 1747168-1748001
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 189
Sequence coverage: 100 %
E-value: 1e-54
NCBI BlastP on this gene
Toce_1746
carbohydrate ABC transporter membrane protein 1, CUT1 family
Accession: ADL08479
Location: 1746219-1747163
BlastP hit with WP_039959892.1
Percentage identity: 35 %
BlastP bit score: 180
Sequence coverage: 94 %
E-value: 2e-50
NCBI BlastP on this gene
Accession: ADL08479
Location: 1746219-1747163
BlastP hit with WP_039959892.1
Percentage identity: 35 %
BlastP bit score: 180
Sequence coverage: 94 %
E-value: 2e-50
NCBI BlastP on this gene
Toce_1745
carbohydrate ABC transporter substrate-binding protein, CUT1 family
Accession: ADL08478
Location: 1744924-1746219
NCBI BlastP on this gene
Accession: ADL08478
Location: 1744924-1746219
NCBI BlastP on this gene
Toce_1744
two component transcriptional regulator, AraC family
Accession: ADL08477
Location: 1741932-1743059
NCBI BlastP on this gene
Accession: ADL08477
Location: 1741932-1743059
NCBI BlastP on this gene
Toce_1742
alpha amylase catalytic region
Accession: ADL08476
Location: 1740161-1741831
NCBI BlastP on this gene
Accession: ADL08476
Location: 1740161-1741831
NCBI BlastP on this gene
Toce_1741
ABC transporter related protein
Accession: ADL08475
Location: 1739025-1740023
NCBI BlastP on this gene
Accession: ADL08475
Location: 1739025-1740023
NCBI BlastP on this gene
Toce_1740
protein of unknown function DUF990
Accession: ADL08474
Location: 1738244-1739044
NCBI BlastP on this gene
Accession: ADL08474
Location: 1738244-1739044
NCBI BlastP on this gene
Toce_1739
protein of unknown function DUF990
Accession: ADL08473
Location: 1737469-1738260
NCBI BlastP on this gene
Accession: ADL08473
Location: 1737469-1738260
NCBI BlastP on this gene
Toce_1738
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP020773 : Staphylococcus lutrae strain ATCC 700373 chromosome Total score: 2.0 Cumulative Blast bit score: 958
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
B5P37_01840
B5P37_01845
B5P37_01850
two-component sensor histidine kinase
Accession: ARJ50151
Location: 359771-360814
NCBI BlastP on this gene
Accession: ARJ50151
Location: 359771-360814
NCBI BlastP on this gene
B5P37_01855
B5P37_01860
B5P37_01865
type I methionyl aminopeptidase
Accession: ARJ50154
Location: 362013-362777
NCBI BlastP on this gene
Accession: ARJ50154
Location: 362013-362777
NCBI BlastP on this gene
B5P37_01870
B5P37_01875
sialidase
Accession: ARJ50156
Location: 364049-365551
BlastP hit with WP_004843631.1
Percentage identity: 48 %
BlastP bit score: 442
Sequence coverage: 65 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: ARJ50156
Location: 364049-365551
BlastP hit with WP_004843631.1
Percentage identity: 48 %
BlastP bit score: 442
Sequence coverage: 65 %
E-value: 1e-143
NCBI BlastP on this gene
B5P37_01880
B5P37_01885
oxidoreductase
Accession: ARJ50158
Location: 367037-368158
BlastP hit with WP_004843633.1
Percentage identity: 67 %
BlastP bit score: 516
Sequence coverage: 98 %
E-value: 1e-179
NCBI BlastP on this gene
Accession: ARJ50158
Location: 367037-368158
BlastP hit with WP_004843633.1
Percentage identity: 67 %
BlastP bit score: 516
Sequence coverage: 98 %
E-value: 1e-179
NCBI BlastP on this gene
B5P37_01890
B5P37_01895
UDP-N-acetylmuramate--alanine ligase
Accession: ARJ50160
Location: 369563-370873
NCBI BlastP on this gene
Accession: ARJ50160
Location: 369563-370873
NCBI BlastP on this gene
B5P37_01900
B5P37_01905
DNA polymerase III subunit epsilon
Accession: ARJ50162
Location: 371835-372392
NCBI BlastP on this gene
Accession: ARJ50162
Location: 371835-372392
NCBI BlastP on this gene
B5P37_01910
B5P37_01915
B5P37_01920
B5P37_01925
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP015626 : Staphylococcus pseudintermedius strain 063228 Total score: 2.0 Cumulative Blast bit score: 955
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS system, galactosamine-specific IIC component
Accession: ANS90142
Location: 1864086-1864982
NCBI BlastP on this gene
Accession: ANS90142
Location: 1864086-1864982
NCBI BlastP on this gene
A6M57_9130
PTS system, mannose-specific IID component
Accession: ANS90143
Location: 1864969-1865862
NCBI BlastP on this gene
Accession: ANS90143
Location: 1864969-1865862
NCBI BlastP on this gene
A6M57_9135
2H phosphoesterase superfamily protein (yjcG)
Accession: ANS90144
Location: 1866435-1866941
NCBI BlastP on this gene
Accession: ANS90144
Location: 1866435-1866941
NCBI BlastP on this gene
A6M57_9140
Putative acetyl esterase YjcH
Accession: ANS90145
Location: 1866995-1867729
NCBI BlastP on this gene
Accession: ANS90145
Location: 1866995-1867729
NCBI BlastP on this gene
A6M57_9145
Sodium/alanine symporter family protein
Accession: ANS90146
Location: 1867832-1869403
NCBI BlastP on this gene
Accession: ANS90146
Location: 1867832-1869403
NCBI BlastP on this gene
A6M57_9150
A6M57_9155
Sialidase
Accession: ANS90148
Location: 1871295-1872746
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 450
Sequence coverage: 68 %
E-value: 4e-147
NCBI BlastP on this gene
Accession: ANS90148
Location: 1871295-1872746
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 450
Sequence coverage: 68 %
E-value: 4e-147
NCBI BlastP on this gene
A6M57_9160
Melibiose carrier protein, Na+/melibiose symporter
Accession: ANS90149
Location: 1872797-1874245
NCBI BlastP on this gene
Accession: ANS90149
Location: 1872797-1874245
NCBI BlastP on this gene
A6M57_9165
Melibiose carrier protein, Na+/melibiose symporter
Accession: ANS90150
Location: 1874302-1874415
NCBI BlastP on this gene
Accession: ANS90150
Location: 1874302-1874415
NCBI BlastP on this gene
A6M57_9170
hypothetical protein
Accession: ANS90151
Location: 1874447-1875565
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: ANS90151
Location: 1874447-1875565
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
A6M57_9175
Enoyl-[acyl-carrier-protein] reductase [NADH]
Accession: ANS90152
Location: 1876070-1876837
NCBI BlastP on this gene
Accession: ANS90152
Location: 1876070-1876837
NCBI BlastP on this gene
A6M57_9180
A6M57_9185
A6M57_9190
ribosomal large subunit pseudouridine synthase D-like protein
Accession: ANS90155
Location: 1880247-1881101
NCBI BlastP on this gene
Accession: ANS90155
Location: 1880247-1881101
NCBI BlastP on this gene
A6M57_9195
A6M57_9200
A6M57_9205
A6M57_9210
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP035741 : Staphylococcus pseudintermedius strain 53_60 chromosome. Total score: 2.0 Cumulative Blast bit score: 953
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS sugar transporter subunit IIC
Accession: QBG72523
Location: 982811-983707
NCBI BlastP on this gene
Accession: QBG72523
Location: 982811-983707
NCBI BlastP on this gene
EW136_04565
PTS system mannose/fructose/sorbose family transporter subunit IID
Accession: QBG72522
Location: 981931-982824
NCBI BlastP on this gene
Accession: QBG72522
Location: 981931-982824
NCBI BlastP on this gene
EW136_04560
EW136_04555
EW136_04550
alanine:cation symporter family protein
Accession: QBG72519
Location: 978391-979962
NCBI BlastP on this gene
Accession: QBG72519
Location: 978391-979962
NCBI BlastP on this gene
EW136_04545
EW136_04540
sialidase
Accession: QBG73939
Location: 975048-976499
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: QBG73939
Location: 975048-976499
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
EW136_04535
EW136_04530
Gfo/Idh/MocA family oxidoreductase
Accession: QBG72516
Location: 972390-973508
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: QBG72516
Location: 972390-973508
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
EW136_04525
enoyl-[acyl-carrier-protein] reductase FabI
Accession: QBG72515
Location: 971116-971886
NCBI BlastP on this gene
Accession: QBG72515
Location: 971116-971886
NCBI BlastP on this gene
fabI
EW136_04515
mgtE
RluA family pseudouridine synthase
Accession: QBG72512
Location: 966855-967709
NCBI BlastP on this gene
Accession: QBG72512
Location: 966855-967709
NCBI BlastP on this gene
EW136_04505
EW136_04500
GTP pyrophosphokinase family protein
Accession: QBG72510
Location: 965399-966037
NCBI BlastP on this gene
Accession: QBG72510
Location: 965399-966037
NCBI BlastP on this gene
EW136_04495
EW136_04490
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP035740 : Staphylococcus pseudintermedius strain 53_88 chromosome. Total score: 2.0 Cumulative Blast bit score: 953
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS sugar transporter subunit IIC
Accession: QBG75759
Location: 2086396-2087292
NCBI BlastP on this gene
Accession: QBG75759
Location: 2086396-2087292
NCBI BlastP on this gene
EW134_09820
PTS system mannose/fructose/sorbose family transporter subunit IID
Accession: QBG75760
Location: 2087279-2088172
NCBI BlastP on this gene
Accession: QBG75760
Location: 2087279-2088172
NCBI BlastP on this gene
EW134_09825
EW134_09830
EW134_09835
alanine:cation symporter family protein
Accession: QBG75763
Location: 2090141-2091712
NCBI BlastP on this gene
Accession: QBG75763
Location: 2090141-2091712
NCBI BlastP on this gene
EW134_09840
EW134_09845
sialidase
Accession: QBG76271
Location: 2093604-2095055
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: QBG76271
Location: 2093604-2095055
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
EW134_09850
EW134_09855
Gfo/Idh/MocA family oxidoreductase
Accession: QBG75766
Location: 2096595-2097713
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: QBG75766
Location: 2096595-2097713
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
EW134_09860
enoyl-[acyl-carrier-protein] reductase FabI
Accession: QBG75767
Location: 2098217-2098987
NCBI BlastP on this gene
Accession: QBG75767
Location: 2098217-2098987
NCBI BlastP on this gene
fabI
EW134_09870
mgtE
RluA family pseudouridine synthase
Accession: QBG75770
Location: 2102394-2103248
NCBI BlastP on this gene
Accession: QBG75770
Location: 2102394-2103248
NCBI BlastP on this gene
EW134_09880
EW134_09885
GTP pyrophosphokinase family protein
Accession: QBG75772
Location: 2104066-2104704
NCBI BlastP on this gene
Accession: QBG75772
Location: 2104066-2104704
NCBI BlastP on this gene
EW134_09890
EW134_09895
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP032682 : Staphylococcus pseudintermedius strain G3C4 chromosome Total score: 2.0 Cumulative Blast bit score: 953
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS sugar transporter subunit IIC
Accession: AYG57105
Location: 2514904-2515800
NCBI BlastP on this gene
Accession: AYG57105
Location: 2514904-2515800
NCBI BlastP on this gene
D8L98_12040
PTS system mannose/fructose/sorbose family transporter subunit IID
Accession: AYG57106
Location: 2515787-2516680
NCBI BlastP on this gene
Accession: AYG57106
Location: 2515787-2516680
NCBI BlastP on this gene
D8L98_12045
D8L98_12050
D8L98_12055
alanine:cation symporter family protein
Accession: AYG57109
Location: 2518649-2520220
NCBI BlastP on this gene
Accession: AYG57109
Location: 2518649-2520220
NCBI BlastP on this gene
D8L98_12060
D8L98_12065
sialidase
Accession: AYG57386
Location: 2522110-2523561
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
Accession: AYG57386
Location: 2522110-2523561
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
D8L98_12070
D8L98_12075
gfo/Idh/MocA family oxidoreductase
Accession: AYG57112
Location: 2525101-2526219
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: AYG57112
Location: 2525101-2526219
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
D8L98_12080
enoyl-[acyl-carrier-protein] reductase FabI
Accession: AYG57113
Location: 2526724-2527494
NCBI BlastP on this gene
Accession: AYG57113
Location: 2526724-2527494
NCBI BlastP on this gene
fabI
D8L98_12090
mgtE
RluA family pseudouridine synthase
Accession: AYG57116
Location: 2530901-2531755
NCBI BlastP on this gene
Accession: AYG57116
Location: 2530901-2531755
NCBI BlastP on this gene
D8L98_12100
D8L98_12105
GTP pyrophosphokinase family protein
Accession: AYG57118
Location: 2532573-2533211
NCBI BlastP on this gene
Accession: AYG57118
Location: 2532573-2533211
NCBI BlastP on this gene
D8L98_12110
D8L98_12115
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP002478 : Staphylococcus pseudintermedius ED99 Total score: 2.0 Cumulative Blast bit score: 953
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
SPSE_1823
SPSE_1824
conserved hypothetical protein
Accession: ADX77075
Location: 1879879-1880385
NCBI BlastP on this gene
Accession: ADX77075
Location: 1879879-1880385
NCBI BlastP on this gene
SPSE_1825
SPSE_1826
sodium:alanine symporter family protein
Accession: ADX77077
Location: 1881276-1882847
NCBI BlastP on this gene
Accession: ADX77077
Location: 1881276-1882847
NCBI BlastP on this gene
SPSE_1827
conserved membrane protein, putative
Accession: ADX77078
Location: 1883066-1884163
NCBI BlastP on this gene
Accession: ADX77078
Location: 1883066-1884163
NCBI BlastP on this gene
SPSE_1828
putative sialidase, truncated
Accession: ADX77079
Location: 1884739-1886190
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: ADX77079
Location: 1884739-1886190
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
SPSE_1829
sugar (Glycoside-Pentoside-Hexuronide) transporter
Accession: ADX77080
Location: 1886241-1887689
NCBI BlastP on this gene
Accession: ADX77080
Location: 1886241-1887689
NCBI BlastP on this gene
gph
putative oxidoreductase
Accession: ADX77081
Location: 1887730-1888848
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: ADX77081
Location: 1887730-1888848
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
SPSE_1831
enoyl-(acyl carrier protein) reductase
Accession: ADX77082
Location: 1889353-1890120
NCBI BlastP on this gene
Accession: ADX77082
Location: 1889353-1890120
NCBI BlastP on this gene
fabI
SPSE_1833
mgtE
ribosomal large subunit pseudouridine synthase D
Accession: ADX77085
Location: 1893530-1894384
NCBI BlastP on this gene
Accession: ADX77085
Location: 1893530-1894384
NCBI BlastP on this gene
SPSE_1835
inorganic polyphosphate/ATP-NAD kinase
Accession: ADX77086
Location: 1894381-1895190
NCBI BlastP on this gene
Accession: ADX77086
Location: 1894381-1895190
NCBI BlastP on this gene
ppnK
relA2
conserved hypothetical protein
Accession: ADX77088
Location: 1895857-1896204
NCBI BlastP on this gene
Accession: ADX77088
Location: 1895857-1896204
NCBI BlastP on this gene
SPSE_1838
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP016073 : Staphylococcus pseudintermedius strain 081661 Total score: 2.0 Cumulative Blast bit score: 952
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS fructose transporter subunit IIC
Accession: ANQ88760
Location: 1958806-1959702
NCBI BlastP on this gene
Accession: ANQ88760
Location: 1958806-1959702
NCBI BlastP on this gene
A9I65_09150
PTS fructose transporter subunit IID
Accession: ANQ89508
Location: 1959815-1960582
NCBI BlastP on this gene
Accession: ANQ89508
Location: 1959815-1960582
NCBI BlastP on this gene
A9I65_09155
A9I65_09160
A9I65_09165
A9I65_09170
A9I65_09175
sialidase
Accession: ANQ89509
Location: 1966012-1967463
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
Accession: ANQ89509
Location: 1966012-1967463
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
A9I65_09180
A9I65_09185
oxidoreductase
Accession: ANQ89510
Location: 1969003-1970109
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
Accession: ANQ89510
Location: 1969003-1970109
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
A9I65_09190
enoyl-[acyl-carrier-protein] reductase
Accession: ANQ88766
Location: 1970626-1971396
NCBI BlastP on this gene
Accession: ANQ88766
Location: 1970626-1971396
NCBI BlastP on this gene
A9I65_09195
A9I65_09200
A9I65_09205
A9I65_09210
A9I65_09215
A9I65_09220
A9I65_09225
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP009120 : Staphylococcus pseudintermedius strain 5912 Total score: 2.0 Cumulative Blast bit score: 952
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS fructose transporter subunit IIC
Accession: ASQ51073
Location: 1882925-1883821
NCBI BlastP on this gene
Accession: ASQ51073
Location: 1882925-1883821
NCBI BlastP on this gene
SPS5912_08900
PTS fructose transporter subunit IID
Accession: ASQ51074
Location: 1883808-1884701
NCBI BlastP on this gene
Accession: ASQ51074
Location: 1883808-1884701
NCBI BlastP on this gene
SPS5912_08905
SPS5912_08910
SPS5912_08915
SPS5912_08920
SPS5912_08925
sialidase
Accession: ASQ51079
Location: 1890133-1891584
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: ASQ51079
Location: 1890133-1891584
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
SPS5912_08930
SPS5912_08935
oxidoreductase
Accession: ASQ51081
Location: 1893124-1894230
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
Accession: ASQ51081
Location: 1893124-1894230
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
SPS5912_08940
SPS5912_08945
SPS5912_08950
SPS5912_08955
SPS5912_08960
inorganic polyphosphate kinase
Accession: ASQ51086
Location: 1899774-1900583
NCBI BlastP on this gene
Accession: ASQ51086
Location: 1899774-1900583
NCBI BlastP on this gene
ppnK
SPS5912_08970
SPS5912_08975
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
AP019372 : Staphylococcus pseudintermedius SP79 DNA Total score: 2.0 Cumulative Blast bit score: 952
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS fructose transporter subunit IIC
Accession: BBH74515
Location: 1852843-1853739
NCBI BlastP on this gene
Accession: BBH74515
Location: 1852843-1853739
NCBI BlastP on this gene
GSP_17090
PTS fructose transporter subunit IID
Accession: BBH74516
Location: 1853726-1854619
NCBI BlastP on this gene
Accession: BBH74516
Location: 1853726-1854619
NCBI BlastP on this gene
GSP_17100
GSP_17110
GSP_17120
GSP_17130
GSP_17140
hypothetical protein
Accession: BBH74521
Location: 1860053-1861537
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: BBH74521
Location: 1860053-1861537
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
GSP_17150
GSP_17160
oxidoreductase
Accession: BBH74523
Location: 1863044-1864162
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
Accession: BBH74523
Location: 1863044-1864162
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
GSP_17170
enoyl-[acyl-carrier-protein] reductase [NADPH] FabI
Accession: BBH74524
Location: 1864667-1865437
NCBI BlastP on this gene
Accession: BBH74524
Location: 1864667-1865437
NCBI BlastP on this gene
fabI
GSP_17190
GSP_17200
GSP_17210
nadK
GSP_17230
GSP_17240
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP002439 : Staphylococcus pseudintermedius HKU10-03 Total score: 2.0 Cumulative Blast bit score: 949
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
YjcG, 2H phosphoesterase superfamily protein
Accession: ADV05227
Location: 725685-726191
NCBI BlastP on this gene
Accession: ADV05227
Location: 725685-726191
NCBI BlastP on this gene
SPSINT_0698
SPSINT_0697
sodium/alanine symporter family protein
Accession: ADV05225
Location: 723223-724794
NCBI BlastP on this gene
Accession: ADV05225
Location: 723223-724794
NCBI BlastP on this gene
SPSINT_0696
SPSINT_0695
Sialidase
Accession: ADV05223
Location: 719882-721333
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 444
Sequence coverage: 68 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: ADV05223
Location: 719882-721333
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 444
Sequence coverage: 68 %
E-value: 2e-144
NCBI BlastP on this gene
SPSINT_0694
Melibiose carrier protein, Na+/melibiose symporter
Accession: ADV05222
Location: 718383-719831
NCBI BlastP on this gene
Accession: ADV05222
Location: 718383-719831
NCBI BlastP on this gene
SPSINT_0693
Putative oxidoreductase
Accession: ADV05221
Location: 717224-718342
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: ADV05221
Location: 717224-718342
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
SPSINT_0692
Enoyl-[acyl-carrier-protein] reductase [NADH]
Accession: ADV05220
Location: 715954-716721
NCBI BlastP on this gene
Accession: ADV05220
Location: 715954-716721
NCBI BlastP on this gene
SPSINT_0691
SPSINT_0690
SPSINT_0689
SPSINT_0688
SPSINT_0687
SPSINT_0686
SPSINT_0685
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP046351 : Staphylococcus fleurettii strain FDAARGOS_682 chromosome Total score: 2.0 Cumulative Blast bit score: 937
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
FOB90_03495
YihY family inner membrane protein
Accession: QGS45805
Location: 694961-696220
NCBI BlastP on this gene
Accession: QGS45805
Location: 694961-696220
NCBI BlastP on this gene
FOB90_03500
FOB90_03505
FOB90_03510
two-component sensor histidine kinase
Accession: QGS45808
Location: 697499-698542
NCBI BlastP on this gene
Accession: QGS45808
Location: 697499-698542
NCBI BlastP on this gene
FOB90_03515
FOB90_03520
FOB90_03525
type I methionyl aminopeptidase
Accession: QGS45811
Location: 699740-700501
NCBI BlastP on this gene
Accession: QGS45811
Location: 699740-700501
NCBI BlastP on this gene
map
aromatic acid exporter family protein
Accession: QGS45812
Location: 700599-701597
NCBI BlastP on this gene
Accession: QGS45812
Location: 700599-701597
NCBI BlastP on this gene
FOB90_03535
sialidase
Accession: FOB90_03540
Location: 701714-703222
BlastP hit with WP_004843631.1
Percentage identity: 49 %
BlastP bit score: 439
Sequence coverage: 66 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: FOB90_03540
Location: 701714-703222
BlastP hit with WP_004843631.1
Percentage identity: 49 %
BlastP bit score: 439
Sequence coverage: 66 %
E-value: 3e-142
NCBI BlastP on this gene
FOB90_03540
FOB90_03545
gfo/Idh/MocA family oxidoreductase
Accession: QGS45814
Location: 704707-705831
BlastP hit with WP_004843633.1
Percentage identity: 64 %
BlastP bit score: 498
Sequence coverage: 98 %
E-value: 2e-172
NCBI BlastP on this gene
Accession: QGS45814
Location: 704707-705831
BlastP hit with WP_004843633.1
Percentage identity: 64 %
BlastP bit score: 498
Sequence coverage: 98 %
E-value: 2e-172
NCBI BlastP on this gene
FOB90_03550
FOB90_03555
DUF1727 domain-containing protein
Accession: QGS45816
Location: 706817-708136
NCBI BlastP on this gene
Accession: QGS45816
Location: 706817-708136
NCBI BlastP on this gene
FOB90_03560
FOB90_03565
DNA polymerase III subunit epsilon
Accession: QGS45818
Location: 709065-709625
NCBI BlastP on this gene
Accession: QGS45818
Location: 709065-709625
NCBI BlastP on this gene
FOB90_03570
dinB
FOB90_03580
DUF3267 domain-containing protein
Accession: QGS45821
Location: 711505-712059
NCBI BlastP on this gene
Accession: QGS45821
Location: 711505-712059
NCBI BlastP on this gene
FOB90_03585
23S rRNA (uracil(1939)-C(5))-methyltransferase RlmD
Accession: QGS45822
Location: 712135-713496
NCBI BlastP on this gene
Accession: QGS45822
Location: 712135-713496
NCBI BlastP on this gene
rlmD
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP040518 : Blautia sp. SC05B48 chromosome Total score: 2.0 Cumulative Blast bit score: 871
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
xylB
LacI family transcriptional regulator
Accession: QCU01232
Location: 516200-517225
NCBI BlastP on this gene
Accession: QCU01232
Location: 516200-517225
NCBI BlastP on this gene
EYS05_02290
class II fructose-bisphosphate aldolase
Accession: QCU01233
Location: 517260-518111
NCBI BlastP on this gene
Accession: QCU01233
Location: 517260-518111
NCBI BlastP on this gene
EYS05_02295
EYS05_02300
EYS05_02305
EYS05_02310
glf
ROK family protein
Accession: QCU01238
Location: 524287-525174
BlastP hit with WP_004843625.1
Percentage identity: 72 %
BlastP bit score: 459
Sequence coverage: 98 %
E-value: 2e-159
NCBI BlastP on this gene
Accession: QCU01238
Location: 524287-525174
BlastP hit with WP_004843625.1
Percentage identity: 72 %
BlastP bit score: 459
Sequence coverage: 98 %
E-value: 2e-159
NCBI BlastP on this gene
EYS05_02320
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QCU01239
Location: 525204-525896
BlastP hit with WP_024854132.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 4e-143
NCBI BlastP on this gene
Accession: QCU01239
Location: 525204-525896
BlastP hit with WP_024854132.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 4e-143
NCBI BlastP on this gene
EYS05_02325
EYS05_02330
EYS05_02335
EYS05_02340
plasmid pRiA4b ORF-3 family protein
Accession: QCU03867
Location: 528809-529231
NCBI BlastP on this gene
Accession: QCU03867
Location: 528809-529231
NCBI BlastP on this gene
EYS05_02345
EYS05_02350
EYS05_02355
EYS05_02360
glf
EYS05_02370
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP022713 : Blautia coccoides strain YL58 genome. Total score: 2.0 Cumulative Blast bit score: 738
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
ADH70_018255
ADH70_018260
ADH70_018265
ADH70_018270
ADH70_018275
ADH70_018280
ADH70_018285
MurR/RpiR family transcriptional regulator
Accession: ASU30572
Location: 3863593-3864408
NCBI BlastP on this gene
Accession: ASU30572
Location: 3863593-3864408
NCBI BlastP on this gene
ADH70_018290
ROK family protein
Accession: ASU30573
Location: 3864478-3865365
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
Accession: ASU30573
Location: 3864478-3865365
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
ADH70_018295
DUF386 domain-containing protein
Accession: ASU30574
Location: 3865539-3865991
NCBI BlastP on this gene
Accession: ASU30574
Location: 3865539-3865991
NCBI BlastP on this gene
ADH70_018300
ADH70_018305
ADH70_018310
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ASU30577
Location: 3868603-3869301
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
Accession: ASU30577
Location: 3868603-3869301
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
ADH70_018315
N-acetylglucosamine-6-phosphate deacetylase
Accession: ASU30578
Location: 3869664-3870767
NCBI BlastP on this gene
Accession: ASU30578
Location: 3869664-3870767
NCBI BlastP on this gene
nagA
GIY-YIG nuclease family protein
Accession: ASU30579
Location: 3870891-3871127
NCBI BlastP on this gene
Accession: ASU30579
Location: 3870891-3871127
NCBI BlastP on this gene
ADH70_018325
ADH70_018330
ADH70_018335
stage II sporulation protein D
Accession: ASU30582
Location: 3874896-3875765
NCBI BlastP on this gene
Accession: ASU30582
Location: 3874896-3875765
NCBI BlastP on this gene
ADH70_018340
16S rRNA (cytosine(1402)-N(4))-methyltransferase
Accession: ASU30583
Location: 3875882-3876445
NCBI BlastP on this gene
Accession: ASU30583
Location: 3875882-3876445
NCBI BlastP on this gene
mraW
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP015405 : Blautia sp. YL58 chromosome Total score: 2.0 Cumulative Blast bit score: 738
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
A4V09_19660
A4V09_19665
A4V09_19670
A4V09_19675
A4V09_19680
A4V09_19685
A4V09_19690
MurR/RpiR family transcriptional regulator
Accession: ANU77767
Location: 4217281-4218096
NCBI BlastP on this gene
Accession: ANU77767
Location: 4217281-4218096
NCBI BlastP on this gene
A4V09_19695
hypothetical protein
Accession: ANU77768
Location: 4218166-4219053
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
Accession: ANU77768
Location: 4218166-4219053
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
A4V09_19700
YhcH/YjgK/YiaL family protein
Accession: ANU77769
Location: 4219227-4219679
NCBI BlastP on this gene
Accession: ANU77769
Location: 4219227-4219679
NCBI BlastP on this gene
A4V09_19705
A4V09_19710
A4V09_19715
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ANU77772
Location: 4222291-4222989
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
Accession: ANU77772
Location: 4222291-4222989
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
A4V09_19720
N-acetylglucosamine-6-phosphate deacetylase
Accession: ANU77773
Location: 4223352-4224455
NCBI BlastP on this gene
Accession: ANU77773
Location: 4223352-4224455
NCBI BlastP on this gene
A4V09_19725
A4V09_19730
A4V09_19735
A4V09_19740
stage II sporulation protein D
Accession: ANU77777
Location: 4228584-4229453
NCBI BlastP on this gene
Accession: ANU77777
Location: 4228584-4229453
NCBI BlastP on this gene
A4V09_19745
16S rRNA (cytosine(1402)-N(4))-methyltransferase
Accession: ANU77778
Location: 4229570-4230133
NCBI BlastP on this gene
Accession: ANU77778
Location: 4229570-4230133
NCBI BlastP on this gene
A4V09_19750
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP048626 : Blautia producta ATCC 27340 = DSM 2950 strain JCM 1471 chromosome Total score: 2.0 Cumulative Blast bit score: 726
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
DUF4430 domain-containing protein
Accession: QIB56116
Location: 3240612-3248192
NCBI BlastP on this gene
Accession: QIB56116
Location: 3240612-3248192
NCBI BlastP on this gene
GXM18_15310
GXM18_15305
GXM18_15300
GXM18_15295
MurR/RpiR family transcriptional regulator
Accession: QIB56112
Location: 3236888-3237703
NCBI BlastP on this gene
Accession: QIB56112
Location: 3236888-3237703
NCBI BlastP on this gene
GXM18_15290
ROK family protein
Accession: QIB56111
Location: 3235958-3236845
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 361
Sequence coverage: 98 %
E-value: 4e-121
NCBI BlastP on this gene
Accession: QIB56111
Location: 3235958-3236845
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 361
Sequence coverage: 98 %
E-value: 4e-121
NCBI BlastP on this gene
GXM18_15285
DUF386 domain-containing protein
Accession: QIB56110
Location: 3235310-3235762
NCBI BlastP on this gene
Accession: QIB56110
Location: 3235310-3235762
NCBI BlastP on this gene
GXM18_15280
GXM18_15275
GXM18_15270
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QIB56107
Location: 3231952-3232650
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
Accession: QIB56107
Location: 3231952-3232650
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
GXM18_15265
N-acetylglucosamine-6-phosphate deacetylase
Accession: QIB56106
Location: 3230486-3231589
NCBI BlastP on this gene
Accession: QIB56106
Location: 3230486-3231589
NCBI BlastP on this gene
nagA
GXM18_15255
GXM18_15250
carbohydrate ABC transporter permease
Accession: QIB56103
Location: 3226632-3227453
NCBI BlastP on this gene
Accession: QIB56103
Location: 3226632-3227453
NCBI BlastP on this gene
GXM18_15245
sugar ABC transporter permease
Accession: QIB56102
Location: 3225723-3226610
NCBI BlastP on this gene
Accession: QIB56102
Location: 3225723-3226610
NCBI BlastP on this gene
GXM18_15240
extracellular solute-binding protein
Accession: QIB56101
Location: 3224282-3225619
NCBI BlastP on this gene
Accession: QIB56101
Location: 3224282-3225619
NCBI BlastP on this gene
GXM18_15235
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP035945 : Blautia producta strain PMF1 chromosome Total score: 2.0 Cumulative Blast bit score: 725
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Multidrug export protein MepA
Accession: QBE96074
Location: 1784878-1786215
NCBI BlastP on this gene
Accession: QBE96074
Location: 1784878-1786215
NCBI BlastP on this gene
mepA_10
PMF13cell1_01611
rbr_2
rbr3A
Peroxide-responsive repressor PerR
Accession: QBE96070
Location: 1780887-1781282
NCBI BlastP on this gene
Accession: QBE96070
Location: 1780887-1781282
NCBI BlastP on this gene
perR
putative HTH-type transcriptional regulator YbbH
Accession: QBE96069
Location: 1779592-1780407
NCBI BlastP on this gene
Accession: QBE96069
Location: 1779592-1780407
NCBI BlastP on this gene
ybbH
Beta-glucoside kinase
Accession: QBE96068
Location: 1778662-1779549
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 360
Sequence coverage: 97 %
E-value: 1e-120
NCBI BlastP on this gene
Accession: QBE96068
Location: 1778662-1779549
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 360
Sequence coverage: 97 %
E-value: 1e-120
NCBI BlastP on this gene
bglK
Toxin-antitoxin biofilm protein TabA
Accession: QBE96067
Location: 1778014-1778466
NCBI BlastP on this gene
Accession: QBE96067
Location: 1778014-1778466
NCBI BlastP on this gene
tabA
sglT
nanA
Putative N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QBE96064
Location: 1774656-1775354
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
Accession: QBE96064
Location: 1774656-1775354
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
nanE
N-acetylglucosamine-6-phosphate deacetylase
Accession: QBE96063
Location: 1773191-1774294
NCBI BlastP on this gene
Accession: QBE96063
Location: 1773191-1774294
NCBI BlastP on this gene
nagA_1
ycdF
bglX_1
L-arabinose transport system permease protein AraQ
Accession: QBE96060
Location: 1769337-1770158
NCBI BlastP on this gene
Accession: QBE96060
Location: 1769337-1770158
NCBI BlastP on this gene
araQ_16
L-arabinose transport system permease protein AraP
Accession: QBE96059
Location: 1768428-1769315
NCBI BlastP on this gene
Accession: QBE96059
Location: 1768428-1769315
NCBI BlastP on this gene
araP_8
Putative binding protein MsmE
Accession: QBE96058
Location: 1766987-1768324
NCBI BlastP on this gene
Accession: QBE96058
Location: 1766987-1768324
NCBI BlastP on this gene
msmE_12
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
LN846931 : Carnobacterium maltaromaticum strain LMA28 megaplasmid Total score: 2.0 Cumulative Blast bit score: 693
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
BN424_mp0041
membrane protein of unknown function
Accession: CRI06582
Location: 27866-28687
NCBI BlastP on this gene
Accession: CRI06582
Location: 27866-28687
NCBI BlastP on this gene
BN424_mp0040
exported protein of unknown function
Accession: CRI06581
Location: 27271-27456
NCBI BlastP on this gene
Accession: CRI06581
Location: 27271-27456
NCBI BlastP on this gene
BN424_mp0039
conserved exported protein of unknown function
Accession: CRI06580
Location: 26748-27245
NCBI BlastP on this gene
Accession: CRI06580
Location: 26748-27245
NCBI BlastP on this gene
BN424_mp0038
BN424_mp0037
membrane protein of unknown function
Accession: CRI06578
Location: 25632-26000
NCBI BlastP on this gene
Accession: CRI06578
Location: 25632-26000
NCBI BlastP on this gene
BN424_mp0036
Peptidoglycan binding domain protein (fragment)
Accession: CRI06577
Location: 23912-25327
NCBI BlastP on this gene
Accession: CRI06577
Location: 23912-25327
NCBI BlastP on this gene
BN424_mp0035
conserved protein of unknown function
Accession: CRI06576
Location: 23349-23882
NCBI BlastP on this gene
Accession: CRI06576
Location: 23349-23882
NCBI BlastP on this gene
BN424_mp0034
conserved protein of unknown function
Accession: CRI06575
Location: 23146-23361
NCBI BlastP on this gene
Accession: CRI06575
Location: 23146-23361
NCBI BlastP on this gene
BN424_mp0033
conserved protein of unknown function
Accession: CRI06574
Location: 21942-22424
NCBI BlastP on this gene
Accession: CRI06574
Location: 21942-22424
NCBI BlastP on this gene
BN424_mp0032
BN424_mp0031
conserved protein of unknown function
Accession: CRI06572
Location: 19884-20786
BlastP hit with WP_004843625.1
Percentage identity: 55 %
BlastP bit score: 337
Sequence coverage: 97 %
E-value: 2e-111
NCBI BlastP on this gene
Accession: CRI06572
Location: 19884-20786
BlastP hit with WP_004843625.1
Percentage identity: 55 %
BlastP bit score: 337
Sequence coverage: 97 %
E-value: 2e-111
NCBI BlastP on this gene
BN424_mp0030
nanA
BN424_mp0028
putative N-acetylmannosamine-6-phosphate 2-epimerase
Accession: CRI06569
Location: 18235-18924
BlastP hit with WP_024854132.1
Percentage identity: 73 %
BlastP bit score: 356
Sequence coverage: 100 %
E-value: 3e-121
NCBI BlastP on this gene
Accession: CRI06569
Location: 18235-18924
BlastP hit with WP_024854132.1
Percentage identity: 73 %
BlastP bit score: 356
Sequence coverage: 100 %
E-value: 3e-121
NCBI BlastP on this gene
nanE
BN424_mp0026
membrane protein of unknown function
Accession: CRI06567
Location: 17573-18055
NCBI BlastP on this gene
Accession: CRI06567
Location: 17573-18055
NCBI BlastP on this gene
BN424_mp0025
conserved protein of unknown function
Accession: CRI06566
Location: 16719-17552
NCBI BlastP on this gene
Accession: CRI06566
Location: 16719-17552
NCBI BlastP on this gene
BN424_mp0024
BN424_mp0023
Sugar ABC superfamily ATP binding cassette transporter, sugar-binding protein
Accession: CRI06564
Location: 14837-16180
NCBI BlastP on this gene
Accession: CRI06564
Location: 14837-16180
NCBI BlastP on this gene
BN424_mp0022
Sugar ABC superfamily ATP binding cassette transporter, membrane protein
Accession: CRI06563
Location: 13983-14819
NCBI BlastP on this gene
Accession: CRI06563
Location: 13983-14819
NCBI BlastP on this gene
BN424_mp0021
putative transport system permease
Accession: CRI06562
Location: 13081-13968
NCBI BlastP on this gene
Accession: CRI06562
Location: 13081-13968
NCBI BlastP on this gene
BN424_mp0020
BN424_mp0019
BN424_mp0018
BN424_mp0017
exported protein of unknown function
Accession: CRI06558
Location: 12031-12201
NCBI BlastP on this gene
Accession: CRI06558
Location: 12031-12201
NCBI BlastP on this gene
BN424_mp0016
BN424_mp0015
conserved protein of unknown function
Accession: CRI06556
Location: 10385-11239
NCBI BlastP on this gene
Accession: CRI06556
Location: 10385-11239
NCBI BlastP on this gene
BN424_mp0014
BN424_mp0013
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP040462 : Enterococcus sp. M190262 plasmid pM190262 Total score: 2.0 Cumulative Blast bit score: 688
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
FE005_16240
FE005_16230
FE005_16225
FE005_16220
FE005_16215
FE005_16210
DUF4062 domain-containing protein
Accession: QCT93524
Location: 97322-98506
NCBI BlastP on this gene
Accession: QCT93524
Location: 97322-98506
NCBI BlastP on this gene
FE005_16205
FE005_16200
type II toxin-antitoxin system death-on-curing family toxin
Accession: QCT93522
Location: 96563-96976
NCBI BlastP on this gene
Accession: QCT93522
Location: 96563-96976
NCBI BlastP on this gene
FE005_16195
FE005_16190
ROK family protein
Accession: QCT93520
Location: 94338-95237
BlastP hit with WP_004843625.1
Percentage identity: 58 %
BlastP bit score: 358
Sequence coverage: 100 %
E-value: 1e-119
NCBI BlastP on this gene
Accession: QCT93520
Location: 94338-95237
BlastP hit with WP_004843625.1
Percentage identity: 58 %
BlastP bit score: 358
Sequence coverage: 100 %
E-value: 1e-119
NCBI BlastP on this gene
FE005_16185
FE005_16180
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QCT93630
Location: 92690-93385
BlastP hit with WP_024854132.1
Percentage identity: 68 %
BlastP bit score: 330
Sequence coverage: 99 %
E-value: 3e-111
NCBI BlastP on this gene
Accession: QCT93630
Location: 92690-93385
BlastP hit with WP_024854132.1
Percentage identity: 68 %
BlastP bit score: 330
Sequence coverage: 99 %
E-value: 3e-111
NCBI BlastP on this gene
FE005_16175
MurR/RpiR family transcriptional regulator
Accession: QCT93518
Location: 91613-92455
NCBI BlastP on this gene
Accession: QCT93518
Location: 91613-92455
NCBI BlastP on this gene
FE005_16170
FE005_16165
extracellular solute-binding protein
Accession: QCT93516
Location: 89772-91130
NCBI BlastP on this gene
Accession: QCT93516
Location: 89772-91130
NCBI BlastP on this gene
FE005_16160
carbohydrate ABC transporter permease
Accession: QCT93515
Location: 88917-89753
NCBI BlastP on this gene
Accession: QCT93515
Location: 88917-89753
NCBI BlastP on this gene
FE005_16155
FE005_16150
FE005_16145
pflB
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP029150 : Glaesserella parasuis strain GZ20170512 chromosome Total score: 2.0 Cumulative Blast bit score: 627
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
spermidine/putrescine ABC transporter substrate-binding protein
Accession: C3363_10695
Location: 2128344-2128600
NCBI BlastP on this gene
Accession: C3363_10695
Location: 2128344-2128600
NCBI BlastP on this gene
C3363_10695
C3363_10700
abortive phage resistance protein
Accession: C3363_10705
Location: 2129146-2129266
NCBI BlastP on this gene
Accession: C3363_10705
Location: 2129146-2129266
NCBI BlastP on this gene
C3363_10705
C3363_10710
C3363_10715
LysR family transcriptional regulator
Accession: QHB45568
Location: 2130433-2131323
NCBI BlastP on this gene
Accession: QHB45568
Location: 2130433-2131323
NCBI BlastP on this gene
C3363_10720
C3363_10725
rplK
C3363_10735
C3363_10740
C3363_10745
bifunctional glycosyl
Location: 2135020-2135301
Location: 2135020-2135301
mrcB
C3363_10755
carbohydrate ABC transporter permease
Accession: C3363_10760
Location: 2136221-2137054
BlastP hit with WP_039959879.1
Percentage identity: 71 %
BlastP bit score: 406
Sequence coverage: 100 %
E-value: 1e-139
NCBI BlastP on this gene
Accession: C3363_10760
Location: 2136221-2137054
BlastP hit with WP_039959879.1
Percentage identity: 71 %
BlastP bit score: 406
Sequence coverage: 100 %
E-value: 1e-139
NCBI BlastP on this gene
C3363_10760
sugar ABC transporter permease
Accession: C3363_10765
Location: 2137056-2137939
NCBI BlastP on this gene
Accession: C3363_10765
Location: 2137056-2137939
NCBI BlastP on this gene
C3363_10765
C3363_10770
C3363_10775
sugar ABC transporter substrate-binding protein
Accession: C3363_10780
Location: 2138740-2139183
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
Accession: C3363_10780
Location: 2138740-2139183
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
C3363_10780
long-chain fatty acid--CoA ligase
Accession: QHB45379
Location: 2139353-2141140
NCBI BlastP on this gene
Accession: QHB45379
Location: 2139353-2141140
NCBI BlastP on this gene
C3363_10785
DNA polymerase III subunit chi
Accession: QHB45380
Location: 2141246-2141683
NCBI BlastP on this gene
Accession: QHB45380
Location: 2141246-2141683
NCBI BlastP on this gene
C3363_10790
C3363_10795
C3363_10800
C3363_10805
copper homeostasis protein CutC
Accession: QHB45383
Location: 2144741-2145472
NCBI BlastP on this gene
Accession: QHB45383
Location: 2144741-2145472
NCBI BlastP on this gene
C3363_10810
aminoimidazole riboside kinase
Accession: QHB45384
Location: 2145634-2146557
NCBI BlastP on this gene
Accession: QHB45384
Location: 2145634-2146557
NCBI BlastP on this gene
C3363_10815
C3363_10820
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP040243 : Glaesserella parasuis strain HPS-1 chromosome Total score: 2.0 Cumulative Blast bit score: 623
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
spermidine/putrescine ABC transporter substrate-binding protein
Accession: FEF05_11060
Location: 2184979-2185235
NCBI BlastP on this gene
Accession: FEF05_11060
Location: 2184979-2185235
NCBI BlastP on this gene
FEF05_11060
FEF05_11065
FEF05_11070
FEF05_11075
FEF05_11080
LysR family transcriptional regulator
Accession: QEM88922
Location: 2187069-2187959
NCBI BlastP on this gene
Accession: QEM88922
Location: 2187069-2187959
NCBI BlastP on this gene
FEF05_11085
rplA
rplK
FEF05_11100
FEF05_11105
FEF05_11110
FEF05_11120
carbohydrate ABC transporter permease
Accession: FEF05_11125
Location: 2192857-2193690
BlastP hit with WP_039959879.1
Percentage identity: 70 %
BlastP bit score: 402
Sequence coverage: 100 %
E-value: 8e-138
NCBI BlastP on this gene
Accession: FEF05_11125
Location: 2192857-2193690
BlastP hit with WP_039959879.1
Percentage identity: 70 %
BlastP bit score: 402
Sequence coverage: 100 %
E-value: 8e-138
NCBI BlastP on this gene
FEF05_11125
sugar ABC transporter permease
Accession: FEF05_11130
Location: 2193692-2194575
NCBI BlastP on this gene
Accession: FEF05_11130
Location: 2193692-2194575
NCBI BlastP on this gene
FEF05_11130
FEF05_11135
extracellular solute-binding protein
Accession: FEF05_11140
Location: 2195418-2195861
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
Accession: FEF05_11140
Location: 2195418-2195861
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
FEF05_11140
long-chain fatty acid--CoA ligase
Accession: QEM88927
Location: 2196031-2197818
NCBI BlastP on this gene
Accession: QEM88927
Location: 2196031-2197818
NCBI BlastP on this gene
FEF05_11145
DNA polymerase III subunit chi
Accession: QEM88928
Location: 2197924-2198361
NCBI BlastP on this gene
Accession: QEM88928
Location: 2197924-2198361
NCBI BlastP on this gene
FEF05_11150
FEF05_11155
galK
FEF05_11165
copper homeostasis protein CutC
Accession: QEM88931
Location: 2201419-2202150
NCBI BlastP on this gene
Accession: QEM88931
Location: 2201419-2202150
NCBI BlastP on this gene
FEF05_11170
aminoimidazole riboside kinase
Accession: QEM88932
Location: 2202312-2203235
NCBI BlastP on this gene
Accession: QEM88932
Location: 2202312-2203235
NCBI BlastP on this gene
FEF05_11175
acpP
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP020832 : Listeria monocytogenes strain PNUSAL000096 chromosome Total score: 2.0 Cumulative Blast bit score: 619
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
PTS mannitol transporter subunit IIA
Accession: ARJ88578
Location: 401783-402229
NCBI BlastP on this gene
Accession: ARJ88578
Location: 401783-402229
NCBI BlastP on this gene
Y281_01980
Y281_01975
PTS mannitol transporter subunit IIBC
Accession: ARJ88576
Location: 399645-401051
NCBI BlastP on this gene
Accession: ARJ88576
Location: 399645-401051
NCBI BlastP on this gene
Y281_01970
Y281_01965
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ARJ88574
Location: 397895-398608
BlastP hit with WP_024854132.1
Percentage identity: 54 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 9e-85
NCBI BlastP on this gene
Accession: ARJ88574
Location: 397895-398608
BlastP hit with WP_024854132.1
Percentage identity: 54 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 9e-85
NCBI BlastP on this gene
Y281_01960
Y281_01955
Y281_01950
Y281_01945
Y281_01940
Y281_01935
Y281_01930
Y281_01925
Y281_01920
Y281_01915
tRNA uridine-5-carboxymethylaminomethyl(34) synthesis enzyme MnmG
Accession: ARJ88566
Location: 390898-392787
NCBI BlastP on this gene
Accession: ARJ88566
Location: 390898-392787
NCBI BlastP on this gene
Y281_01910
Y281_01905
D-alanyl-D-alanine carboxypeptidase
Accession: ARJ88564
Location: 388324-389142
NCBI BlastP on this gene
Accession: ARJ88564
Location: 388324-389142
NCBI BlastP on this gene
Y281_01900
DUF4918 domain-containing protein
Accession: ARJ88563
Location: 387560-388267
NCBI BlastP on this gene
Accession: ARJ88563
Location: 387560-388267
NCBI BlastP on this gene
Y281_01895
TetR family transcriptional regulator
Accession: ARJ88562
Location: 387009-387536
NCBI BlastP on this gene
Accession: ARJ88562
Location: 387009-387536
NCBI BlastP on this gene
Y281_01890
Y281_01885
Y281_01880
Y281_01875
multidrug efflux MFS transporter NorB
Accession: ARJ88558
Location: 382183-383583
NCBI BlastP on this gene
Accession: ARJ88558
Location: 382183-383583
NCBI BlastP on this gene
Y281_01870
Y281_01865
AraC family transcriptional regulator
Accession: ARJ88556
Location: 378444-380786
NCBI BlastP on this gene
Accession: ARJ88556
Location: 378444-380786
NCBI BlastP on this gene
Y281_01860
Y281_00110
Y281_00105
23S rRNA (pseudouridine(1915)-N(3))-methyltransferase RlmH
Accession: ARJ88236
Location: 17414-17893
NCBI BlastP on this gene
Accession: ARJ88236
Location: 17414-17893
NCBI BlastP on this gene
Y281_00100
LysR family transcriptional regulator
Accession: ARJ88235
Location: 16519-17382
NCBI BlastP on this gene
Accession: ARJ88235
Location: 16519-17382
NCBI BlastP on this gene
Y281_00095
Y281_00090
Y281_00085
Y281_00080
PTS lactose transporter subunit IIC
Accession: ARJ88231
Location: 11525-12838
NCBI BlastP on this gene
Accession: ARJ88231
Location: 11525-12838
NCBI BlastP on this gene
Y281_00075
PTS sugar transporter subunit IIB
Accession: ARJ88230
Location: 11230-11520
NCBI BlastP on this gene
Accession: ARJ88230
Location: 11230-11520
NCBI BlastP on this gene
Y281_00070
Y281_00065
Y281_00060
Y281_00055
Y281_00050
low specificity L-threonine aldolase
Accession: ARJ88226
Location: 7189-8256
NCBI BlastP on this gene
Accession: ARJ88226
Location: 7189-8256
NCBI BlastP on this gene
Y281_00045
DNA (cytosine-5-)-methyltransferase
Accession: ARJ88225
Location: 5899-7113
NCBI BlastP on this gene
Accession: ARJ88225
Location: 5899-7113
NCBI BlastP on this gene
Y281_00040
Y281_00035
PTS cellbiose transporter subunit IIC
Accession: ARJ88223
Location: 3436-4221
NCBI BlastP on this gene
Accession: ARJ88223
Location: 3436-4221
NCBI BlastP on this gene
Y281_00030
Y281_00025
Y281_00020
hydroxymethylpyrimidine/phosphomethylpyrimidine kinase
Accession: ARJ88220
Location: 937-1740
NCBI BlastP on this gene
Accession: ARJ88220
Location: 937-1740
NCBI BlastP on this gene
Y281_00015
Y281_00010
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP000234 : Lactobacillus salivarius UCC118 plasmid pMP118 Total score: 2.0 Cumulative Blast bit score: 595
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Hypothetical membrane associated protein
Accession: ABE00728
Location: 208361-209329
NCBI BlastP on this gene
Accession: ABE00728
Location: 208361-209329
NCBI BlastP on this gene
LSL_1924
LSL_1925
Hypothetical membrane associated protein
Accession: ABE00730
Location: 209874-210143
NCBI BlastP on this gene
Accession: ABE00730
Location: 209874-210143
NCBI BlastP on this gene
LSL_1926
avtA
Carbon-nitrogen hydrolase family protein
Accession: ABE00732
Location: 211498-212283
NCBI BlastP on this gene
Accession: ABE00732
Location: 211498-212283
NCBI BlastP on this gene
LSL_1928
Conserved hypothetical membrane protein
Accession: ABE00733
Location: 212426-212926
NCBI BlastP on this gene
Accession: ABE00733
Location: 212426-212926
NCBI BlastP on this gene
LSL_1929
LSL_1930
L-serine dehydratase alpha subunit
Accession: ABE00735
Location: 213275-214156
NCBI BlastP on this gene
Accession: ABE00735
Location: 213275-214156
NCBI BlastP on this gene
sdaA
L-serine dehydratase beta subunit
Accession: ABE00736
Location: 214169-214831
NCBI BlastP on this gene
Accession: ABE00736
Location: 214169-214831
NCBI BlastP on this gene
sdaB
Hypothetical membrane spanning protein
Accession: ABE00737
Location: 215944-217380
NCBI BlastP on this gene
Accession: ABE00737
Location: 215944-217380
NCBI BlastP on this gene
LSL_1933
Putative oxidoreductase
Accession: ABE00738
Location: 217472-218587
BlastP hit with WP_004843633.1
Percentage identity: 69 %
BlastP bit score: 527
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABE00738
Location: 217472-218587
BlastP hit with WP_004843633.1
Percentage identity: 69 %
BlastP bit score: 527
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
LSL_1934
Conserved hypothetical protein
Accession: ABE00739
Location: 218978-219427
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 68
Sequence coverage: 59 %
E-value: 1e-11
NCBI BlastP on this gene
Accession: ABE00739
Location: 218978-219427
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 68
Sequence coverage: 59 %
E-value: 1e-11
NCBI BlastP on this gene
LSL_1935
ndk
nagC
nanA
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ABE00743
Location: 222008-222697
NCBI BlastP on this gene
Accession: ABE00743
Location: 222008-222697
NCBI BlastP on this gene
nanE
Transcriptional regulator, RpiR family
Accession: ABE00744
Location: 222743-223567
NCBI BlastP on this gene
Accession: ABE00744
Location: 222743-223567
NCBI BlastP on this gene
LSL_1940
Hypothetical membrane associated protein
Accession: ABE00745
Location: 223624-224241
NCBI BlastP on this gene
Accession: ABE00745
Location: 223624-224241
NCBI BlastP on this gene
LSL_1941
potE
LSL_1943
rpsN
Tetracycline resistance protein
Accession: ABE00749
Location: 226555-228474
NCBI BlastP on this gene
Accession: ABE00749
Location: 226555-228474
NCBI BlastP on this gene
fusA
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP020814 : Bacillus krulwichiae strain AM31D chromosome Total score: 2.0 Cumulative Blast bit score: 565
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Transcriptional regulatory protein YycF
Accession: ARK29836
Location: 1459140-1459838
NCBI BlastP on this gene
Accession: ARK29836
Location: 1459140-1459838
NCBI BlastP on this gene
yycF_1
Alkaline phosphatase synthesis sensor protein PhoR
Accession: ARK29837
Location: 1459841-1461223
NCBI BlastP on this gene
Accession: ARK29837
Location: 1459841-1461223
NCBI BlastP on this gene
phoR_1
Inner membrane transport protein YdhC
Accession: ARK29838
Location: 1461301-1462503
NCBI BlastP on this gene
Accession: ARK29838
Location: 1461301-1462503
NCBI BlastP on this gene
ydhC
qorB
Putative HTH-type transcriptional regulator YwnA
Accession: ARK29840
Location: 1463813-1464238
NCBI BlastP on this gene
Accession: ARK29840
Location: 1463813-1464238
NCBI BlastP on this gene
ywnA_1
BkAM31D_08195
N-acetylglucosamine-6-phosphate deacetylase
Accession: ARK29842
Location: 1465300-1466484
NCBI BlastP on this gene
Accession: ARK29842
Location: 1465300-1466484
NCBI BlastP on this gene
nagA_1
Sialidase A precursor
Accession: ARK29843
Location: 1466576-1468978
BlastP hit with WP_004843631.1
Percentage identity: 31 %
BlastP bit score: 293
Sequence coverage: 98 %
E-value: 8e-84
NCBI BlastP on this gene
Accession: ARK29843
Location: 1466576-1468978
BlastP hit with WP_004843631.1
Percentage identity: 31 %
BlastP bit score: 293
Sequence coverage: 98 %
E-value: 8e-84
NCBI BlastP on this gene
nanA_1
putative HTH-type transcriptional regulator YbbH
Accession: ARK29844
Location: 1469236-1470162
NCBI BlastP on this gene
Accession: ARK29844
Location: 1469236-1470162
NCBI BlastP on this gene
ybbH_1
Putative N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ARK29845
Location: 1470402-1471091
BlastP hit with WP_024854132.1
Percentage identity: 60 %
BlastP bit score: 272
Sequence coverage: 96 %
E-value: 3e-88
NCBI BlastP on this gene
Accession: ARK29845
Location: 1470402-1471091
BlastP hit with WP_024854132.1
Percentage identity: 60 %
BlastP bit score: 272
Sequence coverage: 96 %
E-value: 3e-88
NCBI BlastP on this gene
nanE
Sialic acid-binding periplasmic protein SiaP precursor
Accession: ARK29846
Location: 1471088-1472119
NCBI BlastP on this gene
Accession: ARK29846
Location: 1471088-1472119
NCBI BlastP on this gene
siaP_2
Sialic acid TRAP transporter permease protein SiaT
Accession: ARK29847
Location: 1472147-1472626
NCBI BlastP on this gene
Accession: ARK29847
Location: 1472147-1472626
NCBI BlastP on this gene
siaT_7
Sialic acid TRAP transporter permease protein SiaT
Accession: ARK29848
Location: 1472627-1472980
NCBI BlastP on this gene
Accession: ARK29848
Location: 1472627-1472980
NCBI BlastP on this gene
siaT_8
Sialic acid TRAP transporter permease protein SiaT
Accession: ARK29849
Location: 1473001-1473903
NCBI BlastP on this gene
Accession: ARK29849
Location: 1473001-1473903
NCBI BlastP on this gene
siaT_9
glkA
sda_2
nanA_2
Glucosamine-6-phosphate deaminase 1
Accession: ARK29853
Location: 1476044-1476781
NCBI BlastP on this gene
Accession: ARK29853
Location: 1476044-1476781
NCBI BlastP on this gene
nagB_1
Inner membrane transport permease YbhR
Accession: ARK29854
Location: 1477031-1478143
NCBI BlastP on this gene
Accession: ARK29854
Location: 1477031-1478143
NCBI BlastP on this gene
ybhR
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP011391 : Faecalibaculum rodentium strain Alo17 Total score: 2.0 Cumulative Blast bit score: 544
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
AALO17_07380
AALO17_07390
AALO17_07400
AALO17_07410
AALO17_07420
AALO17_07430
AALO17_07440
AALO17_07450
yhcH/YjgK/YiaL family protein
Accession: AMK53880
Location: 671957-672430
BlastP hit with WP_004843641.1
Percentage identity: 32 %
BlastP bit score: 67
Sequence coverage: 71 %
E-value: 2e-11
NCBI BlastP on this gene
Accession: AMK53880
Location: 671957-672430
BlastP hit with WP_004843641.1
Percentage identity: 32 %
BlastP bit score: 67
Sequence coverage: 71 %
E-value: 2e-11
NCBI BlastP on this gene
AALO17_07460
solute:sodium symporter (SSS) family transporter
Accession: AMK53881
Location: 672427-673992
NCBI BlastP on this gene
Accession: AMK53881
Location: 672427-673992
NCBI BlastP on this gene
AALO17_07470
N-acetylneuraminate lyase
Accession: AMK53882
Location: 674080-674997
BlastP hit with WP_024854133.1
Percentage identity: 75 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 1e-166
NCBI BlastP on this gene
Accession: AMK53882
Location: 674080-674997
BlastP hit with WP_024854133.1
Percentage identity: 75 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 1e-166
NCBI BlastP on this gene
AALO17_07480
AALO17_07490
AALO17_07500
AALO17_07510
AALO17_07520
AALO17_07530
AALO17_07540
AALO17_07550
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP003700 : Fusobacterium nucleatum subsp. vincentii 3_1_36A2 Total score: 2.0 Cumulative Blast bit score: 434
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
HMPREF0946_01170
HMPREF0946_01169
HMPREF0946_01168
HMPREF0946_01167
hypothetical protein
Accession: EEU33093
Location: 1914239-1914913
BlastP hit with WP_024854132.1
Percentage identity: 58 %
BlastP bit score: 265
Sequence coverage: 99 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: EEU33093
Location: 1914239-1914913
BlastP hit with WP_024854132.1
Percentage identity: 58 %
BlastP bit score: 265
Sequence coverage: 99 %
E-value: 2e-85
NCBI BlastP on this gene
HMPREF0946_01166
HMPREF0946_01165
HMPREF0946_01164
TRAP transporter, DctM subunit
Accession: EEU33090
Location: 1910597-1912450
NCBI BlastP on this gene
Accession: EEU33090
Location: 1910597-1912450
NCBI BlastP on this gene
HMPREF0946_01163
DctP family TRAP transporter solute receptor
Accession: EEU33089
Location: 1909590-1910573
NCBI BlastP on this gene
Accession: EEU33089
Location: 1909590-1910573
NCBI BlastP on this gene
HMPREF0946_01162
HMPREF0946_01161
cyclically-permuted mutarotase
Accession: EEU33087
Location: 1907404-1908552
NCBI BlastP on this gene
Accession: EEU33087
Location: 1907404-1908552
NCBI BlastP on this gene
HMPREF0946_01160
type I restriction-modification system, M subunit
Accession: EEU33086
Location: 1905592-1907154
NCBI BlastP on this gene
Accession: EEU33086
Location: 1905592-1907154
NCBI BlastP on this gene
HMPREF0946_01159
HMPREF0946_01158
HMPREF0946_01157
HsdR family type I site-specific deoxyribonuclease
Accession: EEU33083
Location: 1899625-1902666
NCBI BlastP on this gene
Accession: EEU33083
Location: 1899625-1902666
NCBI BlastP on this gene
HMPREF0946_01156
HMPREF0946_01155
HMPREF0946_01154
HMPREF0946_01153
HMPREF0946_01152
HMPREF0946_01151
hypothetical protein
Accession: EEU33077
Location: 1895241-1895642
BlastP hit with WP_004843624.1
Percentage identity: 62 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 1e-50
NCBI BlastP on this gene
Accession: EEU33077
Location: 1895241-1895642
BlastP hit with WP_004843624.1
Percentage identity: 62 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 1e-50
NCBI BlastP on this gene
HMPREF0946_01150
1-deoxy-D-xylulose-5-phosphate synthase
Accession: EEU33076
Location: 1893480-1895231
NCBI BlastP on this gene
Accession: EEU33076
Location: 1893480-1895231
NCBI BlastP on this gene
HMPREF0946_01149
HMPREF0946_01148
HMPREF0946_01147
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP010855 : Marinovum algicola DG 898 Total score: 2.0 Cumulative Blast bit score: 377
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
type I secretion membrane fusion protein, HlyD family
Accession: AKO95294
Location: 83010-84326
NCBI BlastP on this gene
Accession: AKO95294
Location: 83010-84326
NCBI BlastP on this gene
MALG_00076
MALG_00075
diaminobutyrate acetyltransferase
Accession: AKO95292
Location: 81727-82260
NCBI BlastP on this gene
Accession: AKO95292
Location: 81727-82260
NCBI BlastP on this gene
MALG_00074
diaminobutyrate--2-oxoglutarate aminotransferase
Accession: AKO95291
Location: 80380-81672
NCBI BlastP on this gene
Accession: AKO95291
Location: 80380-81672
NCBI BlastP on this gene
MALG_00073
MALG_00072
MALG_00071
MALG_00070
putative metal-dependent hydrolase of the TIM-barrel fold protein
Accession: AKO95287
Location: 77397-78236
NCBI BlastP on this gene
Accession: AKO95287
Location: 77397-78236
NCBI BlastP on this gene
MALG_00069
L-fuco-beta-pyranose dehydrogenase
Accession: AKO95286
Location: 76457-77395
NCBI BlastP on this gene
Accession: AKO95286
Location: 76457-77395
NCBI BlastP on this gene
MALG_00068
ABC-type sugar transport system, ATPase component
Accession: AKO95285
Location: 75393-76460
NCBI BlastP on this gene
Accession: AKO95285
Location: 75393-76460
NCBI BlastP on this gene
MALG_00067
ABC-type sugar transport system, permease component
Accession: AKO95284
Location: 74491-75378
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 187
Sequence coverage: 87 %
E-value: 3e-53
NCBI BlastP on this gene
Accession: AKO95284
Location: 74491-75378
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 187
Sequence coverage: 87 %
E-value: 3e-53
NCBI BlastP on this gene
MALG_00066
ABC-type sugar transport system, periplasmic component
Accession: AKO95283
Location: 73186-74433
NCBI BlastP on this gene
Accession: AKO95283
Location: 73186-74433
NCBI BlastP on this gene
MALG_00065
ABC-type sugar transport system, permease component
Accession: AKO95282
Location: 72253-73131
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 190
Sequence coverage: 99 %
E-value: 1e-54
NCBI BlastP on this gene
Accession: AKO95282
Location: 72253-73131
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 190
Sequence coverage: 99 %
E-value: 1e-54
NCBI BlastP on this gene
MALG_00064
MALG_00063
NAD-dependent aldehyde dehydrogenase
Accession: AKO95280
Location: 69708-71141
NCBI BlastP on this gene
Accession: AKO95280
Location: 69708-71141
NCBI BlastP on this gene
MALG_00062
2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase
Accession: AKO95279
Location: 68905-69711
NCBI BlastP on this gene
Accession: AKO95279
Location: 68905-69711
NCBI BlastP on this gene
MALG_00061
MALG_00060
MALG_00059
MALG_00058
MALG_00057
MALG_00056
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP034358 : Clostridium septicum strain VAT12 chromosome Total score: 2.0 Cumulative Blast bit score: 351
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
EI377_06060
EI377_06055
EI377_06050
EI377_06045
EI377_06040
EI377_06035
EI377_06030
MurR/RpiR family transcriptional regulator
Accession: EI377_06025
Location: 1258276-1258694
NCBI BlastP on this gene
Accession: EI377_06025
Location: 1258276-1258694
NCBI BlastP on this gene
EI377_06025
DUF386 domain-containing protein
Accession: QAS60333
Location: 1257619-1258071
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
Accession: QAS60333
Location: 1257619-1258071
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
EI377_06020
MurR/RpiR family transcriptional regulator
Accession: QAS60332
Location: 1256560-1257357
NCBI BlastP on this gene
Accession: QAS60332
Location: 1256560-1257357
NCBI BlastP on this gene
EI377_06015
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QAS60331
Location: 1255768-1256412
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
Accession: QAS60331
Location: 1255768-1256412
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
EI377_06010
PTS glucose transporter subunit IIB
Accession: QAS60330
Location: 1254208-1255698
NCBI BlastP on this gene
Accession: QAS60330
Location: 1254208-1255698
NCBI BlastP on this gene
EI377_06005
EI377_06000
EI377_05995
EI377_05990
EI377_05985
EI377_05980
GTP pyrophosphokinase family protein
Accession: QAS60324
Location: 1247356-1248150
NCBI BlastP on this gene
Accession: QAS60324
Location: 1247356-1248150
NCBI BlastP on this gene
EI377_05975
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP023671 : Clostridium septicum strain DSM 7534 chromosome Total score: 2.0 Cumulative Blast bit score: 351
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
CP523_11265
TIGR01906 family membrane protein
Accession: AYE34944
Location: 2458365-2459066
NCBI BlastP on this gene
Accession: AYE34944
Location: 2458365-2459066
NCBI BlastP on this gene
CP523_11260
CP523_11255
CP523_11250
CP523_11245
type I methionyl aminopeptidase
Accession: AYE34940
Location: 2454540-2455409
NCBI BlastP on this gene
Accession: AYE34940
Location: 2454540-2455409
NCBI BlastP on this gene
map
CP523_11235
DUF386 domain-containing protein
Accession: AYE34939
Location: 2453268-2453720
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
Accession: AYE34939
Location: 2453268-2453720
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
CP523_11230
MurR/RpiR family transcriptional regulator
Accession: AYE34938
Location: 2452209-2453006
NCBI BlastP on this gene
Accession: AYE34938
Location: 2452209-2453006
NCBI BlastP on this gene
CP523_11225
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: AYE34937
Location: 2451417-2452061
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
Accession: AYE34937
Location: 2451417-2452061
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
CP523_11220
PTS glucose transporter subunit IIB
Accession: AYE34936
Location: 2449857-2451347
NCBI BlastP on this gene
Accession: AYE34936
Location: 2449857-2451347
NCBI BlastP on this gene
CP523_11215
CP523_11210
CP523_11205
CP523_11200
CP523_11195
CP523_11190
CP523_11185
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP001656 : Paenibacillus sp. JDR-2 chromosome Total score: 2.0 Cumulative Blast bit score: 334
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Mannitol dehydrogenase domain protein
Accession: ACT04826
Location: 7113910-7115061
NCBI BlastP on this gene
Accession: ACT04826
Location: 7113910-7115061
NCBI BlastP on this gene
Pjdr2_6223
phosphoenolpyruvate-protein phosphotransferase
Accession: ACT04825
Location: 7112149-7113867
NCBI BlastP on this gene
Accession: ACT04825
Location: 7112149-7113867
NCBI BlastP on this gene
Pjdr2_6222
Phosphotransferase system, phosphocarrier protein HPr
Accession: ACT04824
Location: 7111886-7112149
NCBI BlastP on this gene
Accession: ACT04824
Location: 7111886-7112149
NCBI BlastP on this gene
Pjdr2_6221
short-chain dehydrogenase/reductase SDR
Accession: ACT04823
Location: 7111082-7111825
NCBI BlastP on this gene
Accession: ACT04823
Location: 7111082-7111825
NCBI BlastP on this gene
Pjdr2_6220
Helix-turn-helix type 11 domain protein
Accession: ACT04822
Location: 7110093-7111055
NCBI BlastP on this gene
Accession: ACT04822
Location: 7110093-7111055
NCBI BlastP on this gene
Pjdr2_6219
drug resistance transporter, EmrB/QacA subfamily
Accession: ACT04821
Location: 7108533-7110047
NCBI BlastP on this gene
Accession: ACT04821
Location: 7108533-7110047
NCBI BlastP on this gene
Pjdr2_6218
Alcohol dehydrogenase zinc-binding domain protein
Accession: ACT04820
Location: 7107262-7108347
NCBI BlastP on this gene
Accession: ACT04820
Location: 7107262-7108347
NCBI BlastP on this gene
Pjdr2_6217
transcriptional regulator, LuxR family
Accession: ACT04819
Location: 7106411-7107100
NCBI BlastP on this gene
Accession: ACT04819
Location: 7106411-7107100
NCBI BlastP on this gene
Pjdr2_6216
conserved hypothetical protein
Accession: ACT04818
Location: 7105930-7106400
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 71
Sequence coverage: 87 %
E-value: 1e-12
NCBI BlastP on this gene
Accession: ACT04818
Location: 7105930-7106400
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 71
Sequence coverage: 87 %
E-value: 1e-12
NCBI BlastP on this gene
Pjdr2_6215
transcriptional regulator, RpiR family
Accession: ACT04817
Location: 7105050-7105904
NCBI BlastP on this gene
Accession: ACT04817
Location: 7105050-7105904
NCBI BlastP on this gene
Pjdr2_6214
Pjdr2_6213
N-acylglucosamine-6-phosphate 2-epimerase
Accession: ACT04815
Location: 7103502-7104182
BlastP hit with WP_024854132.1
Percentage identity: 57 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 6e-85
NCBI BlastP on this gene
Accession: ACT04815
Location: 7103502-7104182
BlastP hit with WP_024854132.1
Percentage identity: 57 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 6e-85
NCBI BlastP on this gene
Pjdr2_6212
binding-protein-dependent transport systems inner membrane component
Accession: ACT04814
Location: 7102622-7103467
NCBI BlastP on this gene
Accession: ACT04814
Location: 7102622-7103467
NCBI BlastP on this gene
Pjdr2_6211
binding-protein-dependent transport systems inner membrane component
Accession: ACT04813
Location: 7101728-7102603
NCBI BlastP on this gene
Accession: ACT04813
Location: 7101728-7102603
NCBI BlastP on this gene
Pjdr2_6210
extracellular solute-binding protein family 1
Accession: ACT04812
Location: 7100245-7101645
NCBI BlastP on this gene
Accession: ACT04812
Location: 7100245-7101645
NCBI BlastP on this gene
Pjdr2_6209
dihydrodipicolinate synthetase
Accession: ACT04811
Location: 7099283-7100194
NCBI BlastP on this gene
Accession: ACT04811
Location: 7099283-7100194
NCBI BlastP on this gene
Pjdr2_6208
Pjdr2_6207
extracellular solute-binding protein family 1
Accession: ACT04809
Location: 7095570-7096961
NCBI BlastP on this gene
Accession: ACT04809
Location: 7095570-7096961
NCBI BlastP on this gene
Pjdr2_6206
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP036150 : Oceanispirochaeta sp. K2 chromosome Total score: 2.0 Cumulative Blast bit score: 149
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
EXM22_16860
Hsp20/alpha crystallin family protein
Accession: QEN09569
Location: 3716834-3717235
NCBI BlastP on this gene
Accession: QEN09569
Location: 3716834-3717235
NCBI BlastP on this gene
EXM22_16865
Hsp20/alpha crystallin family protein
Accession: QEN09570
Location: 3717263-3717682
NCBI BlastP on this gene
Accession: QEN09570
Location: 3717263-3717682
NCBI BlastP on this gene
EXM22_16870
EXM22_16875
EXM22_16880
EXM22_16885
EXM22_16890
DUF386 domain-containing protein
Accession: QEN09575
Location: 3722518-3722976
BlastP hit with WP_004843641.1
Percentage identity: 40 %
BlastP bit score: 75
Sequence coverage: 67 %
E-value: 4e-14
NCBI BlastP on this gene
Accession: QEN09575
Location: 3722518-3722976
BlastP hit with WP_004843641.1
Percentage identity: 40 %
BlastP bit score: 75
Sequence coverage: 67 %
E-value: 4e-14
NCBI BlastP on this gene
EXM22_16895
EXM22_16900
EXM22_16905
ketopantoate reductase family protein
Accession: QEN09578
Location: 3724544-3725326
NCBI BlastP on this gene
Accession: QEN09578
Location: 3724544-3725326
NCBI BlastP on this gene
EXM22_16910
EXM22_16915
EXM22_16920
AraC family transcriptional regulator
Accession: QEN09581
Location: 3727846-3728703
BlastP hit with WP_101883549.1
Percentage identity: 37 %
BlastP bit score: 74
Sequence coverage: 39 %
E-value: 3e-12
NCBI BlastP on this gene
Accession: QEN09581
Location: 3727846-3728703
BlastP hit with WP_101883549.1
Percentage identity: 37 %
BlastP bit score: 74
Sequence coverage: 39 %
E-value: 3e-12
NCBI BlastP on this gene
EXM22_16925
EXM22_16930
EXM22_16935
C4-dicarboxylate ABC transporter
Accession: QEN09584
Location: 3730554-3731555
NCBI BlastP on this gene
Accession: QEN09584
Location: 3730554-3731555
NCBI BlastP on this gene
EXM22_16940
TRAP transporter small permease
Accession: QEN09585
Location: 3731655-3732185
NCBI BlastP on this gene
Accession: QEN09585
Location: 3731655-3732185
NCBI BlastP on this gene
EXM22_16945
TRAP transporter large permease
Accession: QEN09586
Location: 3732198-3733481
NCBI BlastP on this gene
Accession: QEN09586
Location: 3732198-3733481
NCBI BlastP on this gene
EXM22_16950
EXM22_16955
Gfo/Idh/MocA family oxidoreductase
Accession: QEN09588
Location: 3734559-3735707
NCBI BlastP on this gene
Accession: QEN09588
Location: 3734559-3735707
NCBI BlastP on this gene
EXM22_16960
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP040863 : Rodentibacter heylii strain G1 chromosome Total score: 1.0 Cumulative Blast bit score: 560
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
FEE42_02495
FEE42_02500
helix-turn-helix transcriptional regulator
Accession: QIA76305
Location: 493209-493544
NCBI BlastP on this gene
Accession: QIA76305
Location: 493209-493544
NCBI BlastP on this gene
FEE42_02505
SAM-dependent methyltransferase
Accession: QIA76306
Location: 493741-495918
NCBI BlastP on this gene
Accession: QIA76306
Location: 493741-495918
NCBI BlastP on this gene
FEE42_02510
FEE42_02515
FEE42_02520
Gfo/Idh/MocA family oxidoreductase
Accession: QIA76309
Location: 499174-500277
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
Accession: QIA76309
Location: 499174-500277
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
FEE42_02525
Gfo/Idh/MocA family oxidoreductase
Accession: QIA76310
Location: 500298-501368
NCBI BlastP on this gene
Accession: QIA76310
Location: 500298-501368
NCBI BlastP on this gene
FEE42_02530
FadR family transcriptional regulator
Accession: QIA78132
Location: 501361-502065
NCBI BlastP on this gene
Accession: QIA78132
Location: 501361-502065
NCBI BlastP on this gene
FEE42_02535
FEE42_02540
FEE42_02545
FEE42_02550
putative hydroxymethylpyrimidine transporter CytX
Accession: QIA76313
Location: 504812-505996
NCBI BlastP on this gene
Accession: QIA76313
Location: 504812-505996
NCBI BlastP on this gene
cytX
lysine exporter LysO family protein
Accession: QIA78133
Location: 506066-506974
NCBI BlastP on this gene
Accession: QIA78133
Location: 506066-506974
NCBI BlastP on this gene
FEE42_02560
pepN
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
CP013830 : Pasteurellaceae bacterium NI1060 genome. Total score: 1.0 Cumulative Blast bit score: 560
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
RUMGNA_RS11845
RUMGNA_RS11850
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
RUMGNA_RS11875
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
Nucleoside-diphosphate-sugar epimerase
Accession: AOF54289
Location: 2308051-2309154
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOF54289
Location: 2308051-2309154
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
AC062_2203
putative oxidoreductase protein
Accession: AOF54288
Location: 2306960-2308030
NCBI BlastP on this gene
Accession: AOF54288
Location: 2306960-2308030
NCBI BlastP on this gene
AC062_2202
Transcriptional regulator, GntR family
Accession: AOF54287
Location: 2306254-2306967
NCBI BlastP on this gene
Accession: AOF54287
Location: 2306254-2306967
NCBI BlastP on this gene
AC062_2201
Sialic acid transporter (permease) NanT
Accession: AOF54286
Location: 2304853-2306082
NCBI BlastP on this gene
Accession: AOF54286
Location: 2304853-2306082
NCBI BlastP on this gene
AC062_2200
AC062_2199
AC062_2198
AC062_2197
AC062_2196
AC062_2195
AC062_2194
AC062_2193
AC062_2192
Query: [Ruminococcus] gnavus ATCC 29149 R_gnavus-1.0.1_Cont169, whole
451. : CP015378 Fictibacillus phosphorivorans strain G25-29 chromosome Total score: 2.5 Cumulative Blast bit score: 379
DBD-Pfam|MerR,DBD-SUPERFAMILY|0042546
Accession: WP_004843624.1
Location: 1-405
NCBI BlastP on this gene
Accession: WP_004843624.1
Location: 1-405
NCBI BlastP on this gene
RUMGNA_RS11830
RUMGNA_RS11835
RUMGNA_RS11840
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: WP_024854132.1
Location: 2543-3232
NCBI BlastP on this gene
Accession: WP_024854132.1
Location: 2543-3232
NCBI BlastP on this gene
RUMGNA_RS11845
RUMGNA_RS11850
Gfo/Idh/MocA family oxidoreductase
Accession: WP_004843633.1
Location: 5511-6635
NCBI BlastP on this gene
Accession: WP_004843633.1
Location: 5511-6635
NCBI BlastP on this gene
RUMGNA_RS11855
RUMGNA_RS11860
RUMGNA_RS11865
RUMGNA_RS11870
DBD-Pfam|HTH AraC,DBD-Pfam|HTH AraC,DBD-SUPERFAMI
Accession: WP_101883549.1
Location: 9915-10763
NCBI BlastP on this gene
Accession: WP_101883549.1
Location: 9915-10763
NCBI BlastP on this gene
RUMGNA_RS11875
YhcH/YjgK/YiaL family protein
Accession: WP_004843641.1
Location: 10906-11355
NCBI BlastP on this gene
Accession: WP_004843641.1
Location: 10906-11355
NCBI BlastP on this gene
RUMGNA_RS11880
RUMGNA_RS11885
RUMGNA_RS11895
RUMGNA_RS11890
ABE65_003840
ABE65_003835
gamma carbonic anhydrase family protein
Accession: ANC75988
Location: 730374-730889
NCBI BlastP on this gene
Accession: ANC75988
Location: 730374-730889
NCBI BlastP on this gene
ABE65_003830
phenylacetic acid degradation operon negative regulatory protein PaaX
Accession: ANC75987
Location: 729196-730089
NCBI BlastP on this gene
Accession: ANC75987
Location: 729196-730089
NCBI BlastP on this gene
ABE65_003825
ABE65_003820
ABE65_003815
3-hydroxybutyryl-CoA dehydrogenase
Accession: ANC75984
Location: 726178-727050
NCBI BlastP on this gene
Accession: ANC75984
Location: 726178-727050
NCBI BlastP on this gene
ABE65_003810
ABE65_003805
2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase
Accession: ANC75982
Location: 723372-724145
NCBI BlastP on this gene
Accession: ANC75982
Location: 723372-724145
NCBI BlastP on this gene
ABE65_003800
ABE65_003795
ethyl tert-butyl ether degradation protein EthD
Accession: ANC75980
Location: 722285-722590
NCBI BlastP on this gene
Accession: ANC75980
Location: 722285-722590
NCBI BlastP on this gene
ABE65_003790
phenylacetate-CoA oxygenase subunit PaaJ
Accession: ANC75979
Location: 721747-722247
NCBI BlastP on this gene
Accession: ANC75979
Location: 721747-722247
NCBI BlastP on this gene
ABE65_003785
ABE65_003780
1,2-phenylacetyl-CoA epoxidase subunit B
Accession: ANC75977
Location: 720570-720917
NCBI BlastP on this gene
Accession: ANC75977
Location: 720570-720917
NCBI BlastP on this gene
ABE65_003775
1,2-phenylacetyl-CoA epoxidase subunit A
Accession: ANC75976
Location: 719581-720546
NCBI BlastP on this gene
Accession: ANC75976
Location: 719581-720546
NCBI BlastP on this gene
paaA
ABE65_003765
ABE65_003760
ABE65_003755
LysR family transcriptional regulator
Accession: ANC75972
Location: 715168-716046
NCBI BlastP on this gene
Accession: ANC75972
Location: 715168-716046
NCBI BlastP on this gene
ABE65_003750
ABE65_003745
zinc-binding alcohol dehydrogenase
Accession: ANC75970
Location: 713811-714791
NCBI BlastP on this gene
Accession: ANC75970
Location: 713811-714791
NCBI BlastP on this gene
ABE65_003740
ABE65_003735
sugar ABC transporter permease
Accession: ANC75968
Location: 712039-712866
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 100 %
E-value: 1e-57
NCBI BlastP on this gene
Accession: ANC75968
Location: 712039-712866
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 100 %
E-value: 1e-57
NCBI BlastP on this gene
ABE65_003730
ABC transporter permease
Accession: ANC79261
Location: 710680-712008
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 182
Sequence coverage: 82 %
E-value: 4e-50
NCBI BlastP on this gene
Accession: ANC79261
Location: 710680-712008
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 182
Sequence coverage: 82 %
E-value: 4e-50
NCBI BlastP on this gene
ABE65_003725
ABC transporter substrate-binding protein
Accession: ANC75967
Location: 709204-710511
NCBI BlastP on this gene
Accession: ANC75967
Location: 709204-710511
NCBI BlastP on this gene
ABE65_003720
LacI family transcriptional regulator
Accession: ANC75966
Location: 708163-709149
NCBI BlastP on this gene
Accession: ANC75966
Location: 708163-709149
NCBI BlastP on this gene
ABE65_003715
peptide ABC transporter ATP-binding protein
Accession: ANC75965
Location: 707229-707951
NCBI BlastP on this gene
Accession: ANC75965
Location: 707229-707951
NCBI BlastP on this gene
ABE65_003710
glycine/betaine ABC transporter
Accession: ANC79260
Location: 705438-706970
NCBI BlastP on this gene
Accession: ANC79260
Location: 705438-706970
NCBI BlastP on this gene
ABE65_003705
glycine/betaine ABC transporter ATP-binding protein
Accession: ANC75964
Location: 704270-705424
NCBI BlastP on this gene
Accession: ANC75964
Location: 704270-705424
NCBI BlastP on this gene
ABE65_003700
ABE65_003695
TetR family transcriptional regulator
Accession: ANC75962
Location: 701224-701793
NCBI BlastP on this gene
Accession: ANC75962
Location: 701224-701793
NCBI BlastP on this gene
ABE65_003690
ABE65_003685
ABE65_003680
ABE65_003675
ABE65_003670
ABE65_003665
ABE65_003660
ABE65_003655
ABE65_003650
ABE65_003645
ABE65_003640
452. : CP008820 Ochrobactrum anthropi strain OAB chromosome 1 Total score: 2.5 Cumulative Blast bit score: 377
ureC
urease accessory ureE domain protein
Accession: AIK43905
Location: 2682882-2683259
NCBI BlastP on this gene
Accession: AIK43905
Location: 2682882-2683259
NCBI BlastP on this gene
ureE
DR92_2604
urease accessory protein UreG
Accession: AIK45246
Location: 2681427-2682047
NCBI BlastP on this gene
Accession: AIK45246
Location: 2681427-2682047
NCBI BlastP on this gene
ureG
FAD binding domain of DNA photolyase family protein
Accession: AIK43635
Location: 2680020-2681426
NCBI BlastP on this gene
Accession: AIK43635
Location: 2680020-2681426
NCBI BlastP on this gene
DR92_2602
histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein
Accession: AIK43042
Location: 2678063-2679616
NCBI BlastP on this gene
Accession: AIK43042
Location: 2678063-2679616
NCBI BlastP on this gene
DR92_2600
efflux transporter, RND family, MFP subunit
Accession: AIK45085
Location: 2676555-2677682
NCBI BlastP on this gene
Accession: AIK45085
Location: 2676555-2677682
NCBI BlastP on this gene
DR92_2599
acrB/AcrD/AcrF family protein
Accession: AIK44967
Location: 2673392-2676544
NCBI BlastP on this gene
Accession: AIK44967
Location: 2673392-2676544
NCBI BlastP on this gene
DR92_2598
NMT1/THI5 like family protein
Accession: AIK42988
Location: 2672342-2673340
NCBI BlastP on this gene
Accession: AIK42988
Location: 2672342-2673340
NCBI BlastP on this gene
DR92_2597
binding--dependent transport system inner membrane component family protein
Accession: AIK44254
Location: 2671371-2672228
NCBI BlastP on this gene
Accession: AIK44254
Location: 2671371-2672228
NCBI BlastP on this gene
DR92_2596
binding--dependent transport system inner membrane component family protein
Accession: AIK44308
Location: 2670478-2671371
NCBI BlastP on this gene
Accession: AIK44308
Location: 2670478-2671371
NCBI BlastP on this gene
DR92_2595
ABC transporter family protein
Accession: AIK43610
Location: 2669674-2670441
NCBI BlastP on this gene
Accession: AIK43610
Location: 2669674-2670441
NCBI BlastP on this gene
DR92_2594
hydA
amidase, hydantoinase/carbamoylase family protein
Accession: AIK43518
Location: 2666823-2668070
NCBI BlastP on this gene
Accession: AIK43518
Location: 2666823-2668070
NCBI BlastP on this gene
DR92_2592
HTH-type transcriptional regulator RutR
Accession: AIK45142
Location: 2665905-2666603
NCBI BlastP on this gene
Accession: AIK45142
Location: 2665905-2666603
NCBI BlastP on this gene
rutR
DR92_2589
ABC transporter family protein
Accession: AIK44324
Location: 2664440-2665513
NCBI BlastP on this gene
Accession: AIK44324
Location: 2664440-2665513
NCBI BlastP on this gene
DR92_2590
binding--dependent transport system inner membrane component family protein
Accession: AIK42969
Location: 2663592-2664425
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: AIK42969
Location: 2663592-2664425
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
DR92_2588
binding--dependent transport system inner membrane component family protein
Accession: AIK42897
Location: 2662634-2663587
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Accession: AIK42897
Location: 2662634-2663587
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
DR92_2587
bacterial extracellular solute-binding family protein
Accession: AIK44223
Location: 2661300-2662559
NCBI BlastP on this gene
Accession: AIK44223
Location: 2661300-2662559
NCBI BlastP on this gene
DR92_2586
cys/Met metabolism PLP-dependent enzyme family protein
Accession: AIK44980
Location: 2660095-2661264
NCBI BlastP on this gene
Accession: AIK44980
Location: 2660095-2661264
NCBI BlastP on this gene
DR92_2585
bacterial regulatory s, gntR family protein
Accession: AIK44892
Location: 2659382-2660098
NCBI BlastP on this gene
Accession: AIK44892
Location: 2659382-2660098
NCBI BlastP on this gene
DR92_2584
pld1
DR92_2582
dapA
bacterial regulatory s, gntR family protein
Accession: AIK43481
Location: 2655231-2655863
NCBI BlastP on this gene
Accession: AIK43481
Location: 2655231-2655863
NCBI BlastP on this gene
DR92_2580
Tat (twin-arginine translocation) pathway signal sequence domain protein
Accession: AIK43872
Location: 2653820-2655154
NCBI BlastP on this gene
Accession: AIK43872
Location: 2653820-2655154
NCBI BlastP on this gene
DR92_2579
binding--dependent transport system inner membrane component family protein
Accession: AIK43565
Location: 2652885-2653823
NCBI BlastP on this gene
Accession: AIK43565
Location: 2652885-2653823
NCBI BlastP on this gene
DR92_2578
binding--dependent transport system inner membrane component family protein
Accession: AIK44956
Location: 2652057-2652875
NCBI BlastP on this gene
Accession: AIK44956
Location: 2652057-2652875
NCBI BlastP on this gene
DR92_2577
ABC transporter family protein
Accession: AIK43455
Location: 2650983-2652041
NCBI BlastP on this gene
Accession: AIK43455
Location: 2650983-2652041
NCBI BlastP on this gene
DR92_2576
dihydroorotate dehydrogenase family domain protein
Accession: AIK43244
Location: 2649622-2650932
NCBI BlastP on this gene
Accession: AIK43244
Location: 2649622-2650932
NCBI BlastP on this gene
DR92_2575
DR92_2574
eamA-like transporter family protein
Accession: AIK44672
Location: 2646940-2647872
NCBI BlastP on this gene
Accession: AIK44672
Location: 2646940-2647872
NCBI BlastP on this gene
DR92_2573
polyketide cyclase / dehydrase and lipid transport family protein
Accession: AIK44062
Location: 2646394-2646852
NCBI BlastP on this gene
Accession: AIK44062
Location: 2646394-2646852
NCBI BlastP on this gene
DR92_2572
phosphoglucose isomerase family protein
Accession: AIK43576
Location: 2644594-2646243
NCBI BlastP on this gene
Accession: AIK43576
Location: 2644594-2646243
NCBI BlastP on this gene
DR92_2571
succinylglutamate desuccinylase / Aspartoacylase family protein
Accession: AIK43276
Location: 2643450-2644511
NCBI BlastP on this gene
Accession: AIK43276
Location: 2643450-2644511
NCBI BlastP on this gene
DR92_2570
HAD hydrolase, IA, variant 1 family protein
Accession: AIK43377
Location: 2642725-2643453
NCBI BlastP on this gene
Accession: AIK43377
Location: 2642725-2643453
NCBI BlastP on this gene
DR92_2569
453. : CP000758 Ochrobactrum anthropi ATCC 49188 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 377
UreE urease accessory domain protein
Accession: ABS13067
Location: 350134-350646
NCBI BlastP on this gene
Accession: ABS13067
Location: 350134-350646
NCBI BlastP on this gene
Oant_0336
urease accessory protein UreF, putative
Accession: ABS13068
Location: 350618-351304
NCBI BlastP on this gene
Accession: ABS13068
Location: 350618-351304
NCBI BlastP on this gene
Oant_0337
Oant_0338
Deoxyribodipyrimidine photo-lyase
Accession: ABS13070
Location: 351922-353376
NCBI BlastP on this gene
Accession: ABS13070
Location: 351922-353376
NCBI BlastP on this gene
Oant_0339
integral membrane sensor signal transduction histidine kinase
Accession: ABS13071
Location: 353732-355285
NCBI BlastP on this gene
Accession: ABS13071
Location: 353732-355285
NCBI BlastP on this gene
Oant_0340
efflux transporter, RND family, MFP subunit
Accession: ABS13072
Location: 355666-356793
NCBI BlastP on this gene
Accession: ABS13072
Location: 355666-356793
NCBI BlastP on this gene
Oant_0341
Oant_0342
Oant_0343
binding-protein-dependent transport systems inner membrane component
Accession: ABS13075
Location: 361121-361981
NCBI BlastP on this gene
Accession: ABS13075
Location: 361121-361981
NCBI BlastP on this gene
Oant_0344
binding-protein-dependent transport systems inner membrane component
Accession: ABS13076
Location: 361978-362871
NCBI BlastP on this gene
Accession: ABS13076
Location: 361978-362871
NCBI BlastP on this gene
Oant_0345
Oant_0346
Oant_0347
amidase, hydantoinase/carbamoylase family
Accession: ABS13079
Location: 365294-366541
NCBI BlastP on this gene
Accession: ABS13079
Location: 365294-366541
NCBI BlastP on this gene
Oant_0348
transcriptional regulator, TetR family
Accession: ABS13080
Location: 366762-367460
NCBI BlastP on this gene
Accession: ABS13080
Location: 366762-367460
NCBI BlastP on this gene
Oant_0349
Oant_0350
Oant_0351
binding-protein-dependent transport systems inner membrane component
Accession: ABS13083
Location: 368940-369773
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: ABS13083
Location: 368940-369773
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Oant_0352
binding-protein-dependent transport systems inner membrane component
Accession: ABS13084
Location: 369778-370731
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Accession: ABS13084
Location: 369778-370731
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 183
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Oant_0353
extracellular solute-binding protein family 1
Accession: ABS13085
Location: 370806-372065
NCBI BlastP on this gene
Accession: ABS13085
Location: 370806-372065
NCBI BlastP on this gene
Oant_0354
Oant_0355
Oant_0356
Oant_0357
Oant_0358
Oant_0359
transcriptional regulator, GntR family
Accession: ABS13091
Location: 377472-378134
NCBI BlastP on this gene
Accession: ABS13091
Location: 377472-378134
NCBI BlastP on this gene
Oant_0360
extracellular solute-binding protein family 1
Accession: ABS13092
Location: 378211-379545
NCBI BlastP on this gene
Accession: ABS13092
Location: 378211-379545
NCBI BlastP on this gene
Oant_0361
binding-protein-dependent transport systems inner membrane component
Accession: ABS13093
Location: 379542-380480
NCBI BlastP on this gene
Accession: ABS13093
Location: 379542-380480
NCBI BlastP on this gene
Oant_0362
binding-protein-dependent transport systems inner membrane component
Accession: ABS13094
Location: 380490-381308
NCBI BlastP on this gene
Accession: ABS13094
Location: 380490-381308
NCBI BlastP on this gene
Oant_0363
Oant_0364
dihydroorotate dehydrogenase family protein
Accession: ABS13096
Location: 382419-383729
NCBI BlastP on this gene
Accession: ABS13096
Location: 382419-383729
NCBI BlastP on this gene
Oant_0365
FAD-dependent pyridine nucleotide-disulphide oxidoreductase
Accession: ABS13097
Location: 383741-385243
NCBI BlastP on this gene
Accession: ABS13097
Location: 383741-385243
NCBI BlastP on this gene
Oant_0366
protein of unknown function DUF6 transmembrane
Accession: ABS13098
Location: 385479-386411
NCBI BlastP on this gene
Accession: ABS13098
Location: 385479-386411
NCBI BlastP on this gene
Oant_0367
carbon monoxide dehydrogenase subunit G
Accession: ABS13099
Location: 386499-386957
NCBI BlastP on this gene
Accession: ABS13099
Location: 386499-386957
NCBI BlastP on this gene
Oant_0368
Oant_0369
Succinylglutamate desuccinylase/aspartoacylase
Accession: ABS13101
Location: 388840-389901
NCBI BlastP on this gene
Accession: ABS13101
Location: 388840-389901
NCBI BlastP on this gene
Oant_0370
HAD-superfamily hydrolase, subfamily IA, variant 1
Accession: ABS13102
Location: 389898-390626
NCBI BlastP on this gene
Accession: ABS13102
Location: 389898-390626
NCBI BlastP on this gene
Oant_0371
454. : CP047140 Streptomyces sp. Tu 2975 chromosome Total score: 2.5 Cumulative Blast bit score: 376
polynucleotide kinase-phosphatase
Accession: QIP84099
Location: 1968623-1971184
NCBI BlastP on this gene
Accession: QIP84099
Location: 1968623-1971184
NCBI BlastP on this gene
GLX30_08655
3' terminal RNA ribose 2'-O-methyltransferase Hen1
Accession: QIP84100
Location: 1971181-1972641
NCBI BlastP on this gene
Accession: QIP84100
Location: 1971181-1972641
NCBI BlastP on this gene
GLX30_08660
homocysteine S-methyltransferase
Accession: QIP84101
Location: 1972738-1973640
NCBI BlastP on this gene
Accession: QIP84101
Location: 1972738-1973640
NCBI BlastP on this gene
mmuM
LLM class F420-dependent oxidoreductase
Accession: GLX30_08670
Location: 1973654-1974576
NCBI BlastP on this gene
Accession: GLX30_08670
Location: 1973654-1974576
NCBI BlastP on this gene
GLX30_08670
GLX30_08675
nucleotide pyrophosphohydrolase
Accession: QIP84103
Location: 1975310-1975642
NCBI BlastP on this gene
Accession: QIP84103
Location: 1975310-1975642
NCBI BlastP on this gene
GLX30_08680
GLX30_08685
GLX30_08690
GLX30_08700
LacI family transcriptional regulator
Accession: QIP84106
Location: 1985389-1986447
NCBI BlastP on this gene
Accession: QIP84106
Location: 1985389-1986447
NCBI BlastP on this gene
GLX30_08705
glycoside hydrolase family 13 protein
Accession: QIP84107
Location: 1986435-1988084
NCBI BlastP on this gene
Accession: QIP84107
Location: 1986435-1988084
NCBI BlastP on this gene
GLX30_08710
carbohydrate ABC transporter permease
Accession: QIP88438
Location: 1988168-1989022
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 197
Sequence coverage: 99 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: QIP88438
Location: 1988168-1989022
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 197
Sequence coverage: 99 %
E-value: 2e-57
NCBI BlastP on this gene
GLX30_08715
sugar ABC transporter permease
Accession: QIP84108
Location: 1989016-1990026
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
Accession: QIP84108
Location: 1989016-1990026
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
GLX30_08720
extracellular solute-binding protein
Accession: QIP84109
Location: 1990046-1991320
NCBI BlastP on this gene
Accession: QIP84109
Location: 1990046-1991320
NCBI BlastP on this gene
GLX30_08725
LacI family transcriptional regulator
Accession: QIP84110
Location: 1991715-1992761
NCBI BlastP on this gene
Accession: QIP84110
Location: 1991715-1992761
NCBI BlastP on this gene
GLX30_08730
carbohydrate kinase family protein
Accession: QIP84111
Location: 1992758-1993678
NCBI BlastP on this gene
Accession: QIP84111
Location: 1992758-1993678
NCBI BlastP on this gene
GLX30_08735
carbohydrate ABC transporter permease
Accession: QIP88439
Location: 1993773-1994576
NCBI BlastP on this gene
Accession: QIP88439
Location: 1993773-1994576
NCBI BlastP on this gene
GLX30_08740
sugar ABC transporter permease
Accession: QIP84112
Location: 1994680-1995690
NCBI BlastP on this gene
Accession: QIP84112
Location: 1994680-1995690
NCBI BlastP on this gene
GLX30_08745
carbohydrate ABC transporter substrate-binding protein
Accession: QIP84113
Location: 1995797-1997146
NCBI BlastP on this gene
Accession: QIP84113
Location: 1995797-1997146
NCBI BlastP on this gene
GLX30_08750
LacI family transcriptional regulator
Accession: QIP84114
Location: 1997603-1998628
NCBI BlastP on this gene
Accession: QIP84114
Location: 1997603-1998628
NCBI BlastP on this gene
GLX30_08755
GLX30_08760
GLX30_08765
GLX30_08770
GLX30_08775
GLX30_08780
glycosyltransferase family 9 protein
Accession: GLX30_08785
Location: 2006440-2007479
NCBI BlastP on this gene
Accession: GLX30_08785
Location: 2006440-2007479
NCBI BlastP on this gene
GLX30_08785
D-glycero-beta-D-manno-heptose 1-phosphate adenylyltransferase
Accession: QIP84119
Location: 2007551-2008960
NCBI BlastP on this gene
Accession: QIP84119
Location: 2007551-2008960
NCBI BlastP on this gene
rfaE2
SIS domain-containing protein
Accession: QIP84120
Location: 2008957-2009622
NCBI BlastP on this gene
Accession: QIP84120
Location: 2008957-2009622
NCBI BlastP on this gene
GLX30_08795
glycosyltransferase family 1 protein
Accession: QIP84121
Location: 2009781-2011040
NCBI BlastP on this gene
Accession: QIP84121
Location: 2009781-2011040
NCBI BlastP on this gene
GLX30_08800
455. : CP022604 Ochrobactrum quorumnocens strain A44 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 376
CES85_0276
UDP-N-acetylglucosamine 1-carboxyvinyltransferase
Accession: ASV86779
Location: 275947-277239
NCBI BlastP on this gene
Accession: ASV86779
Location: 275947-277239
NCBI BlastP on this gene
murA
CES85_0278
CES85_0280
histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein
Accession: ASV87277
Location: 278837-280405
NCBI BlastP on this gene
Accession: ASV87277
Location: 278837-280405
NCBI BlastP on this gene
CES85_0282
CES85_0283
efflux transporter, RND family, MFP subunit
Accession: ASV85597
Location: 280896-282032
NCBI BlastP on this gene
Accession: ASV85597
Location: 280896-282032
NCBI BlastP on this gene
CES85_0284
CES85_0285
CES85_0286
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV85292
Location: 286302-287159
NCBI BlastP on this gene
Accession: ASV85292
Location: 286302-287159
NCBI BlastP on this gene
CES85_0287
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV85345
Location: 287156-288049
NCBI BlastP on this gene
Accession: ASV85345
Location: 287156-288049
NCBI BlastP on this gene
CES85_0288
CES85_0289
hydA
amidase, hydantoinase/carbamoylase family protein
Accession: ASV86894
Location: 290492-291754
NCBI BlastP on this gene
Accession: ASV86894
Location: 290492-291754
NCBI BlastP on this gene
CES85_0291
bacterial regulatory, tetR family protein
Accession: ASV87540
Location: 291965-292645
NCBI BlastP on this gene
Accession: ASV87540
Location: 291965-292645
NCBI BlastP on this gene
CES85_0292
CES85_0293
CES85_0294
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV87333
Location: 294125-294958
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: ASV87333
Location: 294125-294958
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 2e-56
NCBI BlastP on this gene
CES85_0295
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV87617
Location: 294963-295847
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 182
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
Accession: ASV87617
Location: 294963-295847
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 182
Sequence coverage: 90 %
E-value: 1e-51
NCBI BlastP on this gene
CES85_0296
bacterial extracellular solute-binding family protein
Accession: ASV86975
Location: 295993-297252
NCBI BlastP on this gene
Accession: ASV86975
Location: 295993-297252
NCBI BlastP on this gene
CES85_0297
cys/Met metabolism PLP-dependent enzyme family protein
Accession: ASV87147
Location: 297290-298459
NCBI BlastP on this gene
Accession: ASV87147
Location: 297290-298459
NCBI BlastP on this gene
CES85_0298
CES85_0299
CES85_0300
pld1
CES85_0302
CES85_0303
bacterial regulatory, gntR family protein
Accession: ASV87433
Location: 302767-303429
NCBI BlastP on this gene
Accession: ASV87433
Location: 302767-303429
NCBI BlastP on this gene
CES85_0304
tat (twin-arginine translocation) pathway signal sequence domain protein
Accession: ASV87356
Location: 303521-304855
NCBI BlastP on this gene
Accession: ASV87356
Location: 303521-304855
NCBI BlastP on this gene
CES85_0305
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV87611
Location: 304852-305790
NCBI BlastP on this gene
Accession: ASV87611
Location: 304852-305790
NCBI BlastP on this gene
CES85_0306
binding-protein-dependent transport system inner membrane component family protein
Accession: ASV87061
Location: 305800-306618
NCBI BlastP on this gene
Accession: ASV87061
Location: 305800-306618
NCBI BlastP on this gene
CES85_0307
CES85_0308
dihydroorotate dehydrogenase family domain protein
Accession: ASV85603
Location: 307757-309067
NCBI BlastP on this gene
Accession: ASV85603
Location: 307757-309067
NCBI BlastP on this gene
CES85_0309
pyridine nucleotide-disulfide oxidoreductase family protein
Accession: ASV86381
Location: 309079-310584
NCBI BlastP on this gene
Accession: ASV86381
Location: 309079-310584
NCBI BlastP on this gene
CES85_0310
eamA-like transporter family protein
Accession: ASV86843
Location: 310819-311763
NCBI BlastP on this gene
Accession: ASV86843
Location: 310819-311763
NCBI BlastP on this gene
CES85_0311
polyketide cyclase / dehydrase and lipid transport family protein
Accession: ASV87079
Location: 311864-312337
NCBI BlastP on this gene
Accession: ASV87079
Location: 311864-312337
NCBI BlastP on this gene
CES85_0312
pgi
succinylglutamate desuccinylase / Aspartoacylase family protein
Accession: ASV86254
Location: 314212-315273
NCBI BlastP on this gene
Accession: ASV86254
Location: 314212-315273
NCBI BlastP on this gene
CES85_0314
HAD hydrolase, IA, variant 1 family protein
Accession: ASV86911
Location: 315270-315998
NCBI BlastP on this gene
Accession: ASV86911
Location: 315270-315998
NCBI BlastP on this gene
CES85_0315
456. : CP003359 Halobacteroides halobius DSM 5150 Total score: 2.5 Cumulative Blast bit score: 376
Halha_1537
Halha_1538
Halha_1540
RHS repeat-associated core domain protein
Accession: AGB41480
Location: 1537026-1543274
NCBI BlastP on this gene
Accession: AGB41480
Location: 1537026-1543274
NCBI BlastP on this gene
Halha_1541
Halha_1542
Protein of unknown function (DUF2012)
Accession: AGB41482
Location: 1543870-1544850
NCBI BlastP on this gene
Accession: AGB41482
Location: 1543870-1544850
NCBI BlastP on this gene
Halha_1543
Halha_1544
Halha_1545
Halha_1546
trehalose/maltose hydrolase or phosphorylase
Accession: AGB41486
Location: 1549302-1551617
NCBI BlastP on this gene
Accession: AGB41486
Location: 1549302-1551617
NCBI BlastP on this gene
Halha_1547
ABC-type sugar transport system, periplasmic component
Accession: AGB41487
Location: 1551714-1553000
NCBI BlastP on this gene
Accession: AGB41487
Location: 1551714-1553000
NCBI BlastP on this gene
Halha_1548
ABC-type sugar transport system, permease component
Accession: AGB41488
Location: 1553098-1553940
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 195
Sequence coverage: 101 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: AGB41488
Location: 1553098-1553940
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 195
Sequence coverage: 101 %
E-value: 1e-56
NCBI BlastP on this gene
Halha_1549
permease component of ABC-type sugar transporter
Accession: AGB41489
Location: 1553955-1554890
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 181
Sequence coverage: 95 %
E-value: 7e-51
NCBI BlastP on this gene
Accession: AGB41489
Location: 1553955-1554890
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 181
Sequence coverage: 95 %
E-value: 7e-51
NCBI BlastP on this gene
Halha_1550
Halha_1551
Halha_1552
dehydrogenase of unknown specificity, short-chain alcohol dehydrogenase like protein
Accession: AGB41492
Location: 1558204-1558965
NCBI BlastP on this gene
Accession: AGB41492
Location: 1558204-1558965
NCBI BlastP on this gene
Halha_1553
trehalose/maltose hydrolase or phosphorylase
Accession: AGB41493
Location: 1558955-1561306
NCBI BlastP on this gene
Accession: AGB41493
Location: 1558955-1561306
NCBI BlastP on this gene
Halha_1554
ABC-type sugar transport system, periplasmic component
Accession: AGB41494
Location: 1561401-1562645
NCBI BlastP on this gene
Accession: AGB41494
Location: 1561401-1562645
NCBI BlastP on this gene
Halha_1555
ABC-type sugar transport system, permease component
Accession: AGB41495
Location: 1562700-1563533
NCBI BlastP on this gene
Accession: AGB41495
Location: 1562700-1563533
NCBI BlastP on this gene
Halha_1556
permease component of ABC-type sugar transporter
Accession: AGB41496
Location: 1563533-1564402
NCBI BlastP on this gene
Accession: AGB41496
Location: 1563533-1564402
NCBI BlastP on this gene
Halha_1557
oligoketide cyclase/lipid transport protein
Accession: AGB41497
Location: 1564750-1565202
NCBI BlastP on this gene
Accession: AGB41497
Location: 1564750-1565202
NCBI BlastP on this gene
Halha_1558
Halha_1559
acetylornithine/succinylornithine aminotransferase
Accession: AGB41499
Location: 1566935-1568176
NCBI BlastP on this gene
Accession: AGB41499
Location: 1566935-1568176
NCBI BlastP on this gene
Halha_1560
single-stranded-DNA-specific exonuclease RecJ
Accession: AGB41500
Location: 1568280-1570556
NCBI BlastP on this gene
Accession: AGB41500
Location: 1568280-1570556
NCBI BlastP on this gene
Halha_1561
Halha_1562
DNA/RNA helicase, superfamily I
Accession: AGB41502
Location: 1571397-1573589
NCBI BlastP on this gene
Accession: AGB41502
Location: 1571397-1573589
NCBI BlastP on this gene
Halha_1563
Halha_1564
457. : CP038147 Streptomyces sp. S501 chromosome Total score: 2.5 Cumulative Blast bit score: 375
TetR/AcrR family transcriptional regulator
Accession: QBR06099
Location: 1746075-1746809
NCBI BlastP on this gene
Accession: QBR06099
Location: 1746075-1746809
NCBI BlastP on this gene
D7Y56_09290
5-carboxymethyl-2-hydroxymuconate Delta-isomerase
Accession: QBR06100
Location: 1746794-1747144
NCBI BlastP on this gene
Accession: QBR06100
Location: 1746794-1747144
NCBI BlastP on this gene
D7Y56_09295
pullulanase-type alpha-1,6-glucosidase
Accession: QBR06101
Location: 1747327-1752633
NCBI BlastP on this gene
Accession: QBR06101
Location: 1747327-1752633
NCBI BlastP on this gene
pulA
D7Y56_09305
D7Y56_09310
D7Y56_09315
D7Y56_09320
D7Y56_09325
XRE family transcriptional regulator
Accession: QBR06106
Location: 1757841-1758719
NCBI BlastP on this gene
Accession: QBR06106
Location: 1757841-1758719
NCBI BlastP on this gene
D7Y56_09330
DUF397 domain-containing protein
Accession: QBR06107
Location: 1758716-1758925
NCBI BlastP on this gene
Accession: QBR06107
Location: 1758716-1758925
NCBI BlastP on this gene
D7Y56_09335
MerR family transcriptional regulator
Accession: QBR06108
Location: 1759002-1759913
NCBI BlastP on this gene
Accession: QBR06108
Location: 1759002-1759913
NCBI BlastP on this gene
D7Y56_09340
D7Y56_09345
RNA polymerase sigma-70 factor
Accession: QBR06110
Location: 1760487-1761386
NCBI BlastP on this gene
Accession: QBR06110
Location: 1760487-1761386
NCBI BlastP on this gene
D7Y56_09350
D7Y56_09355
LacI family transcriptional regulator
Accession: QBR06112
Location: 1762163-1763221
NCBI BlastP on this gene
Accession: QBR06112
Location: 1762163-1763221
NCBI BlastP on this gene
D7Y56_09360
glycoside hydrolase family 13 protein
Accession: QBR06113
Location: 1763206-1764882
NCBI BlastP on this gene
Accession: QBR06113
Location: 1763206-1764882
NCBI BlastP on this gene
D7Y56_09365
carbohydrate ABC transporter permease
Accession: QBR06114
Location: 1764981-1765859
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: QBR06114
Location: 1764981-1765859
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
D7Y56_09370
sugar ABC transporter permease
Accession: QBR06115
Location: 1765856-1766872
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: QBR06115
Location: 1765856-1766872
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
D7Y56_09375
extracellular solute-binding protein
Accession: QBR06116
Location: 1766894-1768165
NCBI BlastP on this gene
Accession: QBR06116
Location: 1766894-1768165
NCBI BlastP on this gene
D7Y56_09380
LacI family transcriptional regulator
Accession: QBR06117
Location: 1768526-1769569
NCBI BlastP on this gene
Accession: QBR06117
Location: 1768526-1769569
NCBI BlastP on this gene
D7Y56_09385
inositol phosphorylceramide synthase
Accession: QBR06118
Location: 1769646-1770644
NCBI BlastP on this gene
Accession: QBR06118
Location: 1769646-1770644
NCBI BlastP on this gene
D7Y56_09390
bifunctional [glutamine synthetase]
Accession: QBR06119
Location: 1770729-1773725
NCBI BlastP on this gene
Accession: QBR06119
Location: 1770729-1773725
NCBI BlastP on this gene
D7Y56_09395
putative protein N(5)-glutamine methyltransferase
Accession: QBR06120
Location: 1773885-1774673
NCBI BlastP on this gene
Accession: QBR06120
Location: 1773885-1774673
NCBI BlastP on this gene
D7Y56_09400
D7Y56_09405
D7Y56_09410
type I glutamate--ammonia ligase
Accession: QBR06123
Location: 1777325-1778686
NCBI BlastP on this gene
Accession: QBR06123
Location: 1777325-1778686
NCBI BlastP on this gene
glnA
DUF3105 domain-containing protein
Accession: QBR06124
Location: 1778933-1779607
NCBI BlastP on this gene
Accession: QBR06124
Location: 1778933-1779607
NCBI BlastP on this gene
D7Y56_09420
DUF305 domain-containing protein
Accession: QBR06125
Location: 1779604-1780281
NCBI BlastP on this gene
Accession: QBR06125
Location: 1779604-1780281
NCBI BlastP on this gene
D7Y56_09425
CBS domain-containing protein
Accession: QBR06126
Location: 1780407-1780832
NCBI BlastP on this gene
Accession: QBR06126
Location: 1780407-1780832
NCBI BlastP on this gene
D7Y56_09430
multicopper oxidase family protein
Accession: QBR06127
Location: 1780960-1782537
NCBI BlastP on this gene
Accession: QBR06127
Location: 1780960-1782537
NCBI BlastP on this gene
D7Y56_09435
D7Y56_09440
D7Y56_09445
D7Y56_09450
D7Y56_09455
458. : CP011340 Streptomyces pristinaespiralis strain HCCB 10218 Total score: 2.5 Cumulative Blast bit score: 375
SPRI_1853
SPRI_1854
SPRI_1855
homocysteine methyltransferase
Accession: ALC20162
Location: 2171061-2171963
NCBI BlastP on this gene
Accession: ALC20162
Location: 2171061-2171963
NCBI BlastP on this gene
SPRI_1856
F420-dependent oxidoreductase
Accession: ALC20163
Location: 2171979-2172902
NCBI BlastP on this gene
Accession: ALC20163
Location: 2171979-2172902
NCBI BlastP on this gene
SPRI_1857
SPRI_1858
nucleotide pyrophosphohydrolase
Accession: ALC20165
Location: 2173636-2173968
NCBI BlastP on this gene
Accession: ALC20165
Location: 2173636-2173968
NCBI BlastP on this gene
SPRI_1859
SPRI_1860
SPRI_1861
sulfonate ABC transporter ATP-binding protein
Accession: ALC20168
Location: 2175830-2181247
NCBI BlastP on this gene
Accession: ALC20168
Location: 2175830-2181247
NCBI BlastP on this gene
SPRI_1862
SPRI_1863
LacI family transcriptional regulator
Accession: ALC20170
Location: 2183581-2184630
NCBI BlastP on this gene
Accession: ALC20170
Location: 2183581-2184630
NCBI BlastP on this gene
SPRI_1864
SPRI_1865
ABC transporter permease
Accession: ALC20172
Location: 2186359-2187210
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 196
Sequence coverage: 99 %
E-value: 4e-57
NCBI BlastP on this gene
Accession: ALC20172
Location: 2186359-2187210
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 196
Sequence coverage: 99 %
E-value: 4e-57
NCBI BlastP on this gene
SPRI_1866
ABC transporter permease
Accession: ALC20173
Location: 2187207-2188217
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
Accession: ALC20173
Location: 2187207-2188217
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 179
Sequence coverage: 102 %
E-value: 5e-50
NCBI BlastP on this gene
SPRI_1867
sugar ABC transporter substrate-binding protein
Accession: ALC20174
Location: 2188237-2189511
NCBI BlastP on this gene
Accession: ALC20174
Location: 2188237-2189511
NCBI BlastP on this gene
SPRI_1868
LacI family transcriptional regulator
Accession: ALC20175
Location: 2189906-2190940
NCBI BlastP on this gene
Accession: ALC20175
Location: 2189906-2190940
NCBI BlastP on this gene
SPRI_1869
SPRI_1870
SPRI_1871
sugar ABC transporter permease
Accession: ALC20178
Location: 2193875-2194786
NCBI BlastP on this gene
Accession: ALC20178
Location: 2193875-2194786
NCBI BlastP on this gene
SPRI_1872
SPRI_1873
SPRI_1874
LacI family transcriptional regulator
Accession: ALC20181
Location: 2197701-2198726
NCBI BlastP on this gene
Accession: ALC20181
Location: 2197701-2198726
NCBI BlastP on this gene
SPRI_1875
SPRI_1876
SPRI_1877
SPRI_1878
short-chain dehydrogenase/reductase SDR
Accession: ALC20185
Location: 2204061-2204894
NCBI BlastP on this gene
Accession: ALC20185
Location: 2204061-2204894
NCBI BlastP on this gene
SPRI_1879
2-hydroxycyclohexanecarboxyl-CoA dehydrogenase
Accession: ALC20186
Location: 2204885-2205619
NCBI BlastP on this gene
Accession: ALC20186
Location: 2204885-2205619
NCBI BlastP on this gene
SPRI_1880
SPRI_1881
SPRI_1882
459. : CP003990 Streptomyces sp. PAMC26508 Total score: 2.5 Cumulative Blast bit score: 375
F750_2071
F750_2072
neopullulanase / Maltodextrin glucosidase
Accession: AGJ54568
Location: 2142397-2147703
NCBI BlastP on this gene
Accession: AGJ54568
Location: 2142397-2147703
NCBI BlastP on this gene
F750_2073
F750_2074
F750_2075
F750_2076
F750_2077
F750_2078
F750_2079
F750_2080
F750_2081
MerR-family transcriptional regulator
Accession: AGJ54577
Location: 2154094-2155005
NCBI BlastP on this gene
Accession: AGJ54577
Location: 2154094-2155005
NCBI BlastP on this gene
F750_2082
putative integral membrane protein
Accession: AGJ54578
Location: 2155138-2155476
NCBI BlastP on this gene
Accession: AGJ54578
Location: 2155138-2155476
NCBI BlastP on this gene
F750_2083
F750_2084
F750_2085
maltose operon transcriptional repressor MalR
Accession: AGJ54581
Location: 2157258-2158304
NCBI BlastP on this gene
Accession: AGJ54581
Location: 2157258-2158304
NCBI BlastP on this gene
malR
F750_2087
maltose/maltodextrin ABC transpor permease protein MalG
Accession: AGJ54583
Location: 2160076-2160954
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: AGJ54583
Location: 2160076-2160954
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
malG
maltose/maltodextrin ABC transpor permease protein MalF
Accession: AGJ54584
Location: 2160951-2161967
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: AGJ54584
Location: 2160951-2161967
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
malF
maltose/maltodextrin ABC transpor substrate binding periplasmic protein MalE
Accession: AGJ54585
Location: 2161989-2163260
NCBI BlastP on this gene
Accession: AGJ54585
Location: 2161989-2163260
NCBI BlastP on this gene
malE
F750_2091
maltose operon transcriptional repressor MalR
Accession: AGJ54587
Location: 2163519-2164664
NCBI BlastP on this gene
Accession: AGJ54587
Location: 2163519-2164664
NCBI BlastP on this gene
malR
putative integral membrane protein
Accession: AGJ54588
Location: 2164666-2165739
NCBI BlastP on this gene
Accession: AGJ54588
Location: 2164666-2165739
NCBI BlastP on this gene
F750_2093
glutamate-ammonia-ligase adenylyltransferase
Accession: AGJ54589
Location: 2165824-2168820
NCBI BlastP on this gene
Accession: AGJ54589
Location: 2165824-2168820
NCBI BlastP on this gene
F750_2094
methylase of polypeptide chain release factors
Accession: AGJ54590
Location: 2168980-2169768
NCBI BlastP on this gene
Accession: AGJ54590
Location: 2168980-2169768
NCBI BlastP on this gene
F750_2095
3',5'-cyclic-nucleotide phosphodiesterase
Accession: AGJ54591
Location: 2169983-2170726
NCBI BlastP on this gene
Accession: AGJ54591
Location: 2169983-2170726
NCBI BlastP on this gene
F750_2096
F750_2097
putative transport protein/putative regulator
Accession: AGJ54593
Location: 2171020-2172273
NCBI BlastP on this gene
Accession: AGJ54593
Location: 2171020-2172273
NCBI BlastP on this gene
F750_2098
F750_2099
cytoplasmic membrane protein FsxA
Accession: AGJ54595
Location: 2174029-2174703
NCBI BlastP on this gene
Accession: AGJ54595
Location: 2174029-2174703
NCBI BlastP on this gene
fsxA
F750_2101
F750_2102
F750_2103
NAD synthetase / Glutamine amidotransferase chain of NAD synthetase
Accession: AGJ54599
Location: 2177735-2179495
NCBI BlastP on this gene
Accession: AGJ54599
Location: 2177735-2179495
NCBI BlastP on this gene
F750_2104
RNA polymerase ECF-subfamily sigma factor
Accession: AGJ54600
Location: 2179645-2180313
NCBI BlastP on this gene
Accession: AGJ54600
Location: 2179645-2180313
NCBI BlastP on this gene
F750_2105
F750_2106
putative transmembrane efflux protein
Accession: AGJ54602
Location: 2181466-2182689
NCBI BlastP on this gene
Accession: AGJ54602
Location: 2181466-2182689
NCBI BlastP on this gene
F750_2107
460. : CP002475 Streptomyces pratensis ATCC 33331 Total score: 2.5 Cumulative Blast bit score: 375
Sfla_4616
Sfla_4615
Sfla_4614
alpha-1,6-glucosidase, pullulanase-type
Accession: ADW06014
Location: 5319065-5324371
NCBI BlastP on this gene
Accession: ADW06014
Location: 5319065-5324371
NCBI BlastP on this gene
Sfla_4613
glycoside hydrolase starch-binding protein
Accession: ADW06013
Location: 5317242-5318966
NCBI BlastP on this gene
Accession: ADW06013
Location: 5317242-5318966
NCBI BlastP on this gene
Sfla_4612
alpha/beta hydrolase fold protein
Accession: ADW06012
Location: 5315785-5316660
NCBI BlastP on this gene
Accession: ADW06012
Location: 5315785-5316660
NCBI BlastP on this gene
Sfla_4611
Sfla_4609
ATP-binding region ATPase domain protein
Accession: ADW06010
Location: 5313884-5314321
NCBI BlastP on this gene
Accession: ADW06010
Location: 5313884-5314321
NCBI BlastP on this gene
Sfla_4608
helix-turn-helix domain protein
Accession: ADW06009
Location: 5312841-5313719
NCBI BlastP on this gene
Accession: ADW06009
Location: 5312841-5313719
NCBI BlastP on this gene
Sfla_4607
protein of unknown function DUF397
Accession: ADW06008
Location: 5312635-5312844
NCBI BlastP on this gene
Accession: ADW06008
Location: 5312635-5312844
NCBI BlastP on this gene
Sfla_4606
transcriptional regulator, MerR family
Accession: ADW06007
Location: 5311647-5312558
NCBI BlastP on this gene
Accession: ADW06007
Location: 5311647-5312558
NCBI BlastP on this gene
Sfla_4605
Sfla_4604
GCN5-related N-acetyltransferase
Accession: ADW06005
Location: 5310619-5311176
NCBI BlastP on this gene
Accession: ADW06005
Location: 5310619-5311176
NCBI BlastP on this gene
Sfla_4603
transcriptional regulator, LacI family
Accession: ADW06004
Location: 5309450-5310496
NCBI BlastP on this gene
Accession: ADW06004
Location: 5309450-5310496
NCBI BlastP on this gene
Sfla_4602
alpha amylase catalytic region
Accession: ADW06003
Location: 5307777-5309453
NCBI BlastP on this gene
Accession: ADW06003
Location: 5307777-5309453
NCBI BlastP on this gene
Sfla_4601
binding-protein-dependent transport systems inner membrane component
Accession: ADW06002
Location: 5306800-5307678
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: ADW06002
Location: 5306800-5307678
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 197
Sequence coverage: 101 %
E-value: 2e-57
NCBI BlastP on this gene
Sfla_4600
binding-protein-dependent transport systems inner membrane component
Accession: ADW06001
Location: 5305787-5306803
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: ADW06001
Location: 5305787-5306803
BlastP hit with WP_039959892.1
Percentage identity: 34 %
BlastP bit score: 178
Sequence coverage: 101 %
E-value: 1e-49
NCBI BlastP on this gene
Sfla_4599
extracellular solute-binding protein family 1
Accession: ADW06000
Location: 5304494-5305765
NCBI BlastP on this gene
Accession: ADW06000
Location: 5304494-5305765
NCBI BlastP on this gene
Sfla_4598
transcriptional regulator, LacI family
Accession: ADW05999
Location: 5303090-5304235
NCBI BlastP on this gene
Accession: ADW05999
Location: 5303090-5304235
NCBI BlastP on this gene
Sfla_4597
Sfla_4596
(Glutamate--ammonia-ligase) adenylyltransferase
Accession: ADW05997
Location: 5298934-5301930
NCBI BlastP on this gene
Accession: ADW05997
Location: 5298934-5301930
NCBI BlastP on this gene
Sfla_4595
modification methylase, HemK family
Accession: ADW05996
Location: 5297986-5298774
NCBI BlastP on this gene
Accession: ADW05996
Location: 5297986-5298774
NCBI BlastP on this gene
Sfla_4594
Sfla_4593
major facilitator superfamily MFS 1
Accession: ADW05994
Location: 5295481-5296734
NCBI BlastP on this gene
Accession: ADW05994
Location: 5295481-5296734
NCBI BlastP on this gene
Sfla_4592
glutamine synthetase catalytic region
Accession: ADW05993
Location: 5293973-5295334
NCBI BlastP on this gene
Accession: ADW05993
Location: 5293973-5295334
NCBI BlastP on this gene
Sfla_4591
Protein of unknown function DUF3105
Accession: ADW05992
Location: 5293052-5293726
NCBI BlastP on this gene
Accession: ADW05992
Location: 5293052-5293726
NCBI BlastP on this gene
Sfla_4590
protein of unknown function DUF305
Accession: ADW05991
Location: 5292378-5293055
NCBI BlastP on this gene
Accession: ADW05991
Location: 5292378-5293055
NCBI BlastP on this gene
Sfla_4589
putative signal transduction protein with CBS domains
Accession: ADW05990
Location: 5291827-5292252
NCBI BlastP on this gene
Accession: ADW05990
Location: 5291827-5292252
NCBI BlastP on this gene
Sfla_4588
Sfla_4587
Sfla_4586
RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: ADW05987
Location: 5287451-5288119
NCBI BlastP on this gene
Accession: ADW05987
Location: 5287451-5288119
NCBI BlastP on this gene
Sfla_4585
Sfla_4584
461. : CP035945 Blautia producta strain PMF1 chromosome Total score: 2.5 Cumulative Blast bit score: 373
PMF13cell1_02540
ydjH_2
putative 6-phospho-beta-glucosidase
Accession: QBE96989
Location: 2882534-2883910
NCBI BlastP on this gene
Accession: QBE96989
Location: 2882534-2883910
NCBI BlastP on this gene
licH_2
PMF13cell1_02537
yxeP_1
N-acetyl-alpha-D-glucosaminyl L-malate deacetylase 1
Accession: QBE96986
Location: 2879085-2879762
NCBI BlastP on this gene
Accession: QBE96986
Location: 2879085-2879762
NCBI BlastP on this gene
bshB1
L-arabinose transport system permease protein AraQ
Accession: QBE96985
Location: 2878256-2879071
NCBI BlastP on this gene
Accession: QBE96985
Location: 2878256-2879071
NCBI BlastP on this gene
araQ_30
Lactose transport system permease protein LacF
Accession: QBE96984
Location: 2877320-2878222
NCBI BlastP on this gene
Accession: QBE96984
Location: 2877320-2878222
NCBI BlastP on this gene
lacF_18
Putative ABC transporter substrate-binding protein YesO
Accession: QBE96983
Location: 2875959-2877245
NCBI BlastP on this gene
Accession: QBE96983
Location: 2875959-2877245
NCBI BlastP on this gene
yesO_6
ccpA_6
gutB_1
Bifunctional purine biosynthesis protein PurH
Accession: QBE96980
Location: 2872205-2873383
NCBI BlastP on this gene
Accession: QBE96980
Location: 2872205-2873383
NCBI BlastP on this gene
purH
PMF13cell1_02528
Ribosomal RNA large subunit methyltransferase I
Accession: QBE96978
Location: 2869806-2871005
NCBI BlastP on this gene
Accession: QBE96978
Location: 2869806-2871005
NCBI BlastP on this gene
rlmI
PMF13cell1_02526
PMF13cell1_02525
map_2
PMF13cell1_02523
PMF13cell1_02522
PMF13cell1_02521
Inner membrane ABC transporter permease protein YcjP
Accession: QBE96971
Location: 2864487-2865332
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 2e-54
NCBI BlastP on this gene
Accession: QBE96971
Location: 2864487-2865332
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 2e-54
NCBI BlastP on this gene
ycjP_6
Inner membrane ABC transporter permease protein YcjO
Accession: QBE96970
Location: 2863557-2864474
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 184
Sequence coverage: 100 %
E-value: 3e-52
NCBI BlastP on this gene
Accession: QBE96970
Location: 2863557-2864474
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 184
Sequence coverage: 100 %
E-value: 3e-52
NCBI BlastP on this gene
ycjO_3
PMF13cell1_02518
Multiple sugar-binding protein
Accession: QBE96968
Location: 2860640-2861938
NCBI BlastP on this gene
Accession: QBE96968
Location: 2860640-2861938
NCBI BlastP on this gene
msmE_19
putative sensor-like histidine kinase
Accession: QBE96967
Location: 2858814-2860628
NCBI BlastP on this gene
Accession: QBE96967
Location: 2858814-2860628
NCBI BlastP on this gene
PMF13cell1_02516
putative response regulatory protein
Accession: QBE96966
Location: 2857235-2858809
NCBI BlastP on this gene
Accession: QBE96966
Location: 2857235-2858809
NCBI BlastP on this gene
PMF13cell1_02515
rbr1_2
cmk_4
Gamma-glutamylputrescine oxidoreductase
Accession: QBE96963
Location: 2853801-2855261
NCBI BlastP on this gene
Accession: QBE96963
Location: 2853801-2855261
NCBI BlastP on this gene
puuB
PMF13cell1_02511
Cyclic pyranopterin monophosphate synthase
Accession: QBE96961
Location: 2852210-2853190
NCBI BlastP on this gene
Accession: QBE96961
Location: 2852210-2853190
NCBI BlastP on this gene
moaA_2
rbr1_1
PMF13cell1_02508
Acetolactate synthase large subunit
Accession: QBE96958
Location: 2848882-2850600
NCBI BlastP on this gene
Accession: QBE96958
Location: 2848882-2850600
NCBI BlastP on this gene
ilvB_1
Acetolactate synthase isozyme 3 small subunit
Accession: QBE96957
Location: 2848371-2848868
NCBI BlastP on this gene
Accession: QBE96957
Location: 2848371-2848868
NCBI BlastP on this gene
ilvH_1
Cyclic di-GMP phosphodiesterase Gmr
Accession: QBE96956
Location: 2845844-2848162
NCBI BlastP on this gene
Accession: QBE96956
Location: 2845844-2848162
NCBI BlastP on this gene
gmr_4
PMF13cell1_02504
Adenosine monophosphate-protein transferase SoFic
Accession: QBE96954
Location: 2843889-2844881
NCBI BlastP on this gene
Accession: QBE96954
Location: 2843889-2844881
NCBI BlastP on this gene
fic
462. : CP030862 Streptomyces globosus strain LZH-48 chromosome Total score: 2.5 Cumulative Blast bit score: 373
C0216_24075
C0216_24080
C0216_24085
C0216_24090
C0216_24095
C0216_24100
TetR family transcriptional regulator
Accession: AXE26132
Location: 5433044-5433799
NCBI BlastP on this gene
Accession: AXE26132
Location: 5433044-5433799
NCBI BlastP on this gene
C0216_24105
C0216_24110
DUF3372 domain-containing protein
Accession: AXE26134
Location: 5434344-5439752
NCBI BlastP on this gene
Accession: AXE26134
Location: 5434344-5439752
NCBI BlastP on this gene
C0216_24115
C0216_24120
LacI family transcriptional regulator
Accession: AXE26136
Location: 5441840-5442871
NCBI BlastP on this gene
Accession: AXE26136
Location: 5441840-5442871
NCBI BlastP on this gene
C0216_24125
C0216_24130
ABC transporter permease
Accession: AXE26138
Location: 5444547-5445455
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 91 %
E-value: 2e-57
NCBI BlastP on this gene
Accession: AXE26138
Location: 5444547-5445455
BlastP hit with WP_039959879.1
Percentage identity: 39 %
BlastP bit score: 197
Sequence coverage: 91 %
E-value: 2e-57
NCBI BlastP on this gene
C0216_24135
sugar ABC transporter permease
Accession: AXE26139
Location: 5445458-5446489
BlastP hit with WP_039959892.1
Percentage identity: 32 %
BlastP bit score: 176
Sequence coverage: 102 %
E-value: 1e-48
NCBI BlastP on this gene
Accession: AXE26139
Location: 5445458-5446489
BlastP hit with WP_039959892.1
Percentage identity: 32 %
BlastP bit score: 176
Sequence coverage: 102 %
E-value: 1e-48
NCBI BlastP on this gene
C0216_24140
sugar ABC transporter substrate-binding protein
Accession: AXE26140
Location: 5446498-5447772
NCBI BlastP on this gene
Accession: AXE26140
Location: 5446498-5447772
NCBI BlastP on this gene
C0216_24145
C0216_24150
LacI family transcriptional regulator
Accession: AXE27717
Location: 5449649-5450695
NCBI BlastP on this gene
Accession: AXE27717
Location: 5449649-5450695
NCBI BlastP on this gene
C0216_24155
C0216_24160
GNAT family N-acetyltransferase
Accession: AXE26143
Location: 5451674-5452108
NCBI BlastP on this gene
Accession: AXE26143
Location: 5451674-5452108
NCBI BlastP on this gene
C0216_24165
C0216_24170
DUF397 domain-containing protein
Accession: AXE26145
Location: 5453350-5453607
NCBI BlastP on this gene
Accession: AXE26145
Location: 5453350-5453607
NCBI BlastP on this gene
C0216_24175
C0216_24180
C0216_24185
C0216_24190
C0216_24195
C0216_24200
C0216_24205
C0216_24210
C0216_24215
DUF397 domain-containing protein
Accession: AXE26151
Location: 5464025-5464300
NCBI BlastP on this gene
Accession: AXE26151
Location: 5464025-5464300
NCBI BlastP on this gene
C0216_24220
C0216_24225
C0216_24230
helix-turn-helix domain-containing protein
Accession: AXE26153
Location: 5465925-5466791
NCBI BlastP on this gene
Accession: AXE26153
Location: 5465925-5466791
NCBI BlastP on this gene
C0216_24235
463. : CP015775 Ochrobactrum pseudogrignonense strain K8 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 372
A8A54_00995
UDP-N-acetylglucosamine 1-carboxyvinyltransferase
Accession: ANG95186
Location: 214357-215649
NCBI BlastP on this gene
Accession: ANG95186
Location: 214357-215649
NCBI BlastP on this gene
A8A54_01000
A8A54_01005
A8A54_01020
efflux transporter periplasmic adaptor subunit
Accession: ANG95189
Location: 218866-220014
NCBI BlastP on this gene
Accession: ANG95189
Location: 218866-220014
NCBI BlastP on this gene
A8A54_01025
A8A54_01030
nitrate ABC transporter substrate-binding protein
Accession: ANG95191
Location: 223392-224390
NCBI BlastP on this gene
Accession: ANG95191
Location: 223392-224390
NCBI BlastP on this gene
A8A54_01035
A8A54_01040
A8A54_01045
sulfonate ABC transporter ATP-binding protein
Accession: ANG95194
Location: 226309-227130
NCBI BlastP on this gene
Accession: ANG95194
Location: 226309-227130
NCBI BlastP on this gene
A8A54_01050
A8A54_01055
A8A54_01060
TetR family transcriptional regulator
Accession: ANG95197
Location: 230167-230847
NCBI BlastP on this gene
Accession: ANG95197
Location: 230167-230847
NCBI BlastP on this gene
A8A54_01065
DNA mismatch repair protein MutT
Accession: ANG95198
Location: 230844-231242
NCBI BlastP on this gene
Accession: ANG95198
Location: 230844-231242
NCBI BlastP on this gene
A8A54_01070
glycerol-3-phosphate ABC transporter ATP-binding protein
Accession: ANG95199
Location: 231239-232312
NCBI BlastP on this gene
Accession: ANG95199
Location: 231239-232312
NCBI BlastP on this gene
A8A54_01075
ABC transporter permease
Accession: ANG95200
Location: 232327-233160
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: ANG95200
Location: 232327-233160
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
A8A54_01080
ABC transporter permease
Accession: ANG95201
Location: 233165-234118
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 178
Sequence coverage: 90 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: ANG95201
Location: 233165-234118
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 178
Sequence coverage: 90 %
E-value: 1e-49
NCBI BlastP on this gene
A8A54_01085
ABC transporter substrate-binding protein
Accession: ANG95202
Location: 234194-235453
NCBI BlastP on this gene
Accession: ANG95202
Location: 234194-235453
NCBI BlastP on this gene
A8A54_01090
A8A54_01095
GntR family transcriptional regulator
Accession: ANG95204
Location: 236655-237371
NCBI BlastP on this gene
Accession: ANG95204
Location: 236655-237371
NCBI BlastP on this gene
A8A54_01100
A8A54_01105
A8A54_01110
dihydrodipicolinate synthase family protein
Accession: ANG95207
Location: 240058-240960
NCBI BlastP on this gene
Accession: ANG95207
Location: 240058-240960
NCBI BlastP on this gene
A8A54_01115
GntR family transcriptional regulator
Accession: ANG98310
Location: 241002-241661
NCBI BlastP on this gene
Accession: ANG98310
Location: 241002-241661
NCBI BlastP on this gene
A8A54_01120
ABC transporter substrate-binding protein
Accession: ANG95208
Location: 241739-243073
NCBI BlastP on this gene
Accession: ANG95208
Location: 241739-243073
NCBI BlastP on this gene
A8A54_01125
A8A54_01130
A8A54_01135
sugar ABC transporter ATP-binding protein
Accession: ANG95211
Location: 244852-245910
NCBI BlastP on this gene
Accession: ANG95211
Location: 244852-245910
NCBI BlastP on this gene
A8A54_01140
dihydropyrimidine dehydrogenase subunit B
Accession: ANG95212
Location: 246060-247370
NCBI BlastP on this gene
Accession: ANG95212
Location: 246060-247370
NCBI BlastP on this gene
A8A54_01145
dihydropyrimidine dehydrogenase
Accession: ANG95213
Location: 247382-248884
NCBI BlastP on this gene
Accession: ANG95213
Location: 247382-248884
NCBI BlastP on this gene
A8A54_01150
A8A54_01155
A8A54_01160
A8A54_01165
A8A54_01170
A8A54_01175
464. : CP044970 Ochrobactrum anthropi strain T16R-87 chromosome 1 Total score: 2.5 Cumulative Blast bit score: 371
ureC
urease accessory protein UreE
Accession: QFP62530
Location: 1054703-1055224
NCBI BlastP on this gene
Accession: QFP62530
Location: 1054703-1055224
NCBI BlastP on this gene
ureE
urease accessory protein UreF
Accession: QFP64003
Location: 1054045-1054731
NCBI BlastP on this gene
Accession: QFP64003
Location: 1054045-1054731
NCBI BlastP on this gene
FT787_05165
urease accessory protein UreG
Accession: QFP62529
Location: 1053428-1054048
NCBI BlastP on this gene
Accession: QFP62529
Location: 1053428-1054048
NCBI BlastP on this gene
ureG
deoxyribodipyrimidine photo-lyase
Accession: QFP62528
Location: 1052021-1053427
NCBI BlastP on this gene
Accession: QFP62528
Location: 1052021-1053427
NCBI BlastP on this gene
FT787_05155
HAMP domain-containing histidine kinase
Accession: QFP62527
Location: 1050064-1051617
NCBI BlastP on this gene
Accession: QFP62527
Location: 1050064-1051617
NCBI BlastP on this gene
FT787_05145
efflux RND transporter periplasmic adaptor subunit
Accession: QFP62526
Location: 1048556-1049683
NCBI BlastP on this gene
Accession: QFP62526
Location: 1048556-1049683
NCBI BlastP on this gene
FT787_05140
efflux RND transporter permease subunit
Accession: QFP62525
Location: 1045393-1048545
NCBI BlastP on this gene
Accession: QFP62525
Location: 1045393-1048545
NCBI BlastP on this gene
FT787_05135
ABC transporter substrate-binding protein
Accession: QFP62524
Location: 1044343-1045341
NCBI BlastP on this gene
Accession: QFP62524
Location: 1044343-1045341
NCBI BlastP on this gene
FT787_05130
FT787_05125
FT787_05120
ABC transporter ATP-binding protein
Accession: QFP62521
Location: 1041621-1042442
NCBI BlastP on this gene
Accession: QFP62521
Location: 1041621-1042442
NCBI BlastP on this gene
FT787_05115
hydA
FT787_05105
HTH-type transcriptional regulator RutR
Accession: QFP62518
Location: 1037905-1038603
NCBI BlastP on this gene
Accession: QFP62518
Location: 1037905-1038603
NCBI BlastP on this gene
rutR
NUDIX domain-containing protein
Accession: QFP62517
Location: 1037510-1037908
NCBI BlastP on this gene
Accession: QFP62517
Location: 1037510-1037908
NCBI BlastP on this gene
FT787_05095
sn-glycerol-3-phosphate ABC transporter ATP-binding protein UgpC
Accession: QFP62516
Location: 1036440-1037513
NCBI BlastP on this gene
Accession: QFP62516
Location: 1036440-1037513
NCBI BlastP on this gene
ugpC
carbohydrate ABC transporter permease
Accession: QFP62515
Location: 1035488-1036321
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
Accession: QFP62515
Location: 1035488-1036321
BlastP hit with WP_039959879.1
Percentage identity: 38 %
BlastP bit score: 194
Sequence coverage: 100 %
E-value: 1e-56
NCBI BlastP on this gene
FT787_05085
sugar ABC transporter permease
Accession: QFP62514
Location: 1034524-1035483
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 177
Sequence coverage: 90 %
E-value: 2e-49
NCBI BlastP on this gene
Accession: QFP62514
Location: 1034524-1035483
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 177
Sequence coverage: 90 %
E-value: 2e-49
NCBI BlastP on this gene
FT787_05080
sugar ABC transporter substrate-binding protein
Accession: QFP62513
Location: 1033189-1034448
NCBI BlastP on this gene
Accession: QFP62513
Location: 1033189-1034448
NCBI BlastP on this gene
FT787_05075
FT787_05070
FadR family transcriptional regulator
Accession: QFP62511
Location: 1031271-1031987
NCBI BlastP on this gene
Accession: QFP62511
Location: 1031271-1031987
NCBI BlastP on this gene
FT787_05065
FT787_05060
FT787_05055
FT787_05050
dihydrodipicolinate synthase family protein
Accession: QFP62509
Location: 1027809-1028711
NCBI BlastP on this gene
Accession: QFP62509
Location: 1027809-1028711
NCBI BlastP on this gene
FT787_05045
GntR family transcriptional regulator
Accession: QFP62508
Location: 1027108-1027770
NCBI BlastP on this gene
Accession: QFP62508
Location: 1027108-1027770
NCBI BlastP on this gene
FT787_05040
sugar ABC transporter substrate-binding protein
Accession: QFP62507
Location: 1025697-1027031
NCBI BlastP on this gene
Accession: QFP62507
Location: 1025697-1027031
NCBI BlastP on this gene
FT787_05035
sugar ABC transporter permease
Accession: QFP62506
Location: 1024762-1025700
NCBI BlastP on this gene
Accession: QFP62506
Location: 1024762-1025700
NCBI BlastP on this gene
FT787_05030
carbohydrate ABC transporter permease
Accession: QFP62505
Location: 1023934-1024752
NCBI BlastP on this gene
Accession: QFP62505
Location: 1023934-1024752
NCBI BlastP on this gene
FT787_05025
sn-glycerol-3-phosphate ABC transporter ATP-binding protein UgpC
Accession: QFP62504
Location: 1022860-1023918
NCBI BlastP on this gene
Accession: QFP62504
Location: 1022860-1023918
NCBI BlastP on this gene
ugpC
NAD-dependent dihydropyrimidine dehydrogenase subunit PreA
Accession: QFP62503
Location: 1021513-1022823
NCBI BlastP on this gene
Accession: QFP62503
Location: 1021513-1022823
NCBI BlastP on this gene
preA
NAD(P)-dependent oxidoreductase
Accession: QFP62502
Location: 1019999-1021501
NCBI BlastP on this gene
Accession: QFP62502
Location: 1019999-1021501
NCBI BlastP on this gene
FT787_05010
FT787_05005
carbon monoxide dehydrogenase subunit G
Accession: QFP62500
Location: 1018285-1018743
NCBI BlastP on this gene
Accession: QFP62500
Location: 1018285-1018743
NCBI BlastP on this gene
FT787_05000
glucose-6-phosphate isomerase
Accession: QFP62499
Location: 1016485-1018134
NCBI BlastP on this gene
Accession: QFP62499
Location: 1016485-1018134
NCBI BlastP on this gene
FT787_04995
succinylglutamate desuccinylase/aspartoacylase family protein
Accession: QFP62498
Location: 1015341-1016402
NCBI BlastP on this gene
Accession: QFP62498
Location: 1015341-1016402
NCBI BlastP on this gene
FT787_04990
FT787_04985
465. : CP021332 Maritalea myrionectae strain HL2708#5 plasmid pHL2708Y3 Total score: 2.5 Cumulative Blast bit score: 371
MXMO3_03638
NADPH-dependent FMN reductase ArsH
Accession: AVX06142
Location: 27302-28024
NCBI BlastP on this gene
Accession: AVX06142
Location: 27302-28024
NCBI BlastP on this gene
MXMO3_03639
arsenical-resistance protein Acr3
Accession: AVX06143
Location: 28032-29093
NCBI BlastP on this gene
Accession: AVX06143
Location: 28032-29093
NCBI BlastP on this gene
MXMO3_03640
arsenate reductase (glutaredoxin)
Accession: AVX06144
Location: 29094-29600
NCBI BlastP on this gene
Accession: AVX06144
Location: 29094-29600
NCBI BlastP on this gene
MXMO3_03641
arsenate reductase (glutaredoxin)
Accession: AVX06145
Location: 29590-29940
NCBI BlastP on this gene
Accession: AVX06145
Location: 29590-29940
NCBI BlastP on this gene
MXMO3_03642
MXMO3_03643
MXMO3_03644
glutathione transport system permease protein GsiC
Accession: AVX06148
Location: 32425-33366
NCBI BlastP on this gene
Accession: AVX06148
Location: 32425-33366
NCBI BlastP on this gene
MXMO3_03645
putative peptide transport system permease protein
Accession: AVX06149
Location: 33367-34218
NCBI BlastP on this gene
Accession: AVX06149
Location: 33367-34218
NCBI BlastP on this gene
MXMO3_03646
putative peptide import ATP-binding protein
Accession: AVX06150
Location: 34228-35223
NCBI BlastP on this gene
Accession: AVX06150
Location: 34228-35223
NCBI BlastP on this gene
MXMO3_03647
dipeptide transport ATP-binding protein DppF
Accession: AVX06151
Location: 35220-36206
NCBI BlastP on this gene
Accession: AVX06151
Location: 35220-36206
NCBI BlastP on this gene
MXMO3_03648
MXMO3_03649
MXMO3_03650
ADP-ribosyl-[dinitrogen reductase] hydrolase
Accession: AVX06154
Location: 39690-40097
NCBI BlastP on this gene
Accession: AVX06154
Location: 39690-40097
NCBI BlastP on this gene
MXMO3_03651
HTH-type transcriptional regulator RafR
Accession: AVX06155
Location: 40227-41258
NCBI BlastP on this gene
Accession: AVX06155
Location: 40227-41258
NCBI BlastP on this gene
MXMO3_03652
MXMO3_03653
MXMO3_03654
mannose-1-phosphate guanylyltransferase
Accession: AVX06158
Location: 45326-45682
NCBI BlastP on this gene
Accession: AVX06158
Location: 45326-45682
NCBI BlastP on this gene
MXMO3_03655
MXMO3_03656
inner membrane ABC transporter permease protein
Accession: AVX06160
Location: 46724-47563
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 194
Sequence coverage: 94 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: AVX06160
Location: 46724-47563
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 194
Sequence coverage: 94 %
E-value: 2e-56
NCBI BlastP on this gene
MXMO3_03657
Sn-glycerol-3-phosphate transport system permease protein UgpA
Accession: AVX06161
Location: 47565-48497
BlastP hit with WP_039959892.1
Percentage identity: 37 %
BlastP bit score: 177
Sequence coverage: 82 %
E-value: 1e-49
NCBI BlastP on this gene
Accession: AVX06161
Location: 47565-48497
BlastP hit with WP_039959892.1
Percentage identity: 37 %
BlastP bit score: 177
Sequence coverage: 82 %
E-value: 1e-49
NCBI BlastP on this gene
MXMO3_03658
MXMO3_03659
putative HTH-type transcriptional regulator YcbG
Accession: AVX06163
Location: 49914-50660
NCBI BlastP on this gene
Accession: AVX06163
Location: 49914-50660
NCBI BlastP on this gene
MXMO3_03660
mercuric resistance operon regulatory protein
Accession: AVX06164
Location: 50815-51222
NCBI BlastP on this gene
Accession: AVX06164
Location: 50815-51222
NCBI BlastP on this gene
MXMO3_03661
MXMO3_03662
mercuric transport protein periplasmic component
Accession: AVX06166
Location: 51942-52235
NCBI BlastP on this gene
Accession: AVX06166
Location: 51942-52235
NCBI BlastP on this gene
MXMO3_03663
MXMO3_03664
MXMO3_03665
MXMO3_03666
peroxyureidoacrylate/ureidoacrylate amidohydrolase
Accession: AVX06170
Location: 60594-61235
NCBI BlastP on this gene
Accession: AVX06170
Location: 60594-61235
NCBI BlastP on this gene
MXMO3_03667
MXMO3_03668
high-affinity branched-chain amino acid transport system permease protein BraD
Accession: AVX06172
Location: 62016-62909
NCBI BlastP on this gene
Accession: AVX06172
Location: 62016-62909
NCBI BlastP on this gene
MXMO3_03669
spermidine/putrescine import ATP-binding protein PotA
Accession: AVX06173
Location: 62911-63630
NCBI BlastP on this gene
Accession: AVX06173
Location: 62911-63630
NCBI BlastP on this gene
MXMO3_03670
MXMO3_03671
high-affinity branched-chain amino acid transport system permease protein BraE
Accession: AVX06175
Location: 64836-66614
NCBI BlastP on this gene
Accession: AVX06175
Location: 64836-66614
NCBI BlastP on this gene
MXMO3_03672
MXMO3_03673
MXMO3_03674
466. : CP015736 Shinella sp. HZN7 Total score: 2.5 Cumulative Blast bit score: 371
shn_16790
shn_16795
shn_16800
shn_16805
shn_16810
shn_16815
shn_16820
shn_16825
spermidine/putrescine ABC transporter ATP-binding protein
Accession: ANH05539
Location: 3499244-3500242
NCBI BlastP on this gene
Accession: ANH05539
Location: 3499244-3500242
NCBI BlastP on this gene
shn_16830
shn_16835
shn_16840
shn_16845
LysR family transcriptional regulator
Accession: ANH05543
Location: 3503131-3504066
NCBI BlastP on this gene
Accession: ANH05543
Location: 3503131-3504066
NCBI BlastP on this gene
shn_16850
shn_16855
mandelate racemase/muconate lactonizing protein
Accession: ANH05545
Location: 3505480-3506586
NCBI BlastP on this gene
Accession: ANH05545
Location: 3505480-3506586
NCBI BlastP on this gene
shn_16860
hypothetical protein
Accession: ANH06816
Location: 3506607-3507440
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 187
Sequence coverage: 97 %
E-value: 8e-54
NCBI BlastP on this gene
Accession: ANH06816
Location: 3506607-3507440
BlastP hit with WP_039959879.1
Percentage identity: 34 %
BlastP bit score: 187
Sequence coverage: 97 %
E-value: 8e-54
NCBI BlastP on this gene
shn_16865
hypothetical protein
Accession: ANH05546
Location: 3507497-3508393
BlastP hit with WP_039959892.1
Percentage identity: 38 %
BlastP bit score: 184
Sequence coverage: 83 %
E-value: 3e-52
NCBI BlastP on this gene
Accession: ANH05546
Location: 3507497-3508393
BlastP hit with WP_039959892.1
Percentage identity: 38 %
BlastP bit score: 184
Sequence coverage: 83 %
E-value: 3e-52
NCBI BlastP on this gene
shn_16870
shn_16875
shn_16880
shn_16885
shn_16890
shn_16895
shn_16900
sugar ABC transporter ATP-binding protein
Accession: ANH05553
Location: 3515516-3516598
NCBI BlastP on this gene
Accession: ANH05553
Location: 3515516-3516598
NCBI BlastP on this gene
shn_16905
shn_16910
shn_16915
ABC transporter substrate-binding protein
Accession: ANH06817
Location: 3518399-3519910
NCBI BlastP on this gene
Accession: ANH06817
Location: 3518399-3519910
NCBI BlastP on this gene
shn_16920
shn_16925
shn_16930
dihydrodipicolinate synthase family protein
Accession: shn_16935
Location: 3523645-3524020
NCBI BlastP on this gene
Accession: shn_16935
Location: 3523645-3524020
NCBI BlastP on this gene
shn_16935
GntR family transcriptional regulator
Accession: ANH05558
Location: 3524422-3525141
NCBI BlastP on this gene
Accession: ANH05558
Location: 3524422-3525141
NCBI BlastP on this gene
shn_16940
pyrroline-5-carboxylate reductase
Accession: ANH05559
Location: 3525144-3525926
NCBI BlastP on this gene
Accession: ANH05559
Location: 3525144-3525926
NCBI BlastP on this gene
shn_16945
autoinducer-2 (AI-2) modifying protein LsrG
Accession: shn_16950
Location: 3526030-3526197
NCBI BlastP on this gene
Accession: shn_16950
Location: 3526030-3526197
NCBI BlastP on this gene
shn_16950
shn_16955
FAD/NAD(P)-binding oxidoreductase
Accession: ANH05561
Location: 3526800-3528266
NCBI BlastP on this gene
Accession: ANH05561
Location: 3526800-3528266
NCBI BlastP on this gene
shn_16960
sarcosine oxidase subunit alpha
Accession: ANH05562
Location: 3528263-3528565
NCBI BlastP on this gene
Accession: ANH05562
Location: 3528263-3528565
NCBI BlastP on this gene
shn_16965
467. : LT907981 Streptomyces sp. 2323.1 genome assembly, chromosome: I. Total score: 2.5 Cumulative Blast bit score: 369
alpha-N-acetylglucosaminidase
Accession: SOE14162
Location: 5847539-5850685
NCBI BlastP on this gene
Accession: SOE14162
Location: 5847539-5850685
NCBI BlastP on this gene
SAMN06272775_5123
SAMN06272775_5122
SAMN06272775_5121
transcriptional regulator, TetR family
Accession: SOE14159
Location: 5844169-5844864
NCBI BlastP on this gene
Accession: SOE14159
Location: 5844169-5844864
NCBI BlastP on this gene
SAMN06272775_5120
Acetyltransferase (GNAT) family protein
Accession: SOE14158
Location: 5843600-5844112
NCBI BlastP on this gene
Accession: SOE14158
Location: 5843600-5844112
NCBI BlastP on this gene
SAMN06272775_5119
alpha-1,6-glucosidases, pullulanase-type
Accession: SOE14157
Location: 5840824-5843466
NCBI BlastP on this gene
Accession: SOE14157
Location: 5840824-5843466
NCBI BlastP on this gene
SAMN06272775_5118
SAMN06272775_5117
SAMN06272775_5116
Cupin domain-containing protein
Accession: SOE14153
Location: 5838589-5839062
NCBI BlastP on this gene
Accession: SOE14153
Location: 5838589-5839062
NCBI BlastP on this gene
SAMN06272775_5115
SAMN06272775_5113
SAMN06272775_5112
Transposase DDE domain-containing protein
Accession: SOE14150
Location: 5837176-5837532
NCBI BlastP on this gene
Accession: SOE14150
Location: 5837176-5837532
NCBI BlastP on this gene
SAMN06272775_5111
Histidine kinase-like ATPase domain-containing protein
Accession: SOE14149
Location: 5836135-5836599
NCBI BlastP on this gene
Accession: SOE14149
Location: 5836135-5836599
NCBI BlastP on this gene
SAMN06272775_5110
Helix-turn-helix domain-containing protein
Accession: SOE14148
Location: 5835150-5835980
NCBI BlastP on this gene
Accession: SOE14148
Location: 5835150-5835980
NCBI BlastP on this gene
SAMN06272775_5109
SAMN06272775_5108
SAMN06272775_5107
transcriptional regulator, LacI family
Accession: SOE14145
Location: 5832559-5833617
NCBI BlastP on this gene
Accession: SOE14145
Location: 5832559-5833617
NCBI BlastP on this gene
SAMN06272775_5106
SAMN06272775_5105
arabinogalactan oligomer / maltooligosaccharide transport system permease protein
Accession: SOE14143
Location: 5830084-5830971
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 3e-54
NCBI BlastP on this gene
Accession: SOE14143
Location: 5830084-5830971
BlastP hit with WP_039959879.1
Percentage identity: 37 %
BlastP bit score: 189
Sequence coverage: 99 %
E-value: 3e-54
NCBI BlastP on this gene
SAMN06272775_5104
arabinogalactan oligomer / maltooligosaccharide transport system permease protein
Accession: SOE14142
Location: 5829146-5830081
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 180
Sequence coverage: 102 %
E-value: 2e-50
NCBI BlastP on this gene
Accession: SOE14142
Location: 5829146-5830081
BlastP hit with WP_039959892.1
Percentage identity: 33 %
BlastP bit score: 180
Sequence coverage: 102 %
E-value: 2e-50
NCBI BlastP on this gene
SAMN06272775_5103
carbohydrate ABC transporter substrate-binding protein, CUT1 family
Accession: SOE14140
Location: 5827872-5829149
NCBI BlastP on this gene
Accession: SOE14140
Location: 5827872-5829149
NCBI BlastP on this gene
SAMN06272775_5102
SAMN06272775_5101
transcriptional regulator, LacI family
Accession: SOE14138
Location: 5824905-5825954
NCBI BlastP on this gene
Accession: SOE14138
Location: 5824905-5825954
NCBI BlastP on this gene
SAMN06272775_5100
SAMN06272775_5099
SAMN06272775_5098
SAMN06272775_5097
glutamate-ammonia-ligase adenylyltransferase
Accession: SOE14134
Location: 5819176-5822202
NCBI BlastP on this gene
Accession: SOE14134
Location: 5819176-5822202
NCBI BlastP on this gene
SAMN06272775_5096
SAMN06272775_5095
Pimeloyl-ACP methyl ester carboxylesterase
Accession: SOE14132
Location: 5816298-5817413
NCBI BlastP on this gene
Accession: SOE14132
Location: 5816298-5817413
NCBI BlastP on this gene
SAMN06272775_5094
Multicopper oxidase with three cupredoxin
Accession: SOE14131
Location: 5814307-5815950
NCBI BlastP on this gene
Accession: SOE14131
Location: 5814307-5815950
NCBI BlastP on this gene
SAMN06272775_5093
NAD+ synthase (glutamine-hydrolysing)
Accession: SOE14130
Location: 5812487-5814241
NCBI BlastP on this gene
Accession: SOE14130
Location: 5812487-5814241
NCBI BlastP on this gene
SAMN06272775_5092
MFS transporter, DHA1 family, inner membrane transport protein
Accession: SOE14128
Location: 5811196-5812431
NCBI BlastP on this gene
Accession: SOE14128
Location: 5811196-5812431
NCBI BlastP on this gene
SAMN06272775_5091
vancomycin resistance protein VanJ
Accession: SOE14127
Location: 5809988-5811007
NCBI BlastP on this gene
Accession: SOE14127
Location: 5809988-5811007
NCBI BlastP on this gene
SAMN06272775_5090
468. : CP002131 Thermosediminibacter oceani DSM 16646 Total score: 2.5 Cumulative Blast bit score: 369
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: ADL08500
Location: 1767983-1769248
NCBI BlastP on this gene
Accession: ADL08500
Location: 1767983-1769248
NCBI BlastP on this gene
Toce_1766
Toce_1765
Toce_1764
quorum-sensing autoinducer 2 (AI-2), LuxS
Accession: ADL08497
Location: 1763264-1763719
NCBI BlastP on this gene
Accession: ADL08497
Location: 1763264-1763719
NCBI BlastP on this gene
Toce_1763
Toce_1762
phosphoesterase PA-phosphatase related protein
Accession: ADL08495
Location: 1762122-1762676
NCBI BlastP on this gene
Accession: ADL08495
Location: 1762122-1762676
NCBI BlastP on this gene
Toce_1761
Toce_1760
Toce_1759
ATP-dependent DNA helicase PcrA
Accession: ADL08492
Location: 1758075-1760270
NCBI BlastP on this gene
Accession: ADL08492
Location: 1758075-1760270
NCBI BlastP on this gene
Toce_1758
Toce_1757
conserved hypothetical protein
Accession: ADL08490
Location: 1755530-1755979
NCBI BlastP on this gene
Accession: ADL08490
Location: 1755530-1755979
NCBI BlastP on this gene
Toce_1756
major facilitator superfamily MFS 1
Accession: ADL08489
Location: 1754147-1755400
NCBI BlastP on this gene
Accession: ADL08489
Location: 1754147-1755400
NCBI BlastP on this gene
Toce_1755
conserved hypothetical protein
Accession: ADL08488
Location: 1753494-1754036
NCBI BlastP on this gene
Accession: ADL08488
Location: 1753494-1754036
NCBI BlastP on this gene
Toce_1754
4Fe-4S ferredoxin iron-sulfur binding domain protein
Accession: ADL08487
Location: 1752621-1753376
NCBI BlastP on this gene
Accession: ADL08487
Location: 1752621-1753376
NCBI BlastP on this gene
Toce_1753
conserved hypothetical protein
Accession: ADL08486
Location: 1752164-1752334
NCBI BlastP on this gene
Accession: ADL08486
Location: 1752164-1752334
NCBI BlastP on this gene
Toce_1752
death-on-curing family protein
Accession: ADL08485
Location: 1751781-1752164
NCBI BlastP on this gene
Accession: ADL08485
Location: 1751781-1752164
NCBI BlastP on this gene
Toce_1751
Toce_1750
Peptidoglycan-binding domain 1 protein
Accession: ADL08483
Location: 1749229-1750689
NCBI BlastP on this gene
Accession: ADL08483
Location: 1749229-1750689
NCBI BlastP on this gene
Toce_1749
Toce_1748
RNA polymerase sigma factor, sigma-70 family
Accession: ADL08481
Location: 1748032-1748457
NCBI BlastP on this gene
Accession: ADL08481
Location: 1748032-1748457
NCBI BlastP on this gene
Toce_1747
carbohydrate ABC transporter membrane protein 2, CUT1 family
Accession: ADL08480
Location: 1747168-1748001
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 189
Sequence coverage: 100 %
E-value: 1e-54
NCBI BlastP on this gene
Accession: ADL08480
Location: 1747168-1748001
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 189
Sequence coverage: 100 %
E-value: 1e-54
NCBI BlastP on this gene
Toce_1746
carbohydrate ABC transporter membrane protein 1, CUT1 family
Accession: ADL08479
Location: 1746219-1747163
BlastP hit with WP_039959892.1
Percentage identity: 35 %
BlastP bit score: 180
Sequence coverage: 94 %
E-value: 2e-50
NCBI BlastP on this gene
Accession: ADL08479
Location: 1746219-1747163
BlastP hit with WP_039959892.1
Percentage identity: 35 %
BlastP bit score: 180
Sequence coverage: 94 %
E-value: 2e-50
NCBI BlastP on this gene
Toce_1745
carbohydrate ABC transporter substrate-binding protein, CUT1 family
Accession: ADL08478
Location: 1744924-1746219
NCBI BlastP on this gene
Accession: ADL08478
Location: 1744924-1746219
NCBI BlastP on this gene
Toce_1744
two component transcriptional regulator, AraC family
Accession: ADL08477
Location: 1741932-1743059
NCBI BlastP on this gene
Accession: ADL08477
Location: 1741932-1743059
NCBI BlastP on this gene
Toce_1742
alpha amylase catalytic region
Accession: ADL08476
Location: 1740161-1741831
NCBI BlastP on this gene
Accession: ADL08476
Location: 1740161-1741831
NCBI BlastP on this gene
Toce_1741
ABC transporter related protein
Accession: ADL08475
Location: 1739025-1740023
NCBI BlastP on this gene
Accession: ADL08475
Location: 1739025-1740023
NCBI BlastP on this gene
Toce_1740
protein of unknown function DUF990
Accession: ADL08474
Location: 1738244-1739044
NCBI BlastP on this gene
Accession: ADL08474
Location: 1738244-1739044
NCBI BlastP on this gene
Toce_1739
protein of unknown function DUF990
Accession: ADL08473
Location: 1737469-1738260
NCBI BlastP on this gene
Accession: ADL08473
Location: 1737469-1738260
NCBI BlastP on this gene
Toce_1738
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase
Accession: ADL08472
Location: 1736626-1737432
NCBI BlastP on this gene
Accession: ADL08472
Location: 1736626-1737432
NCBI BlastP on this gene
Toce_1737
Toce_1736
protein of unknown function UPF0027
Accession: ADL08470
Location: 1733988-1735421
NCBI BlastP on this gene
Accession: ADL08470
Location: 1733988-1735421
NCBI BlastP on this gene
Toce_1735
conserved hypothetical protein
Accession: ADL08469
Location: 1731971-1733983
NCBI BlastP on this gene
Accession: ADL08469
Location: 1731971-1733983
NCBI BlastP on this gene
Toce_1734
transcriptional regulator, LacI family
Accession: ADL08468
Location: 1730734-1731765
NCBI BlastP on this gene
Accession: ADL08468
Location: 1730734-1731765
NCBI BlastP on this gene
Toce_1733
Toce_1732
FAD-dependent pyridine nucleotide-disulfide oxidoreductase
Accession: ADL08466
Location: 1726566-1728320
NCBI BlastP on this gene
Accession: ADL08466
Location: 1726566-1728320
NCBI BlastP on this gene
Toce_1731
469. : CP020773 Staphylococcus lutrae strain ATCC 700373 chromosome Total score: 2.0 Cumulative Blast bit score: 958
teichoic acid translocation permease
Accession: ARJ50137
Location: 344892-345680
NCBI BlastP on this gene
Accession: ARJ50137
Location: 344892-345680
NCBI BlastP on this gene
B5P37_01775
teichoic acid ABC transporter ATP-binding protein
Accession: ARJ50138
Location: 345690-347219
NCBI BlastP on this gene
Accession: ARJ50138
Location: 345690-347219
NCBI BlastP on this gene
B5P37_01780
B5P37_01785
B5P37_01790
B5P37_01795
B5P37_01800
B5P37_01805
radical SAM/CxCxxxxC motif protein YfkAB
Accession: ARJ50144
Location: 352348-353517
NCBI BlastP on this gene
Accession: ARJ50144
Location: 352348-353517
NCBI BlastP on this gene
B5P37_01810
B5P37_01815
B5P37_01820
B5P37_01825
B5P37_01830
B5P37_01835
B5P37_01840
B5P37_01845
B5P37_01850
two-component sensor histidine kinase
Accession: ARJ50151
Location: 359771-360814
NCBI BlastP on this gene
Accession: ARJ50151
Location: 359771-360814
NCBI BlastP on this gene
B5P37_01855
B5P37_01860
B5P37_01865
type I methionyl aminopeptidase
Accession: ARJ50154
Location: 362013-362777
NCBI BlastP on this gene
Accession: ARJ50154
Location: 362013-362777
NCBI BlastP on this gene
B5P37_01870
B5P37_01875
sialidase
Accession: ARJ50156
Location: 364049-365551
BlastP hit with WP_004843631.1
Percentage identity: 48 %
BlastP bit score: 442
Sequence coverage: 65 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: ARJ50156
Location: 364049-365551
BlastP hit with WP_004843631.1
Percentage identity: 48 %
BlastP bit score: 442
Sequence coverage: 65 %
E-value: 1e-143
NCBI BlastP on this gene
B5P37_01880
B5P37_01885
oxidoreductase
Accession: ARJ50158
Location: 367037-368158
BlastP hit with WP_004843633.1
Percentage identity: 67 %
BlastP bit score: 516
Sequence coverage: 98 %
E-value: 1e-179
NCBI BlastP on this gene
Accession: ARJ50158
Location: 367037-368158
BlastP hit with WP_004843633.1
Percentage identity: 67 %
BlastP bit score: 516
Sequence coverage: 98 %
E-value: 1e-179
NCBI BlastP on this gene
B5P37_01890
B5P37_01895
UDP-N-acetylmuramate--alanine ligase
Accession: ARJ50160
Location: 369563-370873
NCBI BlastP on this gene
Accession: ARJ50160
Location: 369563-370873
NCBI BlastP on this gene
B5P37_01900
B5P37_01905
DNA polymerase III subunit epsilon
Accession: ARJ50162
Location: 371835-372392
NCBI BlastP on this gene
Accession: ARJ50162
Location: 371835-372392
NCBI BlastP on this gene
B5P37_01910
B5P37_01915
B5P37_01920
B5P37_01925
B5P37_01930
B5P37_01935
B5P37_01940
23S rRNA (uracil(1939)-C(5))-methyltransferase RlmD
Accession: ARJ50169
Location: 379208-380572
NCBI BlastP on this gene
Accession: ARJ50169
Location: 379208-380572
NCBI BlastP on this gene
B5P37_01945
B5P37_01950
aspartyl/glutamyl-tRNA amidotransferase subunit B
Accession: ARJ50171
Location: 382343-383770
NCBI BlastP on this gene
Accession: ARJ50171
Location: 382343-383770
NCBI BlastP on this gene
B5P37_01955
aspartyl/glutamyl-tRNA amidotransferase subunit A
Accession: ARJ50172
Location: 383784-385244
NCBI BlastP on this gene
Accession: ARJ50172
Location: 383784-385244
NCBI BlastP on this gene
gatA
aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C
Accession: ARJ50173
Location: 385247-385549
NCBI BlastP on this gene
Accession: ARJ50173
Location: 385247-385549
NCBI BlastP on this gene
B5P37_01965
B5P37_01970
B5P37_01975
470. : CP015626 Staphylococcus pseudintermedius strain 063228 Total score: 2.0 Cumulative Blast bit score: 955
Serine protease, DegP/HtrA, do-like
Accession: ANS90134
Location: 1852550-1854202
NCBI BlastP on this gene
Accession: ANS90134
Location: 1852550-1854202
NCBI BlastP on this gene
A6M57_9090
Toxic anion resistance protein
Accession: ANS90135
Location: 1854513-1855316
NCBI BlastP on this gene
Accession: ANS90135
Location: 1854513-1855316
NCBI BlastP on this gene
A6M57_9095
Peptide chain release factor 3
Accession: ANS90136
Location: 1855785-1857347
NCBI BlastP on this gene
Accession: ANS90136
Location: 1855785-1857347
NCBI BlastP on this gene
A6M57_9100
A6M57_9105
UDP-N-acetylmuramoylalanyl-D-glutamate--L-lysine ligase
Accession: ANS90138
Location: 1857597-1859081
NCBI BlastP on this gene
Accession: ANS90138
Location: 1857597-1859081
NCBI BlastP on this gene
A6M57_9110
diglucosyldiacylglycerol synthase (LTA membrane anchor synthesis)
Accession: ANS90139
Location: 1859751-1860926
NCBI BlastP on this gene
Accession: ANS90139
Location: 1859751-1860926
NCBI BlastP on this gene
A6M57_9115
A6M57_9120
A6M57_9125
PTS system, galactosamine-specific IIC component
Accession: ANS90142
Location: 1864086-1864982
NCBI BlastP on this gene
Accession: ANS90142
Location: 1864086-1864982
NCBI BlastP on this gene
A6M57_9130
PTS system, mannose-specific IID component
Accession: ANS90143
Location: 1864969-1865862
NCBI BlastP on this gene
Accession: ANS90143
Location: 1864969-1865862
NCBI BlastP on this gene
A6M57_9135
2H phosphoesterase superfamily protein (yjcG)
Accession: ANS90144
Location: 1866435-1866941
NCBI BlastP on this gene
Accession: ANS90144
Location: 1866435-1866941
NCBI BlastP on this gene
A6M57_9140
Putative acetyl esterase YjcH
Accession: ANS90145
Location: 1866995-1867729
NCBI BlastP on this gene
Accession: ANS90145
Location: 1866995-1867729
NCBI BlastP on this gene
A6M57_9145
Sodium/alanine symporter family protein
Accession: ANS90146
Location: 1867832-1869403
NCBI BlastP on this gene
Accession: ANS90146
Location: 1867832-1869403
NCBI BlastP on this gene
A6M57_9150
A6M57_9155
Sialidase
Accession: ANS90148
Location: 1871295-1872746
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 450
Sequence coverage: 68 %
E-value: 4e-147
NCBI BlastP on this gene
Accession: ANS90148
Location: 1871295-1872746
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 450
Sequence coverage: 68 %
E-value: 4e-147
NCBI BlastP on this gene
A6M57_9160
Melibiose carrier protein, Na+/melibiose symporter
Accession: ANS90149
Location: 1872797-1874245
NCBI BlastP on this gene
Accession: ANS90149
Location: 1872797-1874245
NCBI BlastP on this gene
A6M57_9165
Melibiose carrier protein, Na+/melibiose symporter
Accession: ANS90150
Location: 1874302-1874415
NCBI BlastP on this gene
Accession: ANS90150
Location: 1874302-1874415
NCBI BlastP on this gene
A6M57_9170
hypothetical protein
Accession: ANS90151
Location: 1874447-1875565
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: ANS90151
Location: 1874447-1875565
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
A6M57_9175
Enoyl-[acyl-carrier-protein] reductase [NADH]
Accession: ANS90152
Location: 1876070-1876837
NCBI BlastP on this gene
Accession: ANS90152
Location: 1876070-1876837
NCBI BlastP on this gene
A6M57_9180
A6M57_9185
A6M57_9190
ribosomal large subunit pseudouridine synthase D-like protein
Accession: ANS90155
Location: 1880247-1881101
NCBI BlastP on this gene
Accession: ANS90155
Location: 1880247-1881101
NCBI BlastP on this gene
A6M57_9195
A6M57_9200
A6M57_9205
A6M57_9210
A6M57_9215
A6M57_9220
YjbH-like, GTP pyrophosphokinase domain
Accession: ANS90161
Location: 1884369-1885157
NCBI BlastP on this gene
Accession: ANS90161
Location: 1884369-1885157
NCBI BlastP on this gene
A6M57_9225
A6M57_9230
A6M57_9235
A6M57_9240
A6M57_9245
A6M57_9250
Glutamyl endopeptidase precursor, blaSE
Accession: ANS90167
Location: 1891197-1891991
NCBI BlastP on this gene
Accession: ANS90167
Location: 1891197-1891991
NCBI BlastP on this gene
A6M57_9255
Negative regulator of genetic competence MecA
Accession: ANS90168
Location: 1892533-1893231
NCBI BlastP on this gene
Accession: ANS90168
Location: 1892533-1893231
NCBI BlastP on this gene
A6M57_9260
A6M57_9265
A6M57_9270
471. : CP035741 Staphylococcus pseudintermedius strain 53_60 chromosome. Total score: 2.0 Cumulative Blast bit score: 953
TrkH family potassium uptake protein
Accession: QBG72532
Location: 995260-996618
NCBI BlastP on this gene
Accession: QBG72532
Location: 995260-996618
NCBI BlastP on this gene
EW136_04610
EW136_04605
EW136_04600
EW136_04595
EW136_04590
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L- lysine ligase
Accession: QBG72527
Location: 988712-990196
NCBI BlastP on this gene
Accession: QBG72527
Location: 988712-990196
NCBI BlastP on this gene
murE
diglucosyl diacylglycerol synthase
Accession: QBG72526
Location: 986867-988042
NCBI BlastP on this gene
Accession: QBG72526
Location: 986867-988042
NCBI BlastP on this gene
EW136_04580
EW136_04575
EW136_04570
PTS sugar transporter subunit IIC
Accession: QBG72523
Location: 982811-983707
NCBI BlastP on this gene
Accession: QBG72523
Location: 982811-983707
NCBI BlastP on this gene
EW136_04565
PTS system mannose/fructose/sorbose family transporter subunit IID
Accession: QBG72522
Location: 981931-982824
NCBI BlastP on this gene
Accession: QBG72522
Location: 981931-982824
NCBI BlastP on this gene
EW136_04560
EW136_04555
EW136_04550
alanine:cation symporter family protein
Accession: QBG72519
Location: 978391-979962
NCBI BlastP on this gene
Accession: QBG72519
Location: 978391-979962
NCBI BlastP on this gene
EW136_04545
EW136_04540
sialidase
Accession: QBG73939
Location: 975048-976499
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: QBG73939
Location: 975048-976499
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
EW136_04535
EW136_04530
Gfo/Idh/MocA family oxidoreductase
Accession: QBG72516
Location: 972390-973508
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: QBG72516
Location: 972390-973508
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
EW136_04525
enoyl-[acyl-carrier-protein] reductase FabI
Accession: QBG72515
Location: 971116-971886
NCBI BlastP on this gene
Accession: QBG72515
Location: 971116-971886
NCBI BlastP on this gene
fabI
EW136_04515
mgtE
RluA family pseudouridine synthase
Accession: QBG72512
Location: 966855-967709
NCBI BlastP on this gene
Accession: QBG72512
Location: 966855-967709
NCBI BlastP on this gene
EW136_04505
EW136_04500
GTP pyrophosphokinase family protein
Accession: QBG72510
Location: 965399-966037
NCBI BlastP on this gene
Accession: QBG72510
Location: 965399-966037
NCBI BlastP on this gene
EW136_04495
EW136_04490
EW136_04485
EW136_04480
EW136_04475
EW136_04470
pepF
EW136_04460
EW136_04455
EW136_04450
mecA
spxA
trpS
472. : CP035740 Staphylococcus pseudintermedius strain 53_88 chromosome. Total score: 2.0 Cumulative Blast bit score: 953
EW134_09780
EW134_09785
peptide chain release factor 3
Accession: QBG75753
Location: 2078095-2079657
NCBI BlastP on this gene
Accession: QBG75753
Location: 2078095-2079657
NCBI BlastP on this gene
EW134_09790
EW134_09795
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L- lysine ligase
Accession: QBG75755
Location: 2079907-2081391
NCBI BlastP on this gene
Accession: QBG75755
Location: 2079907-2081391
NCBI BlastP on this gene
murE
diglucosyl diacylglycerol synthase
Accession: QBG75756
Location: 2082061-2083236
NCBI BlastP on this gene
Accession: QBG75756
Location: 2082061-2083236
NCBI BlastP on this gene
EW134_09805
EW134_09810
EW134_09815
PTS sugar transporter subunit IIC
Accession: QBG75759
Location: 2086396-2087292
NCBI BlastP on this gene
Accession: QBG75759
Location: 2086396-2087292
NCBI BlastP on this gene
EW134_09820
PTS system mannose/fructose/sorbose family transporter subunit IID
Accession: QBG75760
Location: 2087279-2088172
NCBI BlastP on this gene
Accession: QBG75760
Location: 2087279-2088172
NCBI BlastP on this gene
EW134_09825
EW134_09830
EW134_09835
alanine:cation symporter family protein
Accession: QBG75763
Location: 2090141-2091712
NCBI BlastP on this gene
Accession: QBG75763
Location: 2090141-2091712
NCBI BlastP on this gene
EW134_09840
EW134_09845
sialidase
Accession: QBG76271
Location: 2093604-2095055
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: QBG76271
Location: 2093604-2095055
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
EW134_09850
EW134_09855
Gfo/Idh/MocA family oxidoreductase
Accession: QBG75766
Location: 2096595-2097713
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: QBG75766
Location: 2096595-2097713
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
EW134_09860
enoyl-[acyl-carrier-protein] reductase FabI
Accession: QBG75767
Location: 2098217-2098987
NCBI BlastP on this gene
Accession: QBG75767
Location: 2098217-2098987
NCBI BlastP on this gene
fabI
EW134_09870
mgtE
RluA family pseudouridine synthase
Accession: QBG75770
Location: 2102394-2103248
NCBI BlastP on this gene
Accession: QBG75770
Location: 2102394-2103248
NCBI BlastP on this gene
EW134_09880
EW134_09885
GTP pyrophosphokinase family protein
Accession: QBG75772
Location: 2104066-2104704
NCBI BlastP on this gene
Accession: QBG75772
Location: 2104066-2104704
NCBI BlastP on this gene
EW134_09890
EW134_09895
CYTH domain-containing protein
Accession: QBG75774
Location: 2105199-2105780
NCBI BlastP on this gene
Accession: QBG75774
Location: 2105199-2105780
NCBI BlastP on this gene
EW134_09900
EW134_09905
EW134_09910
EW134_09915
pepF
EW134_09925
EW134_09930
EW134_09935
mecA
transcriptional regulator Spx
Accession: QBG75782
Location: 2115496-2115891
NCBI BlastP on this gene
Accession: QBG75782
Location: 2115496-2115891
NCBI BlastP on this gene
spxA
trpS
473. : CP032682 Staphylococcus pseudintermedius strain G3C4 chromosome Total score: 2.0 Cumulative Blast bit score: 953
D8L98_11995
D8L98_12000
peptide chain release factor 3
Accession: AYG57098
Location: 2506603-2508165
NCBI BlastP on this gene
Accession: AYG57098
Location: 2506603-2508165
NCBI BlastP on this gene
D8L98_12005
D8L98_12010
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L- lysine ligase
Accession: AYG57100
Location: 2508415-2509899
NCBI BlastP on this gene
Accession: AYG57100
Location: 2508415-2509899
NCBI BlastP on this gene
murE
D8L98_12020
diglucosyl diacylglycerol synthase
Accession: AYG57102
Location: 2510569-2511744
NCBI BlastP on this gene
Accession: AYG57102
Location: 2510569-2511744
NCBI BlastP on this gene
D8L98_12025
D8L98_12030
D8L98_12035
PTS sugar transporter subunit IIC
Accession: AYG57105
Location: 2514904-2515800
NCBI BlastP on this gene
Accession: AYG57105
Location: 2514904-2515800
NCBI BlastP on this gene
D8L98_12040
PTS system mannose/fructose/sorbose family transporter subunit IID
Accession: AYG57106
Location: 2515787-2516680
NCBI BlastP on this gene
Accession: AYG57106
Location: 2515787-2516680
NCBI BlastP on this gene
D8L98_12045
D8L98_12050
D8L98_12055
alanine:cation symporter family protein
Accession: AYG57109
Location: 2518649-2520220
NCBI BlastP on this gene
Accession: AYG57109
Location: 2518649-2520220
NCBI BlastP on this gene
D8L98_12060
D8L98_12065
sialidase
Accession: AYG57386
Location: 2522110-2523561
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
Accession: AYG57386
Location: 2522110-2523561
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
D8L98_12070
D8L98_12075
gfo/Idh/MocA family oxidoreductase
Accession: AYG57112
Location: 2525101-2526219
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: AYG57112
Location: 2525101-2526219
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
D8L98_12080
enoyl-[acyl-carrier-protein] reductase FabI
Accession: AYG57113
Location: 2526724-2527494
NCBI BlastP on this gene
Accession: AYG57113
Location: 2526724-2527494
NCBI BlastP on this gene
fabI
D8L98_12090
mgtE
RluA family pseudouridine synthase
Accession: AYG57116
Location: 2530901-2531755
NCBI BlastP on this gene
Accession: AYG57116
Location: 2530901-2531755
NCBI BlastP on this gene
D8L98_12100
D8L98_12105
GTP pyrophosphokinase family protein
Accession: AYG57118
Location: 2532573-2533211
NCBI BlastP on this gene
Accession: AYG57118
Location: 2532573-2533211
NCBI BlastP on this gene
D8L98_12110
D8L98_12115
CYTH domain-containing protein
Accession: AYG57120
Location: 2533706-2534287
NCBI BlastP on this gene
Accession: AYG57120
Location: 2533706-2534287
NCBI BlastP on this gene
D8L98_12120
D8L98_12125
D8L98_12130
D8L98_12135
pepF
D8L98_12145
LPXTG cell wall anchor domain-containing protein
Accession: AYG57126
Location: 2540606-2541265
NCBI BlastP on this gene
Accession: AYG57126
Location: 2540606-2541265
NCBI BlastP on this gene
D8L98_12150
D8L98_12155
mecA
transcriptional regulator Spx
Accession: AYG57128
Location: 2544057-2544452
NCBI BlastP on this gene
Accession: AYG57128
Location: 2544057-2544452
NCBI BlastP on this gene
spxA
trpS
474. : CP002478 Staphylococcus pseudintermedius ED99 Total score: 2.0 Cumulative Blast bit score: 953
htrA
toxic anion resistance protein-like protein
Accession: ADX77066
Location: 1867958-1868761
NCBI BlastP on this gene
Accession: ADX77066
Location: 1867958-1868761
NCBI BlastP on this gene
SPSE_1815
peptide chain release factor 3
Accession: ADX77067
Location: 1869230-1870792
NCBI BlastP on this gene
Accession: ADX77067
Location: 1869230-1870792
NCBI BlastP on this gene
prfC
conserved hypothetical protein
Accession: ADX77068
Location: 1870813-1871052
NCBI BlastP on this gene
Accession: ADX77068
Location: 1870813-1871052
NCBI BlastP on this gene
SPSE_1818
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L- lysine ligase
Accession: ADX77069
Location: 1871042-1872526
NCBI BlastP on this gene
Accession: ADX77069
Location: 1871042-1872526
NCBI BlastP on this gene
murE
diacylglycerol glucosyltransferase
Accession: ADX77070
Location: 1873196-1874371
NCBI BlastP on this gene
Accession: ADX77070
Location: 1873196-1874371
NCBI BlastP on this gene
SPSE_1820
putative glycolipid permease LtaA (Lipoteichoic acid protein A)
Accession: ADX77071
Location: 1874349-1875533
NCBI BlastP on this gene
Accession: ADX77071
Location: 1874349-1875533
NCBI BlastP on this gene
SPSE_1821
SPSE_1822
SPSE_1823
SPSE_1824
conserved hypothetical protein
Accession: ADX77075
Location: 1879879-1880385
NCBI BlastP on this gene
Accession: ADX77075
Location: 1879879-1880385
NCBI BlastP on this gene
SPSE_1825
SPSE_1826
sodium:alanine symporter family protein
Accession: ADX77077
Location: 1881276-1882847
NCBI BlastP on this gene
Accession: ADX77077
Location: 1881276-1882847
NCBI BlastP on this gene
SPSE_1827
conserved membrane protein, putative
Accession: ADX77078
Location: 1883066-1884163
NCBI BlastP on this gene
Accession: ADX77078
Location: 1883066-1884163
NCBI BlastP on this gene
SPSE_1828
putative sialidase, truncated
Accession: ADX77079
Location: 1884739-1886190
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: ADX77079
Location: 1884739-1886190
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
SPSE_1829
sugar (Glycoside-Pentoside-Hexuronide) transporter
Accession: ADX77080
Location: 1886241-1887689
NCBI BlastP on this gene
Accession: ADX77080
Location: 1886241-1887689
NCBI BlastP on this gene
gph
putative oxidoreductase
Accession: ADX77081
Location: 1887730-1888848
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: ADX77081
Location: 1887730-1888848
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
SPSE_1831
enoyl-(acyl carrier protein) reductase
Accession: ADX77082
Location: 1889353-1890120
NCBI BlastP on this gene
Accession: ADX77082
Location: 1889353-1890120
NCBI BlastP on this gene
fabI
SPSE_1833
mgtE
ribosomal large subunit pseudouridine synthase D
Accession: ADX77085
Location: 1893530-1894384
NCBI BlastP on this gene
Accession: ADX77085
Location: 1893530-1894384
NCBI BlastP on this gene
SPSE_1835
inorganic polyphosphate/ATP-NAD kinase
Accession: ADX77086
Location: 1894381-1895190
NCBI BlastP on this gene
Accession: ADX77086
Location: 1894381-1895190
NCBI BlastP on this gene
ppnK
relA2
conserved hypothetical protein
Accession: ADX77088
Location: 1895857-1896204
NCBI BlastP on this gene
Accession: ADX77088
Location: 1895857-1896204
NCBI BlastP on this gene
SPSE_1838
SPSE_1839
protozoan/cyanobacterial globin family protein
Accession: ADX77090
Location: 1897264-1897635
NCBI BlastP on this gene
Accession: ADX77090
Location: 1897264-1897635
NCBI BlastP on this gene
SPSE_1840
SPSE_1841
SPSE_1842
pepF
transcription factor, putative
Accession: ADX77094
Location: 1901989-1902954
NCBI BlastP on this gene
Accession: ADX77094
Location: 1901989-1902954
NCBI BlastP on this gene
SPSE_1844
LPXTG-motif cell wall anchor domain protein
Accession: ADX77095
Location: 1903228-1903887
NCBI BlastP on this gene
Accession: ADX77095
Location: 1903228-1903887
NCBI BlastP on this gene
spsR
SPSE_1846
mecA
spxA
trpS
475. : CP016073 Staphylococcus pseudintermedius strain 081661 Total score: 2.0 Cumulative Blast bit score: 952
A9I65_09105
A9I65_09110
peptide chain release factor 3
Accession: ANQ88753
Location: 1950505-1952067
NCBI BlastP on this gene
Accession: ANQ88753
Location: 1950505-1952067
NCBI BlastP on this gene
A9I65_09115
A9I65_09120
UDP-N-acetylmuramoylalanyl-D-glutamate--L-lysine ligase
Accession: ANQ88755
Location: 1952317-1953801
NCBI BlastP on this gene
Accession: ANQ88755
Location: 1952317-1953801
NCBI BlastP on this gene
A9I65_09125
A9I65_09130
diglucosyl diacylglycerol synthase
Accession: ANQ88757
Location: 1954471-1955646
NCBI BlastP on this gene
Accession: ANQ88757
Location: 1954471-1955646
NCBI BlastP on this gene
A9I65_09135
A9I65_09140
A9I65_09145
PTS fructose transporter subunit IIC
Accession: ANQ88760
Location: 1958806-1959702
NCBI BlastP on this gene
Accession: ANQ88760
Location: 1958806-1959702
NCBI BlastP on this gene
A9I65_09150
PTS fructose transporter subunit IID
Accession: ANQ89508
Location: 1959815-1960582
NCBI BlastP on this gene
Accession: ANQ89508
Location: 1959815-1960582
NCBI BlastP on this gene
A9I65_09155
A9I65_09160
A9I65_09165
A9I65_09170
A9I65_09175
sialidase
Accession: ANQ89509
Location: 1966012-1967463
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
Accession: ANQ89509
Location: 1966012-1967463
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 2e-146
NCBI BlastP on this gene
A9I65_09180
A9I65_09185
oxidoreductase
Accession: ANQ89510
Location: 1969003-1970109
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
Accession: ANQ89510
Location: 1969003-1970109
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
A9I65_09190
enoyl-[acyl-carrier-protein] reductase
Accession: ANQ88766
Location: 1970626-1971396
NCBI BlastP on this gene
Accession: ANQ88766
Location: 1970626-1971396
NCBI BlastP on this gene
A9I65_09195
A9I65_09200
A9I65_09205
A9I65_09210
A9I65_09215
A9I65_09220
A9I65_09225
A9I65_09230
A9I65_09235
A9I65_09240
A9I65_09245
A9I65_09250
A9I65_09255
A9I65_09260
A9I65_09265
A9I65_09270
transcriptional regulator Spx
Accession: ANQ88781
Location: 1987902-1988297
NCBI BlastP on this gene
Accession: ANQ88781
Location: 1987902-1988297
NCBI BlastP on this gene
A9I65_09275
A9I65_09280
476. : CP009120 Staphylococcus pseudintermedius strain 5912 Total score: 2.0 Cumulative Blast bit score: 952
SPS5912_08855
SPS5912_08860
peptide chain release factor 3
Accession: ASQ51066
Location: 1874624-1876186
NCBI BlastP on this gene
Accession: ASQ51066
Location: 1874624-1876186
NCBI BlastP on this gene
SPS5912_08865
SPS5912_08870
UDP-N-acetylmuramoylalanyl-D-glutamate--L-lysine ligase
Accession: ASQ51068
Location: 1876436-1877920
NCBI BlastP on this gene
Accession: ASQ51068
Location: 1876436-1877920
NCBI BlastP on this gene
SPS5912_08875
SPS5912_08880
diacylglycerol glucosyltransferase
Accession: ASQ51070
Location: 1878590-1879765
NCBI BlastP on this gene
Accession: ASQ51070
Location: 1878590-1879765
NCBI BlastP on this gene
SPS5912_08885
SPS5912_08890
SPS5912_08895
PTS fructose transporter subunit IIC
Accession: ASQ51073
Location: 1882925-1883821
NCBI BlastP on this gene
Accession: ASQ51073
Location: 1882925-1883821
NCBI BlastP on this gene
SPS5912_08900
PTS fructose transporter subunit IID
Accession: ASQ51074
Location: 1883808-1884701
NCBI BlastP on this gene
Accession: ASQ51074
Location: 1883808-1884701
NCBI BlastP on this gene
SPS5912_08905
SPS5912_08910
SPS5912_08915
SPS5912_08920
SPS5912_08925
sialidase
Accession: ASQ51079
Location: 1890133-1891584
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: ASQ51079
Location: 1890133-1891584
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
SPS5912_08930
SPS5912_08935
oxidoreductase
Accession: ASQ51081
Location: 1893124-1894230
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
Accession: ASQ51081
Location: 1893124-1894230
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
SPS5912_08940
SPS5912_08945
SPS5912_08950
SPS5912_08955
SPS5912_08960
inorganic polyphosphate kinase
Accession: ASQ51086
Location: 1899774-1900583
NCBI BlastP on this gene
Accession: ASQ51086
Location: 1899774-1900583
NCBI BlastP on this gene
ppnK
SPS5912_08970
SPS5912_08975
SPS5912_08980
SPS5912_08985
SPS5912_08990
SPS5912_08995
SPS5912_09000
SPS5912_09005
SPS5912_09010
trypsin domain-containing protein
Accession: ASQ51096
Location: 1909305-1910291
NCBI BlastP on this gene
Accession: ASQ51096
Location: 1909305-1910291
NCBI BlastP on this gene
SPS5912_09015
competence negative regulator MecA
Accession: ASQ51097
Location: 1910946-1911644
NCBI BlastP on this gene
Accession: ASQ51097
Location: 1910946-1911644
NCBI BlastP on this gene
SPS5912_09020
ArsR family transcriptional regulator
Accession: ASQ51098
Location: 1911978-1912373
NCBI BlastP on this gene
Accession: ASQ51098
Location: 1911978-1912373
NCBI BlastP on this gene
SPS5912_09025
SPS5912_09030
477. : AP019372 Staphylococcus pseudintermedius SP79 DNA Total score: 2.0 Cumulative Blast bit score: 952
serine protease HtrA-like protein
Accession: BBH74506
Location: 1841306-1842958
NCBI BlastP on this gene
Accession: BBH74506
Location: 1841306-1842958
NCBI BlastP on this gene
GSP_17000
GSP_17010
peptide chain release factor 3
Accession: BBH74508
Location: 1844541-1846103
NCBI BlastP on this gene
Accession: BBH74508
Location: 1844541-1846103
NCBI BlastP on this gene
prfC
GSP_17030
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L- lysi ne ligase
Accession: BBH74510
Location: 1846353-1847837
NCBI BlastP on this gene
Accession: BBH74510
Location: 1846353-1847837
NCBI BlastP on this gene
murE
processive diacylglycerol beta-glucosyltransferase
Accession: BBH74511
Location: 1848508-1849683
NCBI BlastP on this gene
Accession: BBH74511
Location: 1848508-1849683
NCBI BlastP on this gene
ugtP
GSP_17060
GSP_17070
GSP_17080
PTS fructose transporter subunit IIC
Accession: BBH74515
Location: 1852843-1853739
NCBI BlastP on this gene
Accession: BBH74515
Location: 1852843-1853739
NCBI BlastP on this gene
GSP_17090
PTS fructose transporter subunit IID
Accession: BBH74516
Location: 1853726-1854619
NCBI BlastP on this gene
Accession: BBH74516
Location: 1853726-1854619
NCBI BlastP on this gene
GSP_17100
GSP_17110
GSP_17120
GSP_17130
GSP_17140
hypothetical protein
Accession: BBH74521
Location: 1860053-1861537
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
Accession: BBH74521
Location: 1860053-1861537
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 448
Sequence coverage: 68 %
E-value: 3e-146
NCBI BlastP on this gene
GSP_17150
GSP_17160
oxidoreductase
Accession: BBH74523
Location: 1863044-1864162
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
Accession: BBH74523
Location: 1863044-1864162
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 504
Sequence coverage: 97 %
E-value: 3e-175
NCBI BlastP on this gene
GSP_17170
enoyl-[acyl-carrier-protein] reductase [NADPH] FabI
Accession: BBH74524
Location: 1864667-1865437
NCBI BlastP on this gene
Accession: BBH74524
Location: 1864667-1865437
NCBI BlastP on this gene
fabI
GSP_17190
GSP_17200
GSP_17210
nadK
GSP_17230
GSP_17240
GSP_17250
GSP_17260
GSP_17270
GSP_17280
GSP_17290
GSP_17300
GSP_17310
GSP_17320
mecA
spxA
trpS
478. : CP002439 Staphylococcus pseudintermedius HKU10-03 Total score: 2.0 Cumulative Blast bit score: 949
SPSINT_0711
SPSINT_0710
Predicted PTS system, galactosamine-specific IIC component
Accession: ADV05237
Location: 738124-739020
NCBI BlastP on this gene
Accession: ADV05237
Location: 738124-739020
NCBI BlastP on this gene
SPSINT_0709
Predicted PTS system, galactosamine-specific IID component
Accession: ADV05236
Location: 737244-738137
NCBI BlastP on this gene
Accession: ADV05236
Location: 737244-738137
NCBI BlastP on this gene
SPSINT_0708
SPSINT_0707
SPSINT_0706
CAAX amino terminal protease family
Accession: ADV05233
Location: 734946-735770
NCBI BlastP on this gene
Accession: ADV05233
Location: 734946-735770
NCBI BlastP on this gene
SPSINT_0705
Retron-type reverse transcriptase
Accession: ADV05232
Location: 732544-733440
NCBI BlastP on this gene
Accession: ADV05232
Location: 732544-733440
NCBI BlastP on this gene
SPSINT_0704
Retron-type reverse transcriptase
Accession: ADV05231
Location: 732200-732334
NCBI BlastP on this gene
Accession: ADV05231
Location: 732200-732334
NCBI BlastP on this gene
SPSINT_0703
SPSINT_0702
ABC-type multidrug transport system, ATPase component
Accession: ADV05229
Location: 730388-731269
NCBI BlastP on this gene
Accession: ADV05229
Location: 730388-731269
NCBI BlastP on this gene
SPSINT_0701
SPSINT_0700
YjcG, 2H phosphoesterase superfamily protein
Accession: ADV05227
Location: 725685-726191
NCBI BlastP on this gene
Accession: ADV05227
Location: 725685-726191
NCBI BlastP on this gene
SPSINT_0698
SPSINT_0697
sodium/alanine symporter family protein
Accession: ADV05225
Location: 723223-724794
NCBI BlastP on this gene
Accession: ADV05225
Location: 723223-724794
NCBI BlastP on this gene
SPSINT_0696
SPSINT_0695
Sialidase
Accession: ADV05223
Location: 719882-721333
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 444
Sequence coverage: 68 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: ADV05223
Location: 719882-721333
BlastP hit with WP_004843631.1
Percentage identity: 47 %
BlastP bit score: 444
Sequence coverage: 68 %
E-value: 2e-144
NCBI BlastP on this gene
SPSINT_0694
Melibiose carrier protein, Na+/melibiose symporter
Accession: ADV05222
Location: 718383-719831
NCBI BlastP on this gene
Accession: ADV05222
Location: 718383-719831
NCBI BlastP on this gene
SPSINT_0693
Putative oxidoreductase
Accession: ADV05221
Location: 717224-718342
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
Accession: ADV05221
Location: 717224-718342
BlastP hit with WP_004843633.1
Percentage identity: 65 %
BlastP bit score: 505
Sequence coverage: 97 %
E-value: 2e-175
NCBI BlastP on this gene
SPSINT_0692
Enoyl-[acyl-carrier-protein] reductase [NADH]
Accession: ADV05220
Location: 715954-716721
NCBI BlastP on this gene
Accession: ADV05220
Location: 715954-716721
NCBI BlastP on this gene
SPSINT_0691
SPSINT_0690
SPSINT_0689
SPSINT_0688
SPSINT_0687
SPSINT_0686
SPSINT_0685
SPSINT_0684
SPSINT_0683
YjbH-like, GTP pyrophosphokinase domain protein
Accession: ADV05211
Location: 707634-708422
NCBI BlastP on this gene
Accession: ADV05211
Location: 707634-708422
NCBI BlastP on this gene
SPSINT_0682
SPSINT_0681
SPSINT_0680
SPSINT_0679
SPSINT_0678
glutamyl endopeptidase precursor
Accession: ADV05206
Location: 701616-702161
NCBI BlastP on this gene
Accession: ADV05206
Location: 701616-702161
NCBI BlastP on this gene
SPSINT_0677
Glutamyl endopeptidase precursor, blaSE
Accession: ADV05205
Location: 700800-701594
NCBI BlastP on this gene
Accession: ADV05205
Location: 700800-701594
NCBI BlastP on this gene
SPSINT_0676
Negative regulator of genetic competence MecA
Accession: ADV05204
Location: 699560-700258
NCBI BlastP on this gene
Accession: ADV05204
Location: 699560-700258
NCBI BlastP on this gene
SPSINT_0675
Arsenate reductase family protein
Accession: ADV05203
Location: 698831-699226
NCBI BlastP on this gene
Accession: ADV05203
Location: 698831-699226
NCBI BlastP on this gene
SPSINT_0674
SPSINT_0673
479. : CP046351 Staphylococcus fleurettii strain FDAARGOS_682 chromosome Total score: 2.0 Cumulative Blast bit score: 937
FOB90_03430
teichoic acid translocation permease
Accession: QGS45792
Location: 682946-683719
NCBI BlastP on this gene
Accession: QGS45792
Location: 682946-683719
NCBI BlastP on this gene
FOB90_03435
ATP-binding cassette domain-containing protein
Accession: QGS45793
Location: 683728-685257
NCBI BlastP on this gene
Accession: QGS45793
Location: 683728-685257
NCBI BlastP on this gene
FOB90_03440
FOB90_03445
recX
monofunctional peptidoglycan glycosyltransferase SgtB
Accession: QGS45796
Location: 686570-687376
NCBI BlastP on this gene
Accession: QGS45796
Location: 686570-687376
NCBI BlastP on this gene
sgtB
FOB90_03460
DJ-1/PfpI/YhbO family deglycase/protease
Accession: QGS45798
Location: 689590-690105
NCBI BlastP on this gene
Accession: QGS45798
Location: 689590-690105
NCBI BlastP on this gene
FOB90_03465
radical SAM/CxCxxxxC motif protein YfkAB
Accession: QGS45799
Location: 690457-691593
NCBI BlastP on this gene
Accession: QGS45799
Location: 690457-691593
NCBI BlastP on this gene
yfkAB
FOB90_03475
FOB90_03480
FOB90_03485
FOB90_03490
FOB90_03495
YihY family inner membrane protein
Accession: QGS45805
Location: 694961-696220
NCBI BlastP on this gene
Accession: QGS45805
Location: 694961-696220
NCBI BlastP on this gene
FOB90_03500
FOB90_03505
FOB90_03510
two-component sensor histidine kinase
Accession: QGS45808
Location: 697499-698542
NCBI BlastP on this gene
Accession: QGS45808
Location: 697499-698542
NCBI BlastP on this gene
FOB90_03515
FOB90_03520
FOB90_03525
type I methionyl aminopeptidase
Accession: QGS45811
Location: 699740-700501
NCBI BlastP on this gene
Accession: QGS45811
Location: 699740-700501
NCBI BlastP on this gene
map
aromatic acid exporter family protein
Accession: QGS45812
Location: 700599-701597
NCBI BlastP on this gene
Accession: QGS45812
Location: 700599-701597
NCBI BlastP on this gene
FOB90_03535
sialidase
Accession: FOB90_03540
Location: 701714-703222
BlastP hit with WP_004843631.1
Percentage identity: 49 %
BlastP bit score: 439
Sequence coverage: 66 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: FOB90_03540
Location: 701714-703222
BlastP hit with WP_004843631.1
Percentage identity: 49 %
BlastP bit score: 439
Sequence coverage: 66 %
E-value: 3e-142
NCBI BlastP on this gene
FOB90_03540
FOB90_03545
gfo/Idh/MocA family oxidoreductase
Accession: QGS45814
Location: 704707-705831
BlastP hit with WP_004843633.1
Percentage identity: 64 %
BlastP bit score: 498
Sequence coverage: 98 %
E-value: 2e-172
NCBI BlastP on this gene
Accession: QGS45814
Location: 704707-705831
BlastP hit with WP_004843633.1
Percentage identity: 64 %
BlastP bit score: 498
Sequence coverage: 98 %
E-value: 2e-172
NCBI BlastP on this gene
FOB90_03550
FOB90_03555
DUF1727 domain-containing protein
Accession: QGS45816
Location: 706817-708136
NCBI BlastP on this gene
Accession: QGS45816
Location: 706817-708136
NCBI BlastP on this gene
FOB90_03560
FOB90_03565
DNA polymerase III subunit epsilon
Accession: QGS45818
Location: 709065-709625
NCBI BlastP on this gene
Accession: QGS45818
Location: 709065-709625
NCBI BlastP on this gene
FOB90_03570
dinB
FOB90_03580
DUF3267 domain-containing protein
Accession: QGS45821
Location: 711505-712059
NCBI BlastP on this gene
Accession: QGS45821
Location: 711505-712059
NCBI BlastP on this gene
FOB90_03585
23S rRNA (uracil(1939)-C(5))-methyltransferase RlmD
Accession: QGS45822
Location: 712135-713496
NCBI BlastP on this gene
Accession: QGS45822
Location: 712135-713496
NCBI BlastP on this gene
rlmD
FOB90_03595
FOB90_03600
Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase subunit GatB
Accession: QGS45825
Location: 715302-716729
NCBI BlastP on this gene
Accession: QGS45825
Location: 715302-716729
NCBI BlastP on this gene
gatB
Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase subunit GatA
Accession: QGS45826
Location: 716743-718203
NCBI BlastP on this gene
Accession: QGS45826
Location: 716743-718203
NCBI BlastP on this gene
gatA
Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase subunit GatC
Accession: QGS45827
Location: 718206-718508
NCBI BlastP on this gene
Accession: QGS45827
Location: 718206-718508
NCBI BlastP on this gene
gatC
putP
CamS family sex pheromone protein
Accession: QGS45829
Location: 720637-721845
NCBI BlastP on this gene
Accession: QGS45829
Location: 720637-721845
NCBI BlastP on this gene
FOB90_03625
ligA
pcrA
480. : CP040518 Blautia sp. SC05B48 chromosome Total score: 2.0 Cumulative Blast bit score: 871
sugar ABC transporter ATP-binding protein
Accession: QCU01223
Location: 504334-505845
NCBI BlastP on this gene
Accession: QCU01223
Location: 504334-505845
NCBI BlastP on this gene
EYS05_02245
EYS05_02250
substrate-binding domain-containing protein
Accession: QCU01225
Location: 506999-508006
NCBI BlastP on this gene
Accession: QCU01225
Location: 506999-508006
NCBI BlastP on this gene
EYS05_02255
EYS05_02260
EYS05_02265
EYS05_02270
pfkB
galactitol-1-phosphate 5-dehydrogenase
Accession: QCU01230
Location: 513628-514680
NCBI BlastP on this gene
Accession: QCU01230
Location: 513628-514680
NCBI BlastP on this gene
EYS05_02280
xylB
LacI family transcriptional regulator
Accession: QCU01232
Location: 516200-517225
NCBI BlastP on this gene
Accession: QCU01232
Location: 516200-517225
NCBI BlastP on this gene
EYS05_02290
class II fructose-bisphosphate aldolase
Accession: QCU01233
Location: 517260-518111
NCBI BlastP on this gene
Accession: QCU01233
Location: 517260-518111
NCBI BlastP on this gene
EYS05_02295
EYS05_02300
EYS05_02305
EYS05_02310
glf
ROK family protein
Accession: QCU01238
Location: 524287-525174
BlastP hit with WP_004843625.1
Percentage identity: 72 %
BlastP bit score: 459
Sequence coverage: 98 %
E-value: 2e-159
NCBI BlastP on this gene
Accession: QCU01238
Location: 524287-525174
BlastP hit with WP_004843625.1
Percentage identity: 72 %
BlastP bit score: 459
Sequence coverage: 98 %
E-value: 2e-159
NCBI BlastP on this gene
EYS05_02320
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QCU01239
Location: 525204-525896
BlastP hit with WP_024854132.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 4e-143
NCBI BlastP on this gene
Accession: QCU01239
Location: 525204-525896
BlastP hit with WP_024854132.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 4e-143
NCBI BlastP on this gene
EYS05_02325
EYS05_02330
EYS05_02335
EYS05_02340
plasmid pRiA4b ORF-3 family protein
Accession: QCU03867
Location: 528809-529231
NCBI BlastP on this gene
Accession: QCU03867
Location: 528809-529231
NCBI BlastP on this gene
EYS05_02345
EYS05_02350
EYS05_02355
EYS05_02360
glf
EYS05_02370
EYS05_02375
NAD-dependent epimerase/dehydratase family protein
Accession: QCU01247
Location: 536112-537149
NCBI BlastP on this gene
Accession: QCU01247
Location: 536112-537149
NCBI BlastP on this gene
EYS05_02380
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: QCU01248
Location: 537202-537930
NCBI BlastP on this gene
Accession: QCU01248
Location: 537202-537930
NCBI BlastP on this gene
EYS05_02385
oligosaccharide repeat unit polymerase
Accession: QCU01249
Location: 538007-539386
NCBI BlastP on this gene
Accession: QCU01249
Location: 538007-539386
NCBI BlastP on this gene
EYS05_02390
EYS05_02395
EYS05_02400
glycosyltransferase family 1 protein
Accession: QCU01252
Location: 541541-542671
NCBI BlastP on this gene
Accession: QCU01252
Location: 541541-542671
NCBI BlastP on this gene
EYS05_02405
EYS05_02410
DUF4422 domain-containing protein
Accession: QCU01254
Location: 543749-544573
NCBI BlastP on this gene
Accession: QCU01254
Location: 543749-544573
NCBI BlastP on this gene
EYS05_02415
EYS05_02420
481. : CP022713 Blautia coccoides strain YL58 genome. Total score: 2.0 Cumulative Blast bit score: 738
uroporphyrinogen decarboxylase
Accession: ASU30557
Location: 3846158-3847150
NCBI BlastP on this gene
Accession: ASU30557
Location: 3846158-3847150
NCBI BlastP on this gene
ADH70_018210
ADH70_018215
ADH70_018220
pgmB
ADH70_018230
ADH70_018235
ADH70_018240
ADH70_018245
ADH70_018250
ADH70_018255
ADH70_018260
ADH70_018265
ADH70_018270
ADH70_018275
ADH70_018280
ADH70_018285
MurR/RpiR family transcriptional regulator
Accession: ASU30572
Location: 3863593-3864408
NCBI BlastP on this gene
Accession: ASU30572
Location: 3863593-3864408
NCBI BlastP on this gene
ADH70_018290
ROK family protein
Accession: ASU30573
Location: 3864478-3865365
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
Accession: ASU30573
Location: 3864478-3865365
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
ADH70_018295
DUF386 domain-containing protein
Accession: ASU30574
Location: 3865539-3865991
NCBI BlastP on this gene
Accession: ASU30574
Location: 3865539-3865991
NCBI BlastP on this gene
ADH70_018300
ADH70_018305
ADH70_018310
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ASU30577
Location: 3868603-3869301
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
Accession: ASU30577
Location: 3868603-3869301
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
ADH70_018315
N-acetylglucosamine-6-phosphate deacetylase
Accession: ASU30578
Location: 3869664-3870767
NCBI BlastP on this gene
Accession: ASU30578
Location: 3869664-3870767
NCBI BlastP on this gene
nagA
GIY-YIG nuclease family protein
Accession: ASU30579
Location: 3870891-3871127
NCBI BlastP on this gene
Accession: ASU30579
Location: 3870891-3871127
NCBI BlastP on this gene
ADH70_018325
ADH70_018330
ADH70_018335
stage II sporulation protein D
Accession: ASU30582
Location: 3874896-3875765
NCBI BlastP on this gene
Accession: ASU30582
Location: 3874896-3875765
NCBI BlastP on this gene
ADH70_018340
16S rRNA (cytosine(1402)-N(4))-methyltransferase
Accession: ASU30583
Location: 3875882-3876445
NCBI BlastP on this gene
Accession: ASU30583
Location: 3875882-3876445
NCBI BlastP on this gene
mraW
ADH70_018350
MATE family efflux transporter
Accession: ASU30585
Location: 3877879-3879240
NCBI BlastP on this gene
Accession: ASU30585
Location: 3877879-3879240
NCBI BlastP on this gene
ADH70_018355
ADH70_018360
RNA polymerase subunit sigma-70
Accession: ADH70_018365
Location: 3880601-3880705
NCBI BlastP on this gene
Accession: ADH70_018365
Location: 3880601-3880705
NCBI BlastP on this gene
ADH70_018365
1-acyl-sn-glycerol-3-phosphate acyltransferase
Accession: ASU30587
Location: 3887677-3888393
NCBI BlastP on this gene
Accession: ASU30587
Location: 3887677-3888393
NCBI BlastP on this gene
ADH70_018425
ADH70_018430
ADH70_018435
482. : CP015405 Blautia sp. YL58 chromosome Total score: 2.0 Cumulative Blast bit score: 738
uroporphyrinogen decarboxylase
Accession: ANU77752
Location: 4199846-4200838
NCBI BlastP on this gene
Accession: ANU77752
Location: 4199846-4200838
NCBI BlastP on this gene
A4V09_19615
A4V09_19620
galactoside O-acetyltransferase
Accession: ANU77754
Location: 4201221-4201862
NCBI BlastP on this gene
Accession: ANU77754
Location: 4201221-4201862
NCBI BlastP on this gene
A4V09_19625
A4V09_19630
A4V09_19635
A4V09_19640
A4V09_19645
A4V09_19650
A4V09_19655
A4V09_19660
A4V09_19665
A4V09_19670
A4V09_19675
A4V09_19680
A4V09_19685
A4V09_19690
MurR/RpiR family transcriptional regulator
Accession: ANU77767
Location: 4217281-4218096
NCBI BlastP on this gene
Accession: ANU77767
Location: 4217281-4218096
NCBI BlastP on this gene
A4V09_19695
hypothetical protein
Accession: ANU77768
Location: 4218166-4219053
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
Accession: ANU77768
Location: 4218166-4219053
BlastP hit with WP_004843625.1
Percentage identity: 61 %
BlastP bit score: 368
Sequence coverage: 98 %
E-value: 9e-124
NCBI BlastP on this gene
A4V09_19700
YhcH/YjgK/YiaL family protein
Accession: ANU77769
Location: 4219227-4219679
NCBI BlastP on this gene
Accession: ANU77769
Location: 4219227-4219679
NCBI BlastP on this gene
A4V09_19705
A4V09_19710
A4V09_19715
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ANU77772
Location: 4222291-4222989
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
Accession: ANU77772
Location: 4222291-4222989
BlastP hit with WP_024854132.1
Percentage identity: 77 %
BlastP bit score: 370
Sequence coverage: 99 %
E-value: 6e-127
NCBI BlastP on this gene
A4V09_19720
N-acetylglucosamine-6-phosphate deacetylase
Accession: ANU77773
Location: 4223352-4224455
NCBI BlastP on this gene
Accession: ANU77773
Location: 4223352-4224455
NCBI BlastP on this gene
A4V09_19725
A4V09_19730
A4V09_19735
A4V09_19740
stage II sporulation protein D
Accession: ANU77777
Location: 4228584-4229453
NCBI BlastP on this gene
Accession: ANU77777
Location: 4228584-4229453
NCBI BlastP on this gene
A4V09_19745
16S rRNA (cytosine(1402)-N(4))-methyltransferase
Accession: ANU77778
Location: 4229570-4230133
NCBI BlastP on this gene
Accession: ANU77778
Location: 4229570-4230133
NCBI BlastP on this gene
A4V09_19750
O-acetylhomoserine aminocarboxypropyltransferase
Accession: ANU77779
Location: 4230137-4231426
NCBI BlastP on this gene
Accession: ANU77779
Location: 4230137-4231426
NCBI BlastP on this gene
A4V09_19755
MATE family efflux transporter
Accession: ANU77780
Location: 4231567-4232928
NCBI BlastP on this gene
Accession: ANU77780
Location: 4231567-4232928
NCBI BlastP on this gene
A4V09_19760
A4V09_19765
RNA polymerase subunit sigma-70
Accession: A4V09_24370
Location: 4234289-4234393
NCBI BlastP on this gene
Accession: A4V09_24370
Location: 4234289-4234393
NCBI BlastP on this gene
A4V09_24370
1-acyl-sn-glycerol-3-phosphate acyltransferase
Accession: ANU77782
Location: 4241365-4242081
NCBI BlastP on this gene
Accession: ANU77782
Location: 4241365-4242081
NCBI BlastP on this gene
A4V09_19825
A4V09_19830
A4V09_19835
483. : CP048626 Blautia producta ATCC 27340 = DSM 2950 strain JCM 1471 chromosome Total score: 2.0 Cumulative Blast bit score: 726
ATP-binding cassette domain-containing protein
Accession: QIB56122
Location: 3255336-3257033
NCBI BlastP on this gene
Accession: QIB56122
Location: 3255336-3257033
NCBI BlastP on this gene
GXM18_15340
energy-coupling factor transporter transmembrane protein EcfT
Accession: QIB56121
Location: 3254470-3255363
NCBI BlastP on this gene
Accession: QIB56121
Location: 3254470-3255363
NCBI BlastP on this gene
GXM18_15335
DUF4430 domain-containing protein
Accession: QIB56120
Location: 3253532-3254464
NCBI BlastP on this gene
Accession: QIB56120
Location: 3253532-3254464
NCBI BlastP on this gene
GXM18_15330
GXM18_15325
GXM18_15320
terpene cyclase/mutase family protein
Accession: QIB56117
Location: 3248220-3250985
NCBI BlastP on this gene
Accession: QIB56117
Location: 3248220-3250985
NCBI BlastP on this gene
GXM18_15315
DUF4430 domain-containing protein
Accession: QIB56116
Location: 3240612-3248192
NCBI BlastP on this gene
Accession: QIB56116
Location: 3240612-3248192
NCBI BlastP on this gene
GXM18_15310
GXM18_15305
GXM18_15300
GXM18_15295
MurR/RpiR family transcriptional regulator
Accession: QIB56112
Location: 3236888-3237703
NCBI BlastP on this gene
Accession: QIB56112
Location: 3236888-3237703
NCBI BlastP on this gene
GXM18_15290
ROK family protein
Accession: QIB56111
Location: 3235958-3236845
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 361
Sequence coverage: 98 %
E-value: 4e-121
NCBI BlastP on this gene
Accession: QIB56111
Location: 3235958-3236845
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 361
Sequence coverage: 98 %
E-value: 4e-121
NCBI BlastP on this gene
GXM18_15285
DUF386 domain-containing protein
Accession: QIB56110
Location: 3235310-3235762
NCBI BlastP on this gene
Accession: QIB56110
Location: 3235310-3235762
NCBI BlastP on this gene
GXM18_15280
GXM18_15275
GXM18_15270
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QIB56107
Location: 3231952-3232650
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
Accession: QIB56107
Location: 3231952-3232650
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
GXM18_15265
N-acetylglucosamine-6-phosphate deacetylase
Accession: QIB56106
Location: 3230486-3231589
NCBI BlastP on this gene
Accession: QIB56106
Location: 3230486-3231589
NCBI BlastP on this gene
nagA
GXM18_15255
GXM18_15250
carbohydrate ABC transporter permease
Accession: QIB56103
Location: 3226632-3227453
NCBI BlastP on this gene
Accession: QIB56103
Location: 3226632-3227453
NCBI BlastP on this gene
GXM18_15245
sugar ABC transporter permease
Accession: QIB56102
Location: 3225723-3226610
NCBI BlastP on this gene
Accession: QIB56102
Location: 3225723-3226610
NCBI BlastP on this gene
GXM18_15240
extracellular solute-binding protein
Accession: QIB56101
Location: 3224282-3225619
NCBI BlastP on this gene
Accession: QIB56101
Location: 3224282-3225619
NCBI BlastP on this gene
GXM18_15235
GXM18_15230
GXM18_15225
GXM18_15220
GIY-YIG nuclease family protein
Accession: QIB56097
Location: 3219673-3219909
NCBI BlastP on this gene
Accession: QIB56097
Location: 3219673-3219909
NCBI BlastP on this gene
GXM18_15215
GXM18_15210
peptidoglycan DD-metalloendopeptidase family protein
Accession: QIB56095
Location: 3216075-3216884
NCBI BlastP on this gene
Accession: QIB56095
Location: 3216075-3216884
NCBI BlastP on this gene
GXM18_15205
SpoIID/LytB domain-containing protein
Accession: QIB56094
Location: 3214948-3215826
NCBI BlastP on this gene
Accession: QIB56094
Location: 3214948-3215826
NCBI BlastP on this gene
GXM18_15200
16S rRNA (cytosine(1402)-N(4))-methyltransferase
Accession: QIB56093
Location: 3214230-3214793
NCBI BlastP on this gene
Accession: QIB56093
Location: 3214230-3214793
NCBI BlastP on this gene
mraW
GXM18_15190
GXM18_15185
484. : CP035945 Blautia producta strain PMF1 chromosome Total score: 2.0 Cumulative Blast bit score: 725
Autoinducer 2 sensor kinase/phosphatase LuxQ
Accession: QBE96085
Location: 1797874-1800723
NCBI BlastP on this gene
Accession: QBE96085
Location: 1797874-1800723
NCBI BlastP on this gene
luxQ_2
PMF13cell1_01622
ams
PMF13cell1_01620
alsK
N-acetyl-D-glucosamine kinase
Accession: QBE96080
Location: 1792410-1793576
NCBI BlastP on this gene
Accession: QBE96080
Location: 1792410-1793576
NCBI BlastP on this gene
nagK
2,2-dialkylglycine decarboxylase
Accession: QBE96079
Location: 1791063-1792361
NCBI BlastP on this gene
Accession: QBE96079
Location: 1791063-1792361
NCBI BlastP on this gene
dgdA
Glucosamine-6-phosphate deaminase 1
Accession: QBE96078
Location: 1790309-1791070
NCBI BlastP on this gene
Accession: QBE96078
Location: 1790309-1791070
NCBI BlastP on this gene
nagB_2
Glycerol-1-phosphate dehydrogenase [NAD(P)+]
Accession: QBE96077
Location: 1789237-1790277
NCBI BlastP on this gene
Accession: QBE96077
Location: 1789237-1790277
NCBI BlastP on this gene
egsA_2
Inner membrane symporter YicJ
Accession: QBE96076
Location: 1787670-1789196
NCBI BlastP on this gene
Accession: QBE96076
Location: 1787670-1789196
NCBI BlastP on this gene
yicJ_2
Glycerol-1-phosphate dehydrogenase [NAD(P)+]
Accession: QBE96075
Location: 1786437-1787477
NCBI BlastP on this gene
Accession: QBE96075
Location: 1786437-1787477
NCBI BlastP on this gene
egsA_1
Multidrug export protein MepA
Accession: QBE96074
Location: 1784878-1786215
NCBI BlastP on this gene
Accession: QBE96074
Location: 1784878-1786215
NCBI BlastP on this gene
mepA_10
PMF13cell1_01611
rbr_2
rbr3A
Peroxide-responsive repressor PerR
Accession: QBE96070
Location: 1780887-1781282
NCBI BlastP on this gene
Accession: QBE96070
Location: 1780887-1781282
NCBI BlastP on this gene
perR
putative HTH-type transcriptional regulator YbbH
Accession: QBE96069
Location: 1779592-1780407
NCBI BlastP on this gene
Accession: QBE96069
Location: 1779592-1780407
NCBI BlastP on this gene
ybbH
Beta-glucoside kinase
Accession: QBE96068
Location: 1778662-1779549
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 360
Sequence coverage: 97 %
E-value: 1e-120
NCBI BlastP on this gene
Accession: QBE96068
Location: 1778662-1779549
BlastP hit with WP_004843625.1
Percentage identity: 59 %
BlastP bit score: 360
Sequence coverage: 97 %
E-value: 1e-120
NCBI BlastP on this gene
bglK
Toxin-antitoxin biofilm protein TabA
Accession: QBE96067
Location: 1778014-1778466
NCBI BlastP on this gene
Accession: QBE96067
Location: 1778014-1778466
NCBI BlastP on this gene
tabA
sglT
nanA
Putative N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QBE96064
Location: 1774656-1775354
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
Accession: QBE96064
Location: 1774656-1775354
BlastP hit with WP_024854132.1
Percentage identity: 76 %
BlastP bit score: 365
Sequence coverage: 99 %
E-value: 1e-124
NCBI BlastP on this gene
nanE
N-acetylglucosamine-6-phosphate deacetylase
Accession: QBE96063
Location: 1773191-1774294
NCBI BlastP on this gene
Accession: QBE96063
Location: 1773191-1774294
NCBI BlastP on this gene
nagA_1
ycdF
bglX_1
L-arabinose transport system permease protein AraQ
Accession: QBE96060
Location: 1769337-1770158
NCBI BlastP on this gene
Accession: QBE96060
Location: 1769337-1770158
NCBI BlastP on this gene
araQ_16
L-arabinose transport system permease protein AraP
Accession: QBE96059
Location: 1768428-1769315
NCBI BlastP on this gene
Accession: QBE96059
Location: 1768428-1769315
NCBI BlastP on this gene
araP_8
Putative binding protein MsmE
Accession: QBE96058
Location: 1766987-1768324
NCBI BlastP on this gene
Accession: QBE96058
Location: 1766987-1768324
NCBI BlastP on this gene
msmE_12
putative response regulatory protein
Accession: QBE96057
Location: 1765070-1766623
NCBI BlastP on this gene
Accession: QBE96057
Location: 1765070-1766623
NCBI BlastP on this gene
PMF13cell1_01595
yehU_3
PMF13cell1_01593
PMF13cell1_01592
PMF13cell1_01591
Gamma-D-glutamyl-L-lysine endopeptidase
Accession: QBE96052
Location: 1759996-1762059
NCBI BlastP on this gene
Accession: QBE96052
Location: 1759996-1762059
NCBI BlastP on this gene
ykfC_1
PMF13cell1_01589
PMF13cell1_01588
PMF13cell1_01587
Ribosomal RNA small subunit methyltransferase H
Accession: QBE96048
Location: 1756978-1757541
NCBI BlastP on this gene
Accession: QBE96048
Location: 1756978-1757541
NCBI BlastP on this gene
rsmH_1
mgl_1
PMF13cell1_01584
485. : LN846931 Carnobacterium maltaromaticum strain LMA28 megaplasmid Total score: 2.0 Cumulative Blast bit score: 693
conserved protein of unknown function
Accession: CRI06599
Location: 40767-41045
NCBI BlastP on this gene
Accession: CRI06599
Location: 40767-41045
NCBI BlastP on this gene
BN424_mp0057
membrane protein of unknown function
Accession: CRI06598
Location: 40220-40540
NCBI BlastP on this gene
Accession: CRI06598
Location: 40220-40540
NCBI BlastP on this gene
BN424_mp0056
BN424_mp0055
putative cell growth regulatory protein
Accession: CRI06596
Location: 39467-39709
NCBI BlastP on this gene
Accession: CRI06596
Location: 39467-39709
NCBI BlastP on this gene
BN424_mp0054
BN424_mp0053
conserved protein of unknown function
Accession: CRI06594
Location: 38722-38934
NCBI BlastP on this gene
Accession: CRI06594
Location: 38722-38934
NCBI BlastP on this gene
BN424_mp0052
tnpA
conserved protein of unknown function
Accession: CRI06592
Location: 34938-35522
NCBI BlastP on this gene
Accession: CRI06592
Location: 34938-35522
NCBI BlastP on this gene
BN424_mp0050
Permease of the major facilitator superfamily (fragment)
Accession: CRI06591
Location: 34501-34923
NCBI BlastP on this gene
Accession: CRI06591
Location: 34501-34923
NCBI BlastP on this gene
BN424_mp0049
BN424_mp0048
BN424_mp0047
membrane protein of unknown function
Accession: CRI06588
Location: 32063-32329
NCBI BlastP on this gene
Accession: CRI06588
Location: 32063-32329
NCBI BlastP on this gene
BN424_mp0046
BN424_mp0045
BN424_mp0044
BN424_mp0043
BN424_mp0042
BN424_mp0041
membrane protein of unknown function
Accession: CRI06582
Location: 27866-28687
NCBI BlastP on this gene
Accession: CRI06582
Location: 27866-28687
NCBI BlastP on this gene
BN424_mp0040
exported protein of unknown function
Accession: CRI06581
Location: 27271-27456
NCBI BlastP on this gene
Accession: CRI06581
Location: 27271-27456
NCBI BlastP on this gene
BN424_mp0039
conserved exported protein of unknown function
Accession: CRI06580
Location: 26748-27245
NCBI BlastP on this gene
Accession: CRI06580
Location: 26748-27245
NCBI BlastP on this gene
BN424_mp0038
BN424_mp0037
membrane protein of unknown function
Accession: CRI06578
Location: 25632-26000
NCBI BlastP on this gene
Accession: CRI06578
Location: 25632-26000
NCBI BlastP on this gene
BN424_mp0036
Peptidoglycan binding domain protein (fragment)
Accession: CRI06577
Location: 23912-25327
NCBI BlastP on this gene
Accession: CRI06577
Location: 23912-25327
NCBI BlastP on this gene
BN424_mp0035
conserved protein of unknown function
Accession: CRI06576
Location: 23349-23882
NCBI BlastP on this gene
Accession: CRI06576
Location: 23349-23882
NCBI BlastP on this gene
BN424_mp0034
conserved protein of unknown function
Accession: CRI06575
Location: 23146-23361
NCBI BlastP on this gene
Accession: CRI06575
Location: 23146-23361
NCBI BlastP on this gene
BN424_mp0033
conserved protein of unknown function
Accession: CRI06574
Location: 21942-22424
NCBI BlastP on this gene
Accession: CRI06574
Location: 21942-22424
NCBI BlastP on this gene
BN424_mp0032
BN424_mp0031
conserved protein of unknown function
Accession: CRI06572
Location: 19884-20786
BlastP hit with WP_004843625.1
Percentage identity: 55 %
BlastP bit score: 337
Sequence coverage: 97 %
E-value: 2e-111
NCBI BlastP on this gene
Accession: CRI06572
Location: 19884-20786
BlastP hit with WP_004843625.1
Percentage identity: 55 %
BlastP bit score: 337
Sequence coverage: 97 %
E-value: 2e-111
NCBI BlastP on this gene
BN424_mp0030
nanA
BN424_mp0028
putative N-acetylmannosamine-6-phosphate 2-epimerase
Accession: CRI06569
Location: 18235-18924
BlastP hit with WP_024854132.1
Percentage identity: 73 %
BlastP bit score: 356
Sequence coverage: 100 %
E-value: 3e-121
NCBI BlastP on this gene
Accession: CRI06569
Location: 18235-18924
BlastP hit with WP_024854132.1
Percentage identity: 73 %
BlastP bit score: 356
Sequence coverage: 100 %
E-value: 3e-121
NCBI BlastP on this gene
nanE
BN424_mp0026
membrane protein of unknown function
Accession: CRI06567
Location: 17573-18055
NCBI BlastP on this gene
Accession: CRI06567
Location: 17573-18055
NCBI BlastP on this gene
BN424_mp0025
conserved protein of unknown function
Accession: CRI06566
Location: 16719-17552
NCBI BlastP on this gene
Accession: CRI06566
Location: 16719-17552
NCBI BlastP on this gene
BN424_mp0024
BN424_mp0023
Sugar ABC superfamily ATP binding cassette transporter, sugar-binding protein
Accession: CRI06564
Location: 14837-16180
NCBI BlastP on this gene
Accession: CRI06564
Location: 14837-16180
NCBI BlastP on this gene
BN424_mp0022
Sugar ABC superfamily ATP binding cassette transporter, membrane protein
Accession: CRI06563
Location: 13983-14819
NCBI BlastP on this gene
Accession: CRI06563
Location: 13983-14819
NCBI BlastP on this gene
BN424_mp0021
putative transport system permease
Accession: CRI06562
Location: 13081-13968
NCBI BlastP on this gene
Accession: CRI06562
Location: 13081-13968
NCBI BlastP on this gene
BN424_mp0020
BN424_mp0019
BN424_mp0018
BN424_mp0017
exported protein of unknown function
Accession: CRI06558
Location: 12031-12201
NCBI BlastP on this gene
Accession: CRI06558
Location: 12031-12201
NCBI BlastP on this gene
BN424_mp0016
BN424_mp0015
conserved protein of unknown function
Accession: CRI06556
Location: 10385-11239
NCBI BlastP on this gene
Accession: CRI06556
Location: 10385-11239
NCBI BlastP on this gene
BN424_mp0014
BN424_mp0013
BN424_mp0012
Cytosine-specific methyltransferase
Accession: CRI06553
Location: 7275-8768
NCBI BlastP on this gene
Accession: CRI06553
Location: 7275-8768
NCBI BlastP on this gene
BN424_mp0011
putative transcriptional regulator, ArpU family
Accession: CRI06552
Location: 6694-7158
NCBI BlastP on this gene
Accession: CRI06552
Location: 6694-7158
NCBI BlastP on this gene
BN424_mp0010
BN424_mp0009
conserved protein of unknown function
Accession: CRI06550
Location: 5791-6051
NCBI BlastP on this gene
Accession: CRI06550
Location: 5791-6051
NCBI BlastP on this gene
BN424_mp0008
BN424_mp0007
BN424_mp0006
BN424_mp0005
BN424_mp0004
BN424_mp0003
BN424_mp0002
BN424_mp0001
486. : CP040462 Enterococcus sp. M190262 plasmid pM190262 Total score: 2.0 Cumulative Blast bit score: 688
FE005_16290
FE005_16285
FE005_16280
deoB
FE005_16270
FE005_16265
FE005_16260
FE005_16255
FE005_16250
FE005_16245
FE005_16240
FE005_16230
FE005_16225
FE005_16220
FE005_16215
FE005_16210
DUF4062 domain-containing protein
Accession: QCT93524
Location: 97322-98506
NCBI BlastP on this gene
Accession: QCT93524
Location: 97322-98506
NCBI BlastP on this gene
FE005_16205
FE005_16200
type II toxin-antitoxin system death-on-curing family toxin
Accession: QCT93522
Location: 96563-96976
NCBI BlastP on this gene
Accession: QCT93522
Location: 96563-96976
NCBI BlastP on this gene
FE005_16195
FE005_16190
ROK family protein
Accession: QCT93520
Location: 94338-95237
BlastP hit with WP_004843625.1
Percentage identity: 58 %
BlastP bit score: 358
Sequence coverage: 100 %
E-value: 1e-119
NCBI BlastP on this gene
Accession: QCT93520
Location: 94338-95237
BlastP hit with WP_004843625.1
Percentage identity: 58 %
BlastP bit score: 358
Sequence coverage: 100 %
E-value: 1e-119
NCBI BlastP on this gene
FE005_16185
FE005_16180
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QCT93630
Location: 92690-93385
BlastP hit with WP_024854132.1
Percentage identity: 68 %
BlastP bit score: 330
Sequence coverage: 99 %
E-value: 3e-111
NCBI BlastP on this gene
Accession: QCT93630
Location: 92690-93385
BlastP hit with WP_024854132.1
Percentage identity: 68 %
BlastP bit score: 330
Sequence coverage: 99 %
E-value: 3e-111
NCBI BlastP on this gene
FE005_16175
MurR/RpiR family transcriptional regulator
Accession: QCT93518
Location: 91613-92455
NCBI BlastP on this gene
Accession: QCT93518
Location: 91613-92455
NCBI BlastP on this gene
FE005_16170
FE005_16165
extracellular solute-binding protein
Accession: QCT93516
Location: 89772-91130
NCBI BlastP on this gene
Accession: QCT93516
Location: 89772-91130
NCBI BlastP on this gene
FE005_16160
carbohydrate ABC transporter permease
Accession: QCT93515
Location: 88917-89753
NCBI BlastP on this gene
Accession: QCT93515
Location: 88917-89753
NCBI BlastP on this gene
FE005_16155
FE005_16150
FE005_16145
pflB
FE005_16135
FE005_16125
FE005_16120
FE005_16115
FE005_16110
ribbon-helix-helix protein, CopG family
Accession: QCT93507
Location: 78131-78394
NCBI BlastP on this gene
Accession: QCT93507
Location: 78131-78394
NCBI BlastP on this gene
FE005_16105
FE005_16100
FE005_16095
FE005_16090
type III secretion system protein PrgN
Accession: QCT93504
Location: 72977-73264
NCBI BlastP on this gene
Accession: QCT93504
Location: 72977-73264
NCBI BlastP on this gene
FE005_16085
487. : CP029150 Glaesserella parasuis strain GZ20170512 chromosome Total score: 2.0 Cumulative Blast bit score: 627
C3363_10635
C3363_10640
protein-ADP-ribose hydrolase
Accession: C3363_10645
Location: 2117861-2118310
NCBI BlastP on this gene
Accession: C3363_10645
Location: 2117861-2118310
NCBI BlastP on this gene
C3363_10645
C3363_10650
type I restriction endonuclease subunit R
Accession: QHB45367
Location: 2118984-2121944
NCBI BlastP on this gene
Accession: QHB45367
Location: 2118984-2121944
NCBI BlastP on this gene
C3363_10655
C3363_10660
DUF1829 domain-containing protein
Accession: QHB45369
Location: 2122399-2123163
NCBI BlastP on this gene
Accession: QHB45369
Location: 2122399-2123163
NCBI BlastP on this gene
C3363_10665
C3363_10670
C3363_10675
type I restriction-modification system subunit M
Accession: QHB45371
Location: 2125766-2127379
NCBI BlastP on this gene
Accession: QHB45371
Location: 2125766-2127379
NCBI BlastP on this gene
C3363_10680
C3363_10685
C3363_10690
spermidine/putrescine ABC transporter substrate-binding protein
Accession: C3363_10695
Location: 2128344-2128600
NCBI BlastP on this gene
Accession: C3363_10695
Location: 2128344-2128600
NCBI BlastP on this gene
C3363_10695
C3363_10700
abortive phage resistance protein
Accession: C3363_10705
Location: 2129146-2129266
NCBI BlastP on this gene
Accession: C3363_10705
Location: 2129146-2129266
NCBI BlastP on this gene
C3363_10705
C3363_10710
C3363_10715
LysR family transcriptional regulator
Accession: QHB45568
Location: 2130433-2131323
NCBI BlastP on this gene
Accession: QHB45568
Location: 2130433-2131323
NCBI BlastP on this gene
C3363_10720
C3363_10725
rplK
C3363_10735
C3363_10740
C3363_10745
bifunctional glycosyl
Location: 2135020-2135301
Location: 2135020-2135301
mrcB
C3363_10755
carbohydrate ABC transporter permease
Accession: C3363_10760
Location: 2136221-2137054
BlastP hit with WP_039959879.1
Percentage identity: 71 %
BlastP bit score: 406
Sequence coverage: 100 %
E-value: 1e-139
NCBI BlastP on this gene
Accession: C3363_10760
Location: 2136221-2137054
BlastP hit with WP_039959879.1
Percentage identity: 71 %
BlastP bit score: 406
Sequence coverage: 100 %
E-value: 1e-139
NCBI BlastP on this gene
C3363_10760
sugar ABC transporter permease
Accession: C3363_10765
Location: 2137056-2137939
NCBI BlastP on this gene
Accession: C3363_10765
Location: 2137056-2137939
NCBI BlastP on this gene
C3363_10765
C3363_10770
C3363_10775
sugar ABC transporter substrate-binding protein
Accession: C3363_10780
Location: 2138740-2139183
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
Accession: C3363_10780
Location: 2138740-2139183
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
C3363_10780
long-chain fatty acid--CoA ligase
Accession: QHB45379
Location: 2139353-2141140
NCBI BlastP on this gene
Accession: QHB45379
Location: 2139353-2141140
NCBI BlastP on this gene
C3363_10785
DNA polymerase III subunit chi
Accession: QHB45380
Location: 2141246-2141683
NCBI BlastP on this gene
Accession: QHB45380
Location: 2141246-2141683
NCBI BlastP on this gene
C3363_10790
C3363_10795
C3363_10800
C3363_10805
copper homeostasis protein CutC
Accession: QHB45383
Location: 2144741-2145472
NCBI BlastP on this gene
Accession: QHB45383
Location: 2144741-2145472
NCBI BlastP on this gene
C3363_10810
aminoimidazole riboside kinase
Accession: QHB45384
Location: 2145634-2146557
NCBI BlastP on this gene
Accession: QHB45384
Location: 2145634-2146557
NCBI BlastP on this gene
C3363_10815
C3363_10820
proS
C3363_10830
C3363_10835
C3363_10840
C3363_10845
C3363_10855
acetyl-CoA carboxylase carboxyl transferase subunit alpha
Accession: C3363_10860
Location: 2156953-2157261
NCBI BlastP on this gene
Accession: C3363_10860
Location: 2156953-2157261
NCBI BlastP on this gene
C3363_10860
electron transport complex subunit A
Accession: QHB45391
Location: 2157465-2158046
NCBI BlastP on this gene
Accession: QHB45391
Location: 2157465-2158046
NCBI BlastP on this gene
C3363_10865
electron transport complex subunit RsxB
Accession: QHB45392
Location: 2158046-2158654
NCBI BlastP on this gene
Accession: QHB45392
Location: 2158046-2158654
NCBI BlastP on this gene
C3363_10870
electron transport complex subunit RsxC
Accession: QHB45393
Location: 2158644-2160533
NCBI BlastP on this gene
Accession: QHB45393
Location: 2158644-2160533
NCBI BlastP on this gene
C3363_10875
488. : CP040243 Glaesserella parasuis strain HPS-1 chromosome Total score: 2.0 Cumulative Blast bit score: 623
iron-containing alcohol dehydrogenase
Accession: FEF05_10995
Location: 2173901-2174562
NCBI BlastP on this gene
Accession: FEF05_10995
Location: 2173901-2174562
NCBI BlastP on this gene
FEF05_10995
FEF05_11000
helix-turn-helix transcriptional regulator
Accession: QEM88915
Location: 2175001-2175366
NCBI BlastP on this gene
Accession: QEM88915
Location: 2175001-2175366
NCBI BlastP on this gene
FEF05_11005
FEF05_11010
FEF05_11015
type I restriction endonuclease subunit R
Accession: QEM88917
Location: 2176574-2179534
NCBI BlastP on this gene
Accession: QEM88917
Location: 2176574-2179534
NCBI BlastP on this gene
FEF05_11020
FEF05_11025
DUF1829 domain-containing protein
Accession: QEM88918
Location: 2179991-2180758
NCBI BlastP on this gene
Accession: QEM88918
Location: 2179991-2180758
NCBI BlastP on this gene
FEF05_11030
FEF05_11035
FEF05_11040
type I restriction-modification system subunit M
Accession: QEM88919
Location: 2182401-2184014
NCBI BlastP on this gene
Accession: QEM88919
Location: 2182401-2184014
NCBI BlastP on this gene
FEF05_11045
FEF05_11050
FEF05_11055
spermidine/putrescine ABC transporter substrate-binding protein
Accession: FEF05_11060
Location: 2184979-2185235
NCBI BlastP on this gene
Accession: FEF05_11060
Location: 2184979-2185235
NCBI BlastP on this gene
FEF05_11060
FEF05_11065
FEF05_11070
FEF05_11075
FEF05_11080
LysR family transcriptional regulator
Accession: QEM88922
Location: 2187069-2187959
NCBI BlastP on this gene
Accession: QEM88922
Location: 2187069-2187959
NCBI BlastP on this gene
FEF05_11085
rplA
rplK
FEF05_11100
FEF05_11105
FEF05_11110
FEF05_11120
carbohydrate ABC transporter permease
Accession: FEF05_11125
Location: 2192857-2193690
BlastP hit with WP_039959879.1
Percentage identity: 70 %
BlastP bit score: 402
Sequence coverage: 100 %
E-value: 8e-138
NCBI BlastP on this gene
Accession: FEF05_11125
Location: 2192857-2193690
BlastP hit with WP_039959879.1
Percentage identity: 70 %
BlastP bit score: 402
Sequence coverage: 100 %
E-value: 8e-138
NCBI BlastP on this gene
FEF05_11125
sugar ABC transporter permease
Accession: FEF05_11130
Location: 2193692-2194575
NCBI BlastP on this gene
Accession: FEF05_11130
Location: 2193692-2194575
NCBI BlastP on this gene
FEF05_11130
FEF05_11135
extracellular solute-binding protein
Accession: FEF05_11140
Location: 2195418-2195861
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
Accession: FEF05_11140
Location: 2195418-2195861
BlastP hit with WP_004843638.1
Percentage identity: 71 %
BlastP bit score: 221
Sequence coverage: 33 %
E-value: 2e-66
NCBI BlastP on this gene
FEF05_11140
long-chain fatty acid--CoA ligase
Accession: QEM88927
Location: 2196031-2197818
NCBI BlastP on this gene
Accession: QEM88927
Location: 2196031-2197818
NCBI BlastP on this gene
FEF05_11145
DNA polymerase III subunit chi
Accession: QEM88928
Location: 2197924-2198361
NCBI BlastP on this gene
Accession: QEM88928
Location: 2197924-2198361
NCBI BlastP on this gene
FEF05_11150
FEF05_11155
galK
FEF05_11165
copper homeostasis protein CutC
Accession: QEM88931
Location: 2201419-2202150
NCBI BlastP on this gene
Accession: QEM88931
Location: 2201419-2202150
NCBI BlastP on this gene
FEF05_11170
aminoimidazole riboside kinase
Accession: QEM88932
Location: 2202312-2203235
NCBI BlastP on this gene
Accession: QEM88932
Location: 2202312-2203235
NCBI BlastP on this gene
FEF05_11175
acpP
proS
autotransporter domain-containing protein
Accession: QEM88935
Location: 2206090-2209719
NCBI BlastP on this gene
Accession: QEM88935
Location: 2206090-2209719
NCBI BlastP on this gene
FEF05_11190
FEF05_11195
FEF05_11200
FEF05_11205
FEF05_11210
pdxY
acetyl-CoA carboxylase carboxyl transferase subunit alpha
Accession: FEF05_11225
Location: 2213184-2213492
NCBI BlastP on this gene
Accession: FEF05_11225
Location: 2213184-2213492
NCBI BlastP on this gene
FEF05_11225
electron transport complex subunit RsxA
Accession: QEM88939
Location: 2213696-2214277
NCBI BlastP on this gene
Accession: QEM88939
Location: 2213696-2214277
NCBI BlastP on this gene
rsxA
electron transport complex subunit RsxB
Accession: QEM88940
Location: 2214277-2214885
NCBI BlastP on this gene
Accession: QEM88940
Location: 2214277-2214885
NCBI BlastP on this gene
rsxB
electron transport complex subunit RsxC
Accession: QEM88941
Location: 2214875-2216875
NCBI BlastP on this gene
Accession: QEM88941
Location: 2214875-2216875
NCBI BlastP on this gene
rsxC
489. : CP020832 Listeria monocytogenes strain PNUSAL000096 chromosome Total score: 2.0 Cumulative Blast bit score: 619
PTS mannitol transporter subunit IIA
Accession: ARJ88578
Location: 401783-402229
NCBI BlastP on this gene
Accession: ARJ88578
Location: 401783-402229
NCBI BlastP on this gene
Y281_01980
Y281_01975
PTS mannitol transporter subunit IIBC
Accession: ARJ88576
Location: 399645-401051
NCBI BlastP on this gene
Accession: ARJ88576
Location: 399645-401051
NCBI BlastP on this gene
Y281_01970
Y281_01965
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ARJ88574
Location: 397895-398608
BlastP hit with WP_024854132.1
Percentage identity: 54 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 9e-85
NCBI BlastP on this gene
Accession: ARJ88574
Location: 397895-398608
BlastP hit with WP_024854132.1
Percentage identity: 54 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 9e-85
NCBI BlastP on this gene
Y281_01960
Y281_01955
Y281_01950
Y281_01945
Y281_01940
Y281_01935
Y281_01930
Y281_01925
Y281_01920
Y281_01915
tRNA uridine-5-carboxymethylaminomethyl(34) synthesis enzyme MnmG
Accession: ARJ88566
Location: 390898-392787
NCBI BlastP on this gene
Accession: ARJ88566
Location: 390898-392787
NCBI BlastP on this gene
Y281_01910
Y281_01905
D-alanyl-D-alanine carboxypeptidase
Accession: ARJ88564
Location: 388324-389142
NCBI BlastP on this gene
Accession: ARJ88564
Location: 388324-389142
NCBI BlastP on this gene
Y281_01900
DUF4918 domain-containing protein
Accession: ARJ88563
Location: 387560-388267
NCBI BlastP on this gene
Accession: ARJ88563
Location: 387560-388267
NCBI BlastP on this gene
Y281_01895
TetR family transcriptional regulator
Accession: ARJ88562
Location: 387009-387536
NCBI BlastP on this gene
Accession: ARJ88562
Location: 387009-387536
NCBI BlastP on this gene
Y281_01890
Y281_01885
Y281_01880
Y281_01875
multidrug efflux MFS transporter NorB
Accession: ARJ88558
Location: 382183-383583
NCBI BlastP on this gene
Accession: ARJ88558
Location: 382183-383583
NCBI BlastP on this gene
Y281_01870
Y281_01865
AraC family transcriptional regulator
Accession: ARJ88556
Location: 378444-380786
NCBI BlastP on this gene
Accession: ARJ88556
Location: 378444-380786
NCBI BlastP on this gene
Y281_01860
Y281_00110
Y281_00105
23S rRNA (pseudouridine(1915)-N(3))-methyltransferase RlmH
Accession: ARJ88236
Location: 17414-17893
NCBI BlastP on this gene
Accession: ARJ88236
Location: 17414-17893
NCBI BlastP on this gene
Y281_00100
LysR family transcriptional regulator
Accession: ARJ88235
Location: 16519-17382
NCBI BlastP on this gene
Accession: ARJ88235
Location: 16519-17382
NCBI BlastP on this gene
Y281_00095
Y281_00090
Y281_00085
Y281_00080
PTS lactose transporter subunit IIC
Accession: ARJ88231
Location: 11525-12838
NCBI BlastP on this gene
Accession: ARJ88231
Location: 11525-12838
NCBI BlastP on this gene
Y281_00075
PTS sugar transporter subunit IIB
Accession: ARJ88230
Location: 11230-11520
NCBI BlastP on this gene
Accession: ARJ88230
Location: 11230-11520
NCBI BlastP on this gene
Y281_00070
Y281_00065
Y281_00060
Y281_00055
Y281_00050
low specificity L-threonine aldolase
Accession: ARJ88226
Location: 7189-8256
NCBI BlastP on this gene
Accession: ARJ88226
Location: 7189-8256
NCBI BlastP on this gene
Y281_00045
DNA (cytosine-5-)-methyltransferase
Accession: ARJ88225
Location: 5899-7113
NCBI BlastP on this gene
Accession: ARJ88225
Location: 5899-7113
NCBI BlastP on this gene
Y281_00040
Y281_00035
PTS cellbiose transporter subunit IIC
Accession: ARJ88223
Location: 3436-4221
NCBI BlastP on this gene
Accession: ARJ88223
Location: 3436-4221
NCBI BlastP on this gene
Y281_00030
Y281_00025
Y281_00020
hydroxymethylpyrimidine/phosphomethylpyrimidine kinase
Accession: ARJ88220
Location: 937-1740
NCBI BlastP on this gene
Accession: ARJ88220
Location: 937-1740
NCBI BlastP on this gene
Y281_00015
Y281_00010
490. : CP000234 Lactobacillus salivarius UCC118 plasmid pMP118 Total score: 2.0 Cumulative Blast bit score: 595
LSL_1906
LSL_1907
LSL_1908
AbpD bacteriocin export accessory protein
Accession: ABE00713
Location: 199638-200792
NCBI BlastP on this gene
Accession: ABE00713
Location: 199638-200792
NCBI BlastP on this gene
abpD
AbpT bacteriocin export accessory protein
Accession: ABE00714
Location: 200809-202968
NCBI BlastP on this gene
Accession: ABE00714
Location: 200809-202968
NCBI BlastP on this gene
abpT
Hypothetical membrane spanning protein
Accession: ABE00715
Location: 203118-203387
NCBI BlastP on this gene
Accession: ABE00715
Location: 203118-203387
NCBI BlastP on this gene
LSL_1911
abpR
AbpK sensory Transduction Histidine Kinase
Accession: ABE00717
Location: 204203-205492
NCBI BlastP on this gene
Accession: ABE00717
Location: 204203-205492
NCBI BlastP on this gene
abpK
abpIP
AbpIM bacteriocin immunity protein
Accession: ABE00719
Location: 205728-205895
NCBI BlastP on this gene
Accession: ABE00719
Location: 205728-205895
NCBI BlastP on this gene
abpIM
Abp118 bacteriocin beta peptide
Accession: ABE00720
Location: 206038-206244
NCBI BlastP on this gene
Accession: ABE00720
Location: 206038-206244
NCBI BlastP on this gene
abp118b
Abp118 bacteriocin alpha peptide
Accession: ABE00721
Location: 206262-206456
NCBI BlastP on this gene
Accession: ABE00721
Location: 206262-206456
NCBI BlastP on this gene
abp118a
LSL_1918
Hypothetical membrane associated protein
Accession: ABE00723
Location: 206824-206985
NCBI BlastP on this gene
Accession: ABE00723
Location: 206824-206985
NCBI BlastP on this gene
LSL_1919
LSL_1920
Nonfunctional salvaricin B precursor
Accession: ABE00725
Location: 207404-207577
NCBI BlastP on this gene
Accession: ABE00725
Location: 207404-207577
NCBI BlastP on this gene
LSL_1921
Hypothetical membrane spanning protein
Accession: ABE00726
Location: 207809-208072
NCBI BlastP on this gene
Accession: ABE00726
Location: 207809-208072
NCBI BlastP on this gene
LSL_1922
LSL_1923
Hypothetical membrane associated protein
Accession: ABE00728
Location: 208361-209329
NCBI BlastP on this gene
Accession: ABE00728
Location: 208361-209329
NCBI BlastP on this gene
LSL_1924
LSL_1925
Hypothetical membrane associated protein
Accession: ABE00730
Location: 209874-210143
NCBI BlastP on this gene
Accession: ABE00730
Location: 209874-210143
NCBI BlastP on this gene
LSL_1926
avtA
Carbon-nitrogen hydrolase family protein
Accession: ABE00732
Location: 211498-212283
NCBI BlastP on this gene
Accession: ABE00732
Location: 211498-212283
NCBI BlastP on this gene
LSL_1928
Conserved hypothetical membrane protein
Accession: ABE00733
Location: 212426-212926
NCBI BlastP on this gene
Accession: ABE00733
Location: 212426-212926
NCBI BlastP on this gene
LSL_1929
LSL_1930
L-serine dehydratase alpha subunit
Accession: ABE00735
Location: 213275-214156
NCBI BlastP on this gene
Accession: ABE00735
Location: 213275-214156
NCBI BlastP on this gene
sdaA
L-serine dehydratase beta subunit
Accession: ABE00736
Location: 214169-214831
NCBI BlastP on this gene
Accession: ABE00736
Location: 214169-214831
NCBI BlastP on this gene
sdaB
Hypothetical membrane spanning protein
Accession: ABE00737
Location: 215944-217380
NCBI BlastP on this gene
Accession: ABE00737
Location: 215944-217380
NCBI BlastP on this gene
LSL_1933
Putative oxidoreductase
Accession: ABE00738
Location: 217472-218587
BlastP hit with WP_004843633.1
Percentage identity: 69 %
BlastP bit score: 527
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABE00738
Location: 217472-218587
BlastP hit with WP_004843633.1
Percentage identity: 69 %
BlastP bit score: 527
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
LSL_1934
Conserved hypothetical protein
Accession: ABE00739
Location: 218978-219427
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 68
Sequence coverage: 59 %
E-value: 1e-11
NCBI BlastP on this gene
Accession: ABE00739
Location: 218978-219427
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 68
Sequence coverage: 59 %
E-value: 1e-11
NCBI BlastP on this gene
LSL_1935
ndk
nagC
nanA
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ABE00743
Location: 222008-222697
NCBI BlastP on this gene
Accession: ABE00743
Location: 222008-222697
NCBI BlastP on this gene
nanE
Transcriptional regulator, RpiR family
Accession: ABE00744
Location: 222743-223567
NCBI BlastP on this gene
Accession: ABE00744
Location: 222743-223567
NCBI BlastP on this gene
LSL_1940
Hypothetical membrane associated protein
Accession: ABE00745
Location: 223624-224241
NCBI BlastP on this gene
Accession: ABE00745
Location: 223624-224241
NCBI BlastP on this gene
LSL_1941
potE
LSL_1943
rpsN
Tetracycline resistance protein
Accession: ABE00749
Location: 226555-228474
NCBI BlastP on this gene
Accession: ABE00749
Location: 226555-228474
NCBI BlastP on this gene
fusA
tktA
mipB
rpe
PTS system, mannose-specific IID component
Accession: ABE00753
Location: 232393-233223
NCBI BlastP on this gene
Accession: ABE00753
Location: 232393-233223
NCBI BlastP on this gene
LSL_1949
PTS system, mannose-specific IIC component
Accession: ABE00754
Location: 233223-233987
NCBI BlastP on this gene
Accession: ABE00754
Location: 233223-233987
NCBI BlastP on this gene
LSL_1950
PTS system, mannose-specific IIAB component
Accession: ABE00755
Location: 234005-234481
NCBI BlastP on this gene
Accession: ABE00755
Location: 234005-234481
NCBI BlastP on this gene
LSL_1951
PTS system, mannose-specific IIAB component
Accession: ABE00756
Location: 234495-234941
NCBI BlastP on this gene
Accession: ABE00756
Location: 234495-234941
NCBI BlastP on this gene
manX
Ribitol-5-phosphate 2-dehydrogenase
Accession: ABE00757
Location: 234955-236028
NCBI BlastP on this gene
Accession: ABE00757
Location: 234955-236028
NCBI BlastP on this gene
tdh
Transcriptional regulator, DeoR family
Accession: ABE00758
Location: 236246-237001
NCBI BlastP on this gene
Accession: ABE00758
Location: 236246-237001
NCBI BlastP on this gene
LSL_1955
Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase
Accession: ABE00759
Location: 237097-239463
NCBI BlastP on this gene
Accession: ABE00759
Location: 237097-239463
NCBI BlastP on this gene
LSL_1956
491. : CP020814 Bacillus krulwichiae strain AM31D chromosome Total score: 2.0 Cumulative Blast bit score: 565
putative cadmium-transporting ATPase
Accession: ARK29826
Location: 1446761-1448908
NCBI BlastP on this gene
Accession: ARK29826
Location: 1446761-1448908
NCBI BlastP on this gene
cadA
BkAM31D_08115
BkAM31D_08125
Putative ABC transporter substrate-binding protein YesO
Accession: ARK29828
Location: 1450705-1452000
NCBI BlastP on this gene
Accession: ARK29828
Location: 1450705-1452000
NCBI BlastP on this gene
yesO
L-arabinose transport system permease protein AraP
Accession: ARK29829
Location: 1452165-1453091
NCBI BlastP on this gene
Accession: ARK29829
Location: 1452165-1453091
NCBI BlastP on this gene
araP
L-arabinose transport system permease protein AraQ
Accession: ARK29830
Location: 1453106-1453951
NCBI BlastP on this gene
Accession: ARK29830
Location: 1453106-1453951
NCBI BlastP on this gene
araQ_2
5-dehydro-2-deoxygluconokinase
Accession: ARK29831
Location: 1453965-1454927
NCBI BlastP on this gene
Accession: ARK29831
Location: 1453965-1454927
NCBI BlastP on this gene
iolC_2
sacC_1
BkAM31D_08155
putative ABC transporter ATP-binding protein YxlF
Accession: ARK29834
Location: 1457174-1458094
NCBI BlastP on this gene
Accession: ARK29834
Location: 1457174-1458094
NCBI BlastP on this gene
yxlF_1
ABC-2 family transporter protein
Accession: ARK29835
Location: 1458087-1459043
NCBI BlastP on this gene
Accession: ARK29835
Location: 1458087-1459043
NCBI BlastP on this gene
BkAM31D_08165
Transcriptional regulatory protein YycF
Accession: ARK29836
Location: 1459140-1459838
NCBI BlastP on this gene
Accession: ARK29836
Location: 1459140-1459838
NCBI BlastP on this gene
yycF_1
Alkaline phosphatase synthesis sensor protein PhoR
Accession: ARK29837
Location: 1459841-1461223
NCBI BlastP on this gene
Accession: ARK29837
Location: 1459841-1461223
NCBI BlastP on this gene
phoR_1
Inner membrane transport protein YdhC
Accession: ARK29838
Location: 1461301-1462503
NCBI BlastP on this gene
Accession: ARK29838
Location: 1461301-1462503
NCBI BlastP on this gene
ydhC
qorB
Putative HTH-type transcriptional regulator YwnA
Accession: ARK29840
Location: 1463813-1464238
NCBI BlastP on this gene
Accession: ARK29840
Location: 1463813-1464238
NCBI BlastP on this gene
ywnA_1
BkAM31D_08195
N-acetylglucosamine-6-phosphate deacetylase
Accession: ARK29842
Location: 1465300-1466484
NCBI BlastP on this gene
Accession: ARK29842
Location: 1465300-1466484
NCBI BlastP on this gene
nagA_1
Sialidase A precursor
Accession: ARK29843
Location: 1466576-1468978
BlastP hit with WP_004843631.1
Percentage identity: 31 %
BlastP bit score: 293
Sequence coverage: 98 %
E-value: 8e-84
NCBI BlastP on this gene
Accession: ARK29843
Location: 1466576-1468978
BlastP hit with WP_004843631.1
Percentage identity: 31 %
BlastP bit score: 293
Sequence coverage: 98 %
E-value: 8e-84
NCBI BlastP on this gene
nanA_1
putative HTH-type transcriptional regulator YbbH
Accession: ARK29844
Location: 1469236-1470162
NCBI BlastP on this gene
Accession: ARK29844
Location: 1469236-1470162
NCBI BlastP on this gene
ybbH_1
Putative N-acetylmannosamine-6-phosphate 2-epimerase
Accession: ARK29845
Location: 1470402-1471091
BlastP hit with WP_024854132.1
Percentage identity: 60 %
BlastP bit score: 272
Sequence coverage: 96 %
E-value: 3e-88
NCBI BlastP on this gene
Accession: ARK29845
Location: 1470402-1471091
BlastP hit with WP_024854132.1
Percentage identity: 60 %
BlastP bit score: 272
Sequence coverage: 96 %
E-value: 3e-88
NCBI BlastP on this gene
nanE
Sialic acid-binding periplasmic protein SiaP precursor
Accession: ARK29846
Location: 1471088-1472119
NCBI BlastP on this gene
Accession: ARK29846
Location: 1471088-1472119
NCBI BlastP on this gene
siaP_2
Sialic acid TRAP transporter permease protein SiaT
Accession: ARK29847
Location: 1472147-1472626
NCBI BlastP on this gene
Accession: ARK29847
Location: 1472147-1472626
NCBI BlastP on this gene
siaT_7
Sialic acid TRAP transporter permease protein SiaT
Accession: ARK29848
Location: 1472627-1472980
NCBI BlastP on this gene
Accession: ARK29848
Location: 1472627-1472980
NCBI BlastP on this gene
siaT_8
Sialic acid TRAP transporter permease protein SiaT
Accession: ARK29849
Location: 1473001-1473903
NCBI BlastP on this gene
Accession: ARK29849
Location: 1473001-1473903
NCBI BlastP on this gene
siaT_9
glkA
sda_2
nanA_2
Glucosamine-6-phosphate deaminase 1
Accession: ARK29853
Location: 1476044-1476781
NCBI BlastP on this gene
Accession: ARK29853
Location: 1476044-1476781
NCBI BlastP on this gene
nagB_1
Inner membrane transport permease YbhR
Accession: ARK29854
Location: 1477031-1478143
NCBI BlastP on this gene
Accession: ARK29854
Location: 1477031-1478143
NCBI BlastP on this gene
ybhR
Daunorubicin/doxorubicin resistance ATP-binding protein DrrA
Accession: ARK29855
Location: 1478145-1479074
NCBI BlastP on this gene
Accession: ARK29855
Location: 1478145-1479074
NCBI BlastP on this gene
drrA_1
Putative glutamine amidotransferase
Accession: ARK29856
Location: 1479278-1480006
NCBI BlastP on this gene
Accession: ARK29856
Location: 1479278-1480006
NCBI BlastP on this gene
BkAM31D_08270
BkAM31D_08275
Putative ring-cleaving dioxygenase MhqO
Accession: ARK29858
Location: 1481065-1481922
NCBI BlastP on this gene
Accession: ARK29858
Location: 1481065-1481922
NCBI BlastP on this gene
mhqO
flr
mhqD_1
catD_1
NAD(P)H dehydrogenase (quinone)
Accession: ARK29862
Location: 1483903-1484562
NCBI BlastP on this gene
Accession: ARK29862
Location: 1483903-1484562
NCBI BlastP on this gene
BkAM31D_08300
BkAM31D_08305
BkAM31D_08310
yiaA
putative HTH-type transcriptional regulator YurK
Accession: ARK29866
Location: 1486079-1486462
NCBI BlastP on this gene
Accession: ARK29866
Location: 1486079-1486462
NCBI BlastP on this gene
yurK
ABC transporter ATP-binding protein YtrB
Accession: ARK29867
Location: 1486452-1487330
NCBI BlastP on this gene
Accession: ARK29867
Location: 1486452-1487330
NCBI BlastP on this gene
ytrB_2
BkAM31D_08330
RNA polymerase factor sigma-70
Accession: ARK29869
Location: 1488151-1488654
NCBI BlastP on this gene
Accession: ARK29869
Location: 1488151-1488654
NCBI BlastP on this gene
BkAM31D_08335
Quercetin 2,3-dioxygenase
Location: 1489135-1490012
Location: 1489135-1490012
yhhW
putative rhodanese-related sulfurtransferase
Accession: ARK29870
Location: 1490105-1491064
NCBI BlastP on this gene
Accession: ARK29870
Location: 1490105-1491064
NCBI BlastP on this gene
BkAM31D_08350
492. : CP011391 Faecalibaculum rodentium strain Alo17 Total score: 2.0 Cumulative Blast bit score: 544
AALO17_07250
AALO17_07260
nucleotidyltransferase family protein
Accession: AMK53861
Location: 653817-654818
NCBI BlastP on this gene
Accession: AMK53861
Location: 653817-654818
NCBI BlastP on this gene
AALO17_07270
AALO17_07280
HisJ family histidinol phosphate phosphatase
Accession: AMK53863
Location: 655767-656552
NCBI BlastP on this gene
Accession: AMK53863
Location: 655767-656552
NCBI BlastP on this gene
AALO17_07290
AALO17_07300
AALO17_07310
AALO17_07320
AALO17_07330
AALO17_07340
AALO17_07350
AALO17_07360
single-stranded DNA-binding protein
Accession: AMK53871
Location: 663040-663507
NCBI BlastP on this gene
Accession: AMK53871
Location: 663040-663507
NCBI BlastP on this gene
AALO17_07370
AALO17_07380
AALO17_07390
AALO17_07400
AALO17_07410
AALO17_07420
AALO17_07430
AALO17_07440
AALO17_07450
yhcH/YjgK/YiaL family protein
Accession: AMK53880
Location: 671957-672430
BlastP hit with WP_004843641.1
Percentage identity: 32 %
BlastP bit score: 67
Sequence coverage: 71 %
E-value: 2e-11
NCBI BlastP on this gene
Accession: AMK53880
Location: 671957-672430
BlastP hit with WP_004843641.1
Percentage identity: 32 %
BlastP bit score: 67
Sequence coverage: 71 %
E-value: 2e-11
NCBI BlastP on this gene
AALO17_07460
solute:sodium symporter (SSS) family transporter
Accession: AMK53881
Location: 672427-673992
NCBI BlastP on this gene
Accession: AMK53881
Location: 672427-673992
NCBI BlastP on this gene
AALO17_07470
N-acetylneuraminate lyase
Accession: AMK53882
Location: 674080-674997
BlastP hit with WP_024854133.1
Percentage identity: 75 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 1e-166
NCBI BlastP on this gene
Accession: AMK53882
Location: 674080-674997
BlastP hit with WP_024854133.1
Percentage identity: 75 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 1e-166
NCBI BlastP on this gene
AALO17_07480
AALO17_07490
AALO17_07500
AALO17_07510
AALO17_07520
AALO17_07530
AALO17_07540
AALO17_07550
macrolide ABC transporter ATP-binding protein
Accession: AMK53890
Location: 682736-683440
NCBI BlastP on this gene
Accession: AMK53890
Location: 682736-683440
NCBI BlastP on this gene
AALO17_07560
DegV family EDD domain-containing protein
Accession: AMK53891
Location: 683557-684420
NCBI BlastP on this gene
Accession: AMK53891
Location: 683557-684420
NCBI BlastP on this gene
AALO17_07570
AALO17_07580
AALO17_07590
AALO17_07600
AALO17_07610
AALO17_07620
AALO17_07630
AALO17_07640
AALO17_07650
AALO17_07660
AALO17_07670
primary replicative DNA helicase
Accession: AMK53902
Location: 691705-693054
NCBI BlastP on this gene
Accession: AMK53902
Location: 691705-693054
NCBI BlastP on this gene
AALO17_07680
AALO17_07690
uracil phosphoribosyltransferase
Accession: AMK53904
Location: 694012-694632
NCBI BlastP on this gene
Accession: AMK53904
Location: 694012-694632
NCBI BlastP on this gene
AALO17_07700
AALO17_07710
AALO17_07720
493. : CP003700 Fusobacterium nucleatum subsp. vincentii 3_1_36A2 Total score: 2.0 Cumulative Blast bit score: 434
HMPREF0946_01187
HMPREF0946_01186
HMPREF0946_01185
rod shape-determining protein MreC
Accession: EEU33111
Location: 1929902-1930768
NCBI BlastP on this gene
Accession: EEU33111
Location: 1929902-1930768
NCBI BlastP on this gene
HMPREF0946_01184
HMPREF0946_01183
HMPREF0946_01182
PTS system, fructose subfamily, IIA component
Accession: EEU33108
Location: 1928148-1928621
NCBI BlastP on this gene
Accession: EEU33108
Location: 1928148-1928621
NCBI BlastP on this gene
HMPREF0946_01181
transcriptional repressor NrdR
Accession: EEU33107
Location: 1927680-1928129
NCBI BlastP on this gene
Accession: EEU33107
Location: 1927680-1928129
NCBI BlastP on this gene
HMPREF0946_01180
methionyl-tRNA formyltransferase
Accession: EEU33106
Location: 1926727-1927659
NCBI BlastP on this gene
Accession: EEU33106
Location: 1926727-1927659
NCBI BlastP on this gene
HMPREF0946_01179
HMPREF0946_01178
UPF0735 ACT domain-containing protein
Accession: EEU33104
Location: 1925405-1925866
NCBI BlastP on this gene
Accession: EEU33104
Location: 1925405-1925866
NCBI BlastP on this gene
HMPREF0946_01177
HMPREF0946_01176
HMPREF0946_01175
HMPREF0946_01174
adenine phosphoribosyltransferase
Accession: EEU33100
Location: 1921897-1922409
NCBI BlastP on this gene
Accession: EEU33100
Location: 1921897-1922409
NCBI BlastP on this gene
HMPREF0946_01173
HMPREF0946_01172
queuine tRNA-ribosyltransferase
Accession: EEU33098
Location: 1918568-1919689
NCBI BlastP on this gene
Accession: EEU33098
Location: 1918568-1919689
NCBI BlastP on this gene
HMPREF0946_01171
HMPREF0946_01170
HMPREF0946_01169
HMPREF0946_01168
HMPREF0946_01167
hypothetical protein
Accession: EEU33093
Location: 1914239-1914913
BlastP hit with WP_024854132.1
Percentage identity: 58 %
BlastP bit score: 265
Sequence coverage: 99 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: EEU33093
Location: 1914239-1914913
BlastP hit with WP_024854132.1
Percentage identity: 58 %
BlastP bit score: 265
Sequence coverage: 99 %
E-value: 2e-85
NCBI BlastP on this gene
HMPREF0946_01166
HMPREF0946_01165
HMPREF0946_01164
TRAP transporter, DctM subunit
Accession: EEU33090
Location: 1910597-1912450
NCBI BlastP on this gene
Accession: EEU33090
Location: 1910597-1912450
NCBI BlastP on this gene
HMPREF0946_01163
DctP family TRAP transporter solute receptor
Accession: EEU33089
Location: 1909590-1910573
NCBI BlastP on this gene
Accession: EEU33089
Location: 1909590-1910573
NCBI BlastP on this gene
HMPREF0946_01162
HMPREF0946_01161
cyclically-permuted mutarotase
Accession: EEU33087
Location: 1907404-1908552
NCBI BlastP on this gene
Accession: EEU33087
Location: 1907404-1908552
NCBI BlastP on this gene
HMPREF0946_01160
type I restriction-modification system, M subunit
Accession: EEU33086
Location: 1905592-1907154
NCBI BlastP on this gene
Accession: EEU33086
Location: 1905592-1907154
NCBI BlastP on this gene
HMPREF0946_01159
HMPREF0946_01158
HMPREF0946_01157
HsdR family type I site-specific deoxyribonuclease
Accession: EEU33083
Location: 1899625-1902666
NCBI BlastP on this gene
Accession: EEU33083
Location: 1899625-1902666
NCBI BlastP on this gene
HMPREF0946_01156
HMPREF0946_01155
HMPREF0946_01154
HMPREF0946_01153
HMPREF0946_01152
HMPREF0946_01151
hypothetical protein
Accession: EEU33077
Location: 1895241-1895642
BlastP hit with WP_004843624.1
Percentage identity: 62 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 1e-50
NCBI BlastP on this gene
Accession: EEU33077
Location: 1895241-1895642
BlastP hit with WP_004843624.1
Percentage identity: 62 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 1e-50
NCBI BlastP on this gene
HMPREF0946_01150
1-deoxy-D-xylulose-5-phosphate synthase
Accession: EEU33076
Location: 1893480-1895231
NCBI BlastP on this gene
Accession: EEU33076
Location: 1893480-1895231
NCBI BlastP on this gene
HMPREF0946_01149
HMPREF0946_01148
HMPREF0946_01147
HMPREF0946_01146
pyridoxal biosynthesis lyase pdxS
Accession: EEU33072
Location: 1890839-1891681
NCBI BlastP on this gene
Accession: EEU33072
Location: 1890839-1891681
NCBI BlastP on this gene
HMPREF0946_01145
HMPREF0946_01144
UDP-N-acetylmuramoyl-tripeptide-D-alanyl-D- alanine ligase
Accession: EEU33070
Location: 1887339-1889162
NCBI BlastP on this gene
Accession: EEU33070
Location: 1887339-1889162
NCBI BlastP on this gene
HMPREF0946_01143
phospho-N-acetylmuramoyl-pentapeptide- transferase
Accession: EEU33069
Location: 1885990-1887075
NCBI BlastP on this gene
Accession: EEU33069
Location: 1885990-1887075
NCBI BlastP on this gene
HMPREF0946_01142
UDP-N-acetylmuramoylalanine-D-glutamate ligase
Accession: EEU33068
Location: 1884692-1885990
NCBI BlastP on this gene
Accession: EEU33068
Location: 1884692-1885990
NCBI BlastP on this gene
HMPREF0946_01141
undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase
Accession: EEU33067
Location: 1883618-1884682
NCBI BlastP on this gene
Accession: EEU33067
Location: 1883618-1884682
NCBI BlastP on this gene
HMPREF0946_01140
UDP-N-acetylmuramate-L-alanine ligase
Accession: EEU33066
Location: 1882231-1883613
NCBI BlastP on this gene
Accession: EEU33066
Location: 1882231-1883613
NCBI BlastP on this gene
HMPREF0946_01139
UDP-N-acetylenolpyruvoylglucosamine reductase
Accession: EEU33065
Location: 1881389-1882234
NCBI BlastP on this gene
Accession: EEU33065
Location: 1881389-1882234
NCBI BlastP on this gene
HMPREF0946_01138
HMPREF0946_01137
HMPREF0946_01136
HMPREF0946_01135
HMPREF0946_01134
AspT/YidE/YbjL antiporter duplication domain-containing protein
Accession: EEU33060
Location: 1875751-1877169
NCBI BlastP on this gene
Accession: EEU33060
Location: 1875751-1877169
NCBI BlastP on this gene
HMPREF0946_01133
494. : CP010855 Marinovum algicola DG 898 Total score: 2.0 Cumulative Blast bit score: 377
MALG_00084
MALG_00082
putative acyl-CoA transferase/carnitine dehydratase
Accession: AKO95299
Location: 91669-92862
NCBI BlastP on this gene
Accession: AKO95299
Location: 91669-92862
NCBI BlastP on this gene
MALG_00081
collagen triple helix repeat protein-containing protein
Accession: AKO95298
Location: 88234-91224
NCBI BlastP on this gene
Accession: AKO95298
Location: 88234-91224
NCBI BlastP on this gene
MALG_00080
MALG_00079
MALG_00078
MALG_00077
type I secretion membrane fusion protein, HlyD family
Accession: AKO95294
Location: 83010-84326
NCBI BlastP on this gene
Accession: AKO95294
Location: 83010-84326
NCBI BlastP on this gene
MALG_00076
MALG_00075
diaminobutyrate acetyltransferase
Accession: AKO95292
Location: 81727-82260
NCBI BlastP on this gene
Accession: AKO95292
Location: 81727-82260
NCBI BlastP on this gene
MALG_00074
diaminobutyrate--2-oxoglutarate aminotransferase
Accession: AKO95291
Location: 80380-81672
NCBI BlastP on this gene
Accession: AKO95291
Location: 80380-81672
NCBI BlastP on this gene
MALG_00073
MALG_00072
MALG_00071
MALG_00070
putative metal-dependent hydrolase of the TIM-barrel fold protein
Accession: AKO95287
Location: 77397-78236
NCBI BlastP on this gene
Accession: AKO95287
Location: 77397-78236
NCBI BlastP on this gene
MALG_00069
L-fuco-beta-pyranose dehydrogenase
Accession: AKO95286
Location: 76457-77395
NCBI BlastP on this gene
Accession: AKO95286
Location: 76457-77395
NCBI BlastP on this gene
MALG_00068
ABC-type sugar transport system, ATPase component
Accession: AKO95285
Location: 75393-76460
NCBI BlastP on this gene
Accession: AKO95285
Location: 75393-76460
NCBI BlastP on this gene
MALG_00067
ABC-type sugar transport system, permease component
Accession: AKO95284
Location: 74491-75378
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 187
Sequence coverage: 87 %
E-value: 3e-53
NCBI BlastP on this gene
Accession: AKO95284
Location: 74491-75378
BlastP hit with WP_039959892.1
Percentage identity: 36 %
BlastP bit score: 187
Sequence coverage: 87 %
E-value: 3e-53
NCBI BlastP on this gene
MALG_00066
ABC-type sugar transport system, periplasmic component
Accession: AKO95283
Location: 73186-74433
NCBI BlastP on this gene
Accession: AKO95283
Location: 73186-74433
NCBI BlastP on this gene
MALG_00065
ABC-type sugar transport system, permease component
Accession: AKO95282
Location: 72253-73131
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 190
Sequence coverage: 99 %
E-value: 1e-54
NCBI BlastP on this gene
Accession: AKO95282
Location: 72253-73131
BlastP hit with WP_039959879.1
Percentage identity: 36 %
BlastP bit score: 190
Sequence coverage: 99 %
E-value: 1e-54
NCBI BlastP on this gene
MALG_00064
MALG_00063
NAD-dependent aldehyde dehydrogenase
Accession: AKO95280
Location: 69708-71141
NCBI BlastP on this gene
Accession: AKO95280
Location: 69708-71141
NCBI BlastP on this gene
MALG_00062
2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase
Accession: AKO95279
Location: 68905-69711
NCBI BlastP on this gene
Accession: AKO95279
Location: 68905-69711
NCBI BlastP on this gene
MALG_00061
MALG_00060
MALG_00059
MALG_00058
MALG_00057
MALG_00056
Beta-galactosidase/beta-glucuronidase
Accession: AKO95273
Location: 62659-64440
NCBI BlastP on this gene
Accession: AKO95273
Location: 62659-64440
NCBI BlastP on this gene
MALG_00055
tripartite ATP-independent periplasmic transporter solute receptor, DctP family
Accession: AKO95272
Location: 61674-62651
NCBI BlastP on this gene
Accession: AKO95272
Location: 61674-62651
NCBI BlastP on this gene
MALG_00054
TRAP-type C4-dicarboxylate transport system, small permease component
Accession: AKO95271
Location: 60953-61456
NCBI BlastP on this gene
Accession: AKO95271
Location: 60953-61456
NCBI BlastP on this gene
MALG_00053
MALG_00052
Alpha-glucosidase, family 31 of glycosyl hydrolase
Accession: AKO95269
Location: 57278-59641
NCBI BlastP on this gene
Accession: AKO95269
Location: 57278-59641
NCBI BlastP on this gene
MALG_00051
Mannitol-1-phosphate/altronate dehydrogenase
Accession: AKO95268
Location: 55878-57281
NCBI BlastP on this gene
Accession: AKO95268
Location: 55878-57281
NCBI BlastP on this gene
MALG_00050
MALG_00049
495. : CP034358 Clostridium septicum strain VAT12 chromosome Total score: 2.0 Cumulative Blast bit score: 351
AraC family transcriptional regulator
Accession: QAS60346
Location: 1277972-1278643
NCBI BlastP on this gene
Accession: QAS60346
Location: 1277972-1278643
NCBI BlastP on this gene
EI377_06125
EI377_06120
EI377_06115
EI377_06110
YccF domain-containing protein
Accession: QAS60343
Location: 1274703-1275065
NCBI BlastP on this gene
Accession: QAS60343
Location: 1274703-1275065
NCBI BlastP on this gene
EI377_06105
TIGR02206 family membrane protein
Accession: QAS60342
Location: 1273867-1274628
NCBI BlastP on this gene
Accession: QAS60342
Location: 1273867-1274628
NCBI BlastP on this gene
EI377_06100
EI377_06095
EI377_06090
EI377_06085
EI377_06080
EI377_06075
TIGR01906 family membrane protein
Accession: QAS60338
Location: 1267472-1268173
NCBI BlastP on this gene
Accession: QAS60338
Location: 1267472-1268173
NCBI BlastP on this gene
EI377_06070
EI377_06065
EI377_06060
EI377_06055
EI377_06050
EI377_06045
EI377_06040
EI377_06035
EI377_06030
MurR/RpiR family transcriptional regulator
Accession: EI377_06025
Location: 1258276-1258694
NCBI BlastP on this gene
Accession: EI377_06025
Location: 1258276-1258694
NCBI BlastP on this gene
EI377_06025
DUF386 domain-containing protein
Accession: QAS60333
Location: 1257619-1258071
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
Accession: QAS60333
Location: 1257619-1258071
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
EI377_06020
MurR/RpiR family transcriptional regulator
Accession: QAS60332
Location: 1256560-1257357
NCBI BlastP on this gene
Accession: QAS60332
Location: 1256560-1257357
NCBI BlastP on this gene
EI377_06015
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: QAS60331
Location: 1255768-1256412
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
Accession: QAS60331
Location: 1255768-1256412
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
EI377_06010
PTS glucose transporter subunit IIB
Accession: QAS60330
Location: 1254208-1255698
NCBI BlastP on this gene
Accession: QAS60330
Location: 1254208-1255698
NCBI BlastP on this gene
EI377_06005
EI377_06000
EI377_05995
EI377_05990
EI377_05985
EI377_05980
GTP pyrophosphokinase family protein
Accession: QAS60324
Location: 1247356-1248150
NCBI BlastP on this gene
Accession: QAS60324
Location: 1247356-1248150
NCBI BlastP on this gene
EI377_05975
DUF2225 domain-containing protein
Accession: QAS60323
Location: 1246401-1247171
NCBI BlastP on this gene
Accession: QAS60323
Location: 1246401-1247171
NCBI BlastP on this gene
EI377_05970
EI377_05965
EI377_05960
EI377_05955
beta-channel forming cytolysin
Accession: QAS60319
Location: 1243349-1244299
NCBI BlastP on this gene
Accession: QAS60319
Location: 1243349-1244299
NCBI BlastP on this gene
EI377_05950
EI377_05945
HAMP domain-containing histidine kinase
Accession: QAS60317
Location: 1241664-1242563
NCBI BlastP on this gene
Accession: QAS60317
Location: 1241664-1242563
NCBI BlastP on this gene
EI377_05940
EI377_05935
ATP-binding cassette domain-containing protein
Accession: QAS60315
Location: 1239977-1240900
NCBI BlastP on this gene
Accession: QAS60315
Location: 1239977-1240900
NCBI BlastP on this gene
EI377_05930
response regulator transcription factor
Accession: QAS60314
Location: 1239192-1239887
NCBI BlastP on this gene
Accession: QAS60314
Location: 1239192-1239887
NCBI BlastP on this gene
EI377_05925
EI377_05920
ArsR family transcriptional regulator
Accession: QAS60312
Location: 1236154-1236432
NCBI BlastP on this gene
Accession: QAS60312
Location: 1236154-1236432
NCBI BlastP on this gene
EI377_05915
496. : CP023671 Clostridium septicum strain DSM 7534 chromosome Total score: 2.0 Cumulative Blast bit score: 351
CP523_11340
CP523_11335
ABC transporter ATP-binding protein
Accession: AYE34956
Location: 2471482-2472267
NCBI BlastP on this gene
Accession: AYE34956
Location: 2471482-2472267
NCBI BlastP on this gene
CP523_11330
iron ABC transporter permease
Accession: AYE34955
Location: 2470485-2471492
NCBI BlastP on this gene
Accession: AYE34955
Location: 2470485-2471492
NCBI BlastP on this gene
CP523_11325
ABC transporter substrate-binding protein
Accession: AYE34954
Location: 2469466-2470461
NCBI BlastP on this gene
Accession: AYE34954
Location: 2469466-2470461
NCBI BlastP on this gene
CP523_11320
CP523_11315
CP523_11310
AraC family transcriptional regulator
Accession: AYE34951
Location: 2467155-2467826
NCBI BlastP on this gene
Accession: AYE34951
Location: 2467155-2467826
NCBI BlastP on this gene
CP523_11305
CP523_11300
CP523_11295
TIGR02206 family membrane protein
Accession: AYE34948
Location: 2464684-2465445
NCBI BlastP on this gene
Accession: AYE34948
Location: 2464684-2465445
NCBI BlastP on this gene
CP523_11290
CP523_11285
CP523_11280
CP523_11275
CP523_11270
CP523_11265
TIGR01906 family membrane protein
Accession: AYE34944
Location: 2458365-2459066
NCBI BlastP on this gene
Accession: AYE34944
Location: 2458365-2459066
NCBI BlastP on this gene
CP523_11260
CP523_11255
CP523_11250
CP523_11245
type I methionyl aminopeptidase
Accession: AYE34940
Location: 2454540-2455409
NCBI BlastP on this gene
Accession: AYE34940
Location: 2454540-2455409
NCBI BlastP on this gene
map
CP523_11235
DUF386 domain-containing protein
Accession: AYE34939
Location: 2453268-2453720
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
Accession: AYE34939
Location: 2453268-2453720
BlastP hit with WP_004843641.1
Percentage identity: 31 %
BlastP bit score: 76
Sequence coverage: 99 %
E-value: 2e-14
NCBI BlastP on this gene
CP523_11230
MurR/RpiR family transcriptional regulator
Accession: AYE34938
Location: 2452209-2453006
NCBI BlastP on this gene
Accession: AYE34938
Location: 2452209-2453006
NCBI BlastP on this gene
CP523_11225
N-acetylmannosamine-6-phosphate 2-epimerase
Accession: AYE34937
Location: 2451417-2452061
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
Accession: AYE34937
Location: 2451417-2452061
BlastP hit with WP_024854132.1
Percentage identity: 63 %
BlastP bit score: 275
Sequence coverage: 95 %
E-value: 2e-89
NCBI BlastP on this gene
CP523_11220
PTS glucose transporter subunit IIB
Accession: AYE34936
Location: 2449857-2451347
NCBI BlastP on this gene
Accession: AYE34936
Location: 2449857-2451347
NCBI BlastP on this gene
CP523_11215
CP523_11210
CP523_11205
CP523_11200
CP523_11195
CP523_11190
CP523_11185
DUF2225 domain-containing protein
Accession: AYE34929
Location: 2442050-2442820
NCBI BlastP on this gene
Accession: AYE34929
Location: 2442050-2442820
NCBI BlastP on this gene
CP523_11180
CP523_11175
CP523_11170
CP523_11165
beta-channel forming cytolysin
Accession: AYE34925
Location: 2438998-2439948
NCBI BlastP on this gene
Accession: AYE34925
Location: 2438998-2439948
NCBI BlastP on this gene
CP523_11160
CP523_11155
CP523_11150
CP523_11145
bacitracin ABC transporter ATP-binding protein
Accession: AYE34921
Location: 2435626-2436549
NCBI BlastP on this gene
Accession: AYE34921
Location: 2435626-2436549
NCBI BlastP on this gene
CP523_11140
DNA-binding response regulator
Accession: AYE34920
Location: 2434841-2435536
NCBI BlastP on this gene
Accession: AYE34920
Location: 2434841-2435536
NCBI BlastP on this gene
CP523_11135
CP523_11130
ArsR family transcriptional regulator
Accession: AYE34918
Location: 2431803-2432081
NCBI BlastP on this gene
Accession: AYE34918
Location: 2431803-2432081
NCBI BlastP on this gene
CP523_11125
497. : CP001656 Paenibacillus sp. JDR-2 chromosome Total score: 2.0 Cumulative Blast bit score: 334
Pjdr2_6233
Pjdr2_6232
Pjdr2_6231
Pjdr2_6230
transcriptional regulator, TetR family
Accession: ACT04832
Location: 7121537-7122178
NCBI BlastP on this gene
Accession: ACT04832
Location: 7121537-7122178
NCBI BlastP on this gene
Pjdr2_6229
drug resistance transporter, EmrB/QacA subfamily
Accession: ACT04831
Location: 7119956-7121389
NCBI BlastP on this gene
Accession: ACT04831
Location: 7119956-7121389
NCBI BlastP on this gene
Pjdr2_6228
Pjdr2_6227
PTS system, mannitol-specific IIC subunit
Accession: ACT04829
Location: 7117641-7119107
NCBI BlastP on this gene
Accession: ACT04829
Location: 7117641-7119107
NCBI BlastP on this gene
Pjdr2_6226
PTS modulated transcriptional regulator, MtlR family
Accession: ACT04828
Location: 7115492-7117555
NCBI BlastP on this gene
Accession: ACT04828
Location: 7115492-7117555
NCBI BlastP on this gene
Pjdr2_6225
Protein-N(pi)-phosphohistidine--sugar phosphotransferase
Accession: ACT04827
Location: 7115061-7115495
NCBI BlastP on this gene
Accession: ACT04827
Location: 7115061-7115495
NCBI BlastP on this gene
Pjdr2_6224
Mannitol dehydrogenase domain protein
Accession: ACT04826
Location: 7113910-7115061
NCBI BlastP on this gene
Accession: ACT04826
Location: 7113910-7115061
NCBI BlastP on this gene
Pjdr2_6223
phosphoenolpyruvate-protein phosphotransferase
Accession: ACT04825
Location: 7112149-7113867
NCBI BlastP on this gene
Accession: ACT04825
Location: 7112149-7113867
NCBI BlastP on this gene
Pjdr2_6222
Phosphotransferase system, phosphocarrier protein HPr
Accession: ACT04824
Location: 7111886-7112149
NCBI BlastP on this gene
Accession: ACT04824
Location: 7111886-7112149
NCBI BlastP on this gene
Pjdr2_6221
short-chain dehydrogenase/reductase SDR
Accession: ACT04823
Location: 7111082-7111825
NCBI BlastP on this gene
Accession: ACT04823
Location: 7111082-7111825
NCBI BlastP on this gene
Pjdr2_6220
Helix-turn-helix type 11 domain protein
Accession: ACT04822
Location: 7110093-7111055
NCBI BlastP on this gene
Accession: ACT04822
Location: 7110093-7111055
NCBI BlastP on this gene
Pjdr2_6219
drug resistance transporter, EmrB/QacA subfamily
Accession: ACT04821
Location: 7108533-7110047
NCBI BlastP on this gene
Accession: ACT04821
Location: 7108533-7110047
NCBI BlastP on this gene
Pjdr2_6218
Alcohol dehydrogenase zinc-binding domain protein
Accession: ACT04820
Location: 7107262-7108347
NCBI BlastP on this gene
Accession: ACT04820
Location: 7107262-7108347
NCBI BlastP on this gene
Pjdr2_6217
transcriptional regulator, LuxR family
Accession: ACT04819
Location: 7106411-7107100
NCBI BlastP on this gene
Accession: ACT04819
Location: 7106411-7107100
NCBI BlastP on this gene
Pjdr2_6216
conserved hypothetical protein
Accession: ACT04818
Location: 7105930-7106400
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 71
Sequence coverage: 87 %
E-value: 1e-12
NCBI BlastP on this gene
Accession: ACT04818
Location: 7105930-7106400
BlastP hit with WP_004843641.1
Percentage identity: 35 %
BlastP bit score: 71
Sequence coverage: 87 %
E-value: 1e-12
NCBI BlastP on this gene
Pjdr2_6215
transcriptional regulator, RpiR family
Accession: ACT04817
Location: 7105050-7105904
NCBI BlastP on this gene
Accession: ACT04817
Location: 7105050-7105904
NCBI BlastP on this gene
Pjdr2_6214
Pjdr2_6213
N-acylglucosamine-6-phosphate 2-epimerase
Accession: ACT04815
Location: 7103502-7104182
BlastP hit with WP_024854132.1
Percentage identity: 57 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 6e-85
NCBI BlastP on this gene
Accession: ACT04815
Location: 7103502-7104182
BlastP hit with WP_024854132.1
Percentage identity: 57 %
BlastP bit score: 263
Sequence coverage: 96 %
E-value: 6e-85
NCBI BlastP on this gene
Pjdr2_6212
binding-protein-dependent transport systems inner membrane component
Accession: ACT04814
Location: 7102622-7103467
NCBI BlastP on this gene
Accession: ACT04814
Location: 7102622-7103467
NCBI BlastP on this gene
Pjdr2_6211
binding-protein-dependent transport systems inner membrane component
Accession: ACT04813
Location: 7101728-7102603
NCBI BlastP on this gene
Accession: ACT04813
Location: 7101728-7102603
NCBI BlastP on this gene
Pjdr2_6210
extracellular solute-binding protein family 1
Accession: ACT04812
Location: 7100245-7101645
NCBI BlastP on this gene
Accession: ACT04812
Location: 7100245-7101645
NCBI BlastP on this gene
Pjdr2_6209
dihydrodipicolinate synthetase
Accession: ACT04811
Location: 7099283-7100194
NCBI BlastP on this gene
Accession: ACT04811
Location: 7099283-7100194
NCBI BlastP on this gene
Pjdr2_6208
Pjdr2_6207
extracellular solute-binding protein family 1
Accession: ACT04809
Location: 7095570-7096961
NCBI BlastP on this gene
Accession: ACT04809
Location: 7095570-7096961
NCBI BlastP on this gene
Pjdr2_6206
binding-protein-dependent transport systems inner membrane component
Accession: ACT04808
Location: 7094557-7095453
NCBI BlastP on this gene
Accession: ACT04808
Location: 7094557-7095453
NCBI BlastP on this gene
Pjdr2_6205
binding-protein-dependent transport systems inner membrane component
Accession: ACT04807
Location: 7093709-7094545
NCBI BlastP on this gene
Accession: ACT04807
Location: 7093709-7094545
NCBI BlastP on this gene
Pjdr2_6204
two component transcriptional regulator, AraC family
Accession: ACT04806
Location: 7091964-7093598
NCBI BlastP on this gene
Accession: ACT04806
Location: 7091964-7093598
NCBI BlastP on this gene
Pjdr2_6203
conserved hypothetical protein
Accession: ACT04805
Location: 7091449-7091814
NCBI BlastP on this gene
Accession: ACT04805
Location: 7091449-7091814
NCBI BlastP on this gene
Pjdr2_6202
RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: ACT04804
Location: 7090551-7091429
NCBI BlastP on this gene
Accession: ACT04804
Location: 7090551-7091429
NCBI BlastP on this gene
Pjdr2_6201
aminoglycoside phosphotransferase
Accession: ACT04803
Location: 7089575-7090453
NCBI BlastP on this gene
Accession: ACT04803
Location: 7089575-7090453
NCBI BlastP on this gene
Pjdr2_6200
transcriptional regulator, TetR family
Accession: ACT04802
Location: 7088852-7089421
NCBI BlastP on this gene
Accession: ACT04802
Location: 7088852-7089421
NCBI BlastP on this gene
Pjdr2_6199
short-chain dehydrogenase/reductase SDR
Accession: ACT04801
Location: 7087880-7088722
NCBI BlastP on this gene
Accession: ACT04801
Location: 7087880-7088722
NCBI BlastP on this gene
Pjdr2_6198
Pjdr2_6197
transcriptional regulator, HxlR family
Accession: ACT04799
Location: 7086943-7087296
NCBI BlastP on this gene
Accession: ACT04799
Location: 7086943-7087296
NCBI BlastP on this gene
Pjdr2_6196
Pjdr2_6195
498. : CP036150 Oceanispirochaeta sp. K2 chromosome Total score: 2.0 Cumulative Blast bit score: 149
WYL domain-containing protein
Accession: QEN09558
Location: 3702649-3703656
NCBI BlastP on this gene
Accession: QEN09558
Location: 3702649-3703656
NCBI BlastP on this gene
EXM22_16805
EXM22_16810
EXM22_16815
DUF2971 domain-containing protein
Accession: QEN09561
Location: 3705110-3706000
NCBI BlastP on this gene
Accession: QEN09561
Location: 3705110-3706000
NCBI BlastP on this gene
EXM22_16820
EXM22_16825
EXM22_16830
extracellular solute-binding protein
Accession: QEN09564
Location: 3710227-3711540
NCBI BlastP on this gene
Accession: QEN09564
Location: 3710227-3711540
NCBI BlastP on this gene
EXM22_16835
sugar ABC transporter permease
Accession: QEN09565
Location: 3711554-3712441
NCBI BlastP on this gene
Accession: QEN09565
Location: 3711554-3712441
NCBI BlastP on this gene
EXM22_16840
carbohydrate ABC transporter permease
Accession: QEN09961
Location: 3712453-3713295
NCBI BlastP on this gene
Accession: QEN09961
Location: 3712453-3713295
NCBI BlastP on this gene
EXM22_16845
copper homeostasis protein CutC
Accession: QEN09566
Location: 3713299-3714045
NCBI BlastP on this gene
Accession: QEN09566
Location: 3713299-3714045
NCBI BlastP on this gene
EXM22_16850
ROK family transcriptional regulator
Accession: QEN09567
Location: 3714072-3715256
NCBI BlastP on this gene
Accession: QEN09567
Location: 3714072-3715256
NCBI BlastP on this gene
EXM22_16855
EXM22_16860
Hsp20/alpha crystallin family protein
Accession: QEN09569
Location: 3716834-3717235
NCBI BlastP on this gene
Accession: QEN09569
Location: 3716834-3717235
NCBI BlastP on this gene
EXM22_16865
Hsp20/alpha crystallin family protein
Accession: QEN09570
Location: 3717263-3717682
NCBI BlastP on this gene
Accession: QEN09570
Location: 3717263-3717682
NCBI BlastP on this gene
EXM22_16870
EXM22_16875
EXM22_16880
EXM22_16885
EXM22_16890
DUF386 domain-containing protein
Accession: QEN09575
Location: 3722518-3722976
BlastP hit with WP_004843641.1
Percentage identity: 40 %
BlastP bit score: 75
Sequence coverage: 67 %
E-value: 4e-14
NCBI BlastP on this gene
Accession: QEN09575
Location: 3722518-3722976
BlastP hit with WP_004843641.1
Percentage identity: 40 %
BlastP bit score: 75
Sequence coverage: 67 %
E-value: 4e-14
NCBI BlastP on this gene
EXM22_16895
EXM22_16900
EXM22_16905
ketopantoate reductase family protein
Accession: QEN09578
Location: 3724544-3725326
NCBI BlastP on this gene
Accession: QEN09578
Location: 3724544-3725326
NCBI BlastP on this gene
EXM22_16910
EXM22_16915
EXM22_16920
AraC family transcriptional regulator
Accession: QEN09581
Location: 3727846-3728703
BlastP hit with WP_101883549.1
Percentage identity: 37 %
BlastP bit score: 74
Sequence coverage: 39 %
E-value: 3e-12
NCBI BlastP on this gene
Accession: QEN09581
Location: 3727846-3728703
BlastP hit with WP_101883549.1
Percentage identity: 37 %
BlastP bit score: 74
Sequence coverage: 39 %
E-value: 3e-12
NCBI BlastP on this gene
EXM22_16925
EXM22_16930
EXM22_16935
C4-dicarboxylate ABC transporter
Accession: QEN09584
Location: 3730554-3731555
NCBI BlastP on this gene
Accession: QEN09584
Location: 3730554-3731555
NCBI BlastP on this gene
EXM22_16940
TRAP transporter small permease
Accession: QEN09585
Location: 3731655-3732185
NCBI BlastP on this gene
Accession: QEN09585
Location: 3731655-3732185
NCBI BlastP on this gene
EXM22_16945
TRAP transporter large permease
Accession: QEN09586
Location: 3732198-3733481
NCBI BlastP on this gene
Accession: QEN09586
Location: 3732198-3733481
NCBI BlastP on this gene
EXM22_16950
EXM22_16955
Gfo/Idh/MocA family oxidoreductase
Accession: QEN09588
Location: 3734559-3735707
NCBI BlastP on this gene
Accession: QEN09588
Location: 3734559-3735707
NCBI BlastP on this gene
EXM22_16960
glucosamine-6-phosphate deaminase
Accession: QEN09589
Location: 3735709-3736506
NCBI BlastP on this gene
Accession: QEN09589
Location: 3735709-3736506
NCBI BlastP on this gene
EXM22_16965
mannitol dehydrogenase family protein
Accession: QEN09590
Location: 3736503-3738104
NCBI BlastP on this gene
Accession: QEN09590
Location: 3736503-3738104
NCBI BlastP on this gene
EXM22_16970
ABC transporter ATP-binding protein
Accession: QEN09591
Location: 3738130-3739089
NCBI BlastP on this gene
Accession: QEN09591
Location: 3738130-3739089
NCBI BlastP on this gene
EXM22_16975
EXM22_16980
EXM22_16985
DUF4340 domain-containing protein
Accession: QEN09594
Location: 3741189-3742550
NCBI BlastP on this gene
Accession: QEN09594
Location: 3741189-3742550
NCBI BlastP on this gene
EXM22_16990
GntR family transcriptional regulator
Accession: QEN09595
Location: 3742550-3743302
NCBI BlastP on this gene
Accession: QEN09595
Location: 3742550-3743302
NCBI BlastP on this gene
EXM22_16995
EXM22_17000
EXM22_17005
EXM22_17010
499. : CP040863 Rodentibacter heylii strain G1 chromosome Total score: 1.0 Cumulative Blast bit score: 560
FEE42_02440
FEE42_02445
FEE42_02450
FEE42_02455
FEE42_02460
FEE42_02465
FEE42_02470
helix-turn-helix transcriptional regulator
Accession: QIA76299
Location: 484929-485264
NCBI BlastP on this gene
Accession: QIA76299
Location: 484929-485264
NCBI BlastP on this gene
FEE42_02475
FEE42_02480
site-specific DNA-methyltransferase
Accession: QIA76301
Location: 485552-487171
NCBI BlastP on this gene
Accession: QIA76301
Location: 485552-487171
NCBI BlastP on this gene
FEE42_02485
FEE42_02490
FEE42_02495
FEE42_02500
helix-turn-helix transcriptional regulator
Accession: QIA76305
Location: 493209-493544
NCBI BlastP on this gene
Accession: QIA76305
Location: 493209-493544
NCBI BlastP on this gene
FEE42_02505
SAM-dependent methyltransferase
Accession: QIA76306
Location: 493741-495918
NCBI BlastP on this gene
Accession: QIA76306
Location: 493741-495918
NCBI BlastP on this gene
FEE42_02510
FEE42_02515
FEE42_02520
Gfo/Idh/MocA family oxidoreductase
Accession: QIA76309
Location: 499174-500277
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
Accession: QIA76309
Location: 499174-500277
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
FEE42_02525
Gfo/Idh/MocA family oxidoreductase
Accession: QIA76310
Location: 500298-501368
NCBI BlastP on this gene
Accession: QIA76310
Location: 500298-501368
NCBI BlastP on this gene
FEE42_02530
FadR family transcriptional regulator
Accession: QIA78132
Location: 501361-502065
NCBI BlastP on this gene
Accession: QIA78132
Location: 501361-502065
NCBI BlastP on this gene
FEE42_02535
FEE42_02540
FEE42_02545
FEE42_02550
putative hydroxymethylpyrimidine transporter CytX
Accession: QIA76313
Location: 504812-505996
NCBI BlastP on this gene
Accession: QIA76313
Location: 504812-505996
NCBI BlastP on this gene
cytX
lysine exporter LysO family protein
Accession: QIA78133
Location: 506066-506974
NCBI BlastP on this gene
Accession: QIA78133
Location: 506066-506974
NCBI BlastP on this gene
FEE42_02560
pepN
5-(carboxyamino)imidazole ribonucleotide mutase
Accession: QIA76315
Location: 510293-510790
NCBI BlastP on this gene
Accession: QIA76315
Location: 510293-510790
NCBI BlastP on this gene
purE
5-(carboxyamino)imidazole ribonucleotide synthase
Accession: QIA76316
Location: 510860-511948
NCBI BlastP on this gene
Accession: QIA76316
Location: 510860-511948
NCBI BlastP on this gene
purK
aspartate/tyrosine/aromatic aminotransferase
Accession: QIA76317
Location: 512088-513278
NCBI BlastP on this gene
Accession: QIA76317
Location: 512088-513278
NCBI BlastP on this gene
FEE42_02580
FEE42_02585
ureA
ureB
ureC
ureE
FEE42_02610
ureG
FEE42_02620
cdd
500. : CP013830 Pasteurellaceae bacterium NI1060 genome. Total score: 1.0 Cumulative Blast bit score: 560
Nucleoside-diphosphate-sugar epimerase
Accession: AOF54289
Location: 2308051-2309154
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOF54289
Location: 2308051-2309154
BlastP hit with WP_004843633.1
Percentage identity: 74 %
BlastP bit score: 560
Sequence coverage: 97 %
E-value: 0.0
NCBI BlastP on this gene
AC062_2203
putative oxidoreductase protein
Accession: AOF54288
Location: 2306960-2308030
NCBI BlastP on this gene
Accession: AOF54288
Location: 2306960-2308030
NCBI BlastP on this gene
AC062_2202
Transcriptional regulator, GntR family
Accession: AOF54287
Location: 2306254-2306967
NCBI BlastP on this gene
Accession: AOF54287
Location: 2306254-2306967
NCBI BlastP on this gene
AC062_2201
Sialic acid transporter (permease) NanT
Accession: AOF54286
Location: 2304853-2306082
NCBI BlastP on this gene
Accession: AOF54286
Location: 2304853-2306082
NCBI BlastP on this gene
AC062_2200
AC062_2199
AC062_2198
AC062_2197
AC062_2196
AC062_2195
AC062_2194
AC062_2193
AC062_2192
AC062_2191
Phage tail assembly protein I
Accession: AOF54277
Location: 2295975-2296589
NCBI BlastP on this gene
Accession: AOF54277
Location: 2295975-2296589
NCBI BlastP on this gene
AC062_2190
Putative antirepressor protein
Accession: AOF54276
Location: 2295773-2295919
NCBI BlastP on this gene
Accession: AOF54276
Location: 2295773-2295919
NCBI BlastP on this gene
AC062_2189
Phage tail length tape-measure protein 1
Accession: AOF54275
Location: 2292340-2295615
NCBI BlastP on this gene
Accession: AOF54275
Location: 2292340-2295615
NCBI BlastP on this gene
AC062_2188
AC062_2187
AC062_2186
Diaminopimelate decarboxylase
Accession: AOF54272
Location: 2291401-2291793
NCBI BlastP on this gene
Accession: AOF54272
Location: 2291401-2291793
NCBI BlastP on this gene
AC062_2185
AC062_2184
AC062_2183
AC062_2182
AC062_2181
AC062_2180
AC062_2179
Phage tail completion protein
Accession: AOF54265
Location: 2288770-2289300
NCBI BlastP on this gene
Accession: AOF54265
Location: 2288770-2289300
NCBI BlastP on this gene
AC062_2178
AC062_2177
AC062_2176

Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.