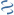MultiGeneBlast hits

Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002582 : Clostridium lentocellum DSM 5427 Total score: 2.5 Cumulative Blast bit score: 812
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
metal-dependent phosphohydrolase HD sub domain
Accession: ADZ85143
Location: 3781070-3781555
NCBI BlastP on this gene
Accession: ADZ85143
Location: 3781070-3781555
NCBI BlastP on this gene
Clole_3456
NADPH-dependent FMN reductase
Accession: ADZ85142
Location: 3780549-3781031
NCBI BlastP on this gene
Accession: ADZ85142
Location: 3780549-3781031
NCBI BlastP on this gene
Clole_3455
cation diffusion facilitator family transporter
Accession: ADZ85141
Location: 3779386-3780273
NCBI BlastP on this gene
Accession: ADZ85141
Location: 3779386-3780273
NCBI BlastP on this gene
Clole_3454
phenazine biosynthesis protein PhzF family
Accession: ADZ85140
Location: 3778411-3779289
NCBI BlastP on this gene
Accession: ADZ85140
Location: 3778411-3779289
NCBI BlastP on this gene
Clole_3453
transcriptional regulator, GntR family with UTRA sensor domain
Accession: ADZ85139
Location: 3777431-3778168
NCBI BlastP on this gene
Accession: ADZ85139
Location: 3777431-3778168
NCBI BlastP on this gene
Clole_3452
PTS system, trehalose-specific IIBC subunit
Accession: ADZ85138
Location: 3775822-3777243
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 311
Sequence coverage: 85 %
E-value: 6e-96
NCBI BlastP on this gene
Accession: ADZ85138
Location: 3775822-3777243
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 311
Sequence coverage: 85 %
E-value: 6e-96
NCBI BlastP on this gene
Clole_3451
alpha,alpha-phosphotrehalase
Accession: ADZ85137
Location: 3774083-3775720
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 501
Sequence coverage: 98 %
E-value: 3e-168
NCBI BlastP on this gene
Accession: ADZ85137
Location: 3774083-3775720
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 501
Sequence coverage: 98 %
E-value: 3e-168
NCBI BlastP on this gene
Clole_3450
Clole_3449
Clole_3448
Clole_3447
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP042243 : Crassaminicella sp. SY095 chromosome Total score: 2.5 Cumulative Blast bit score: 810
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
LL-diaminopimelate aminotransferase
Accession: QEK12819
Location: 2167749-2168981
NCBI BlastP on this gene
Accession: QEK12819
Location: 2167749-2168981
NCBI BlastP on this gene
FQB35_11045
ATP-dependent Clp protease ATP-binding subunit
Accession: QEK12818
Location: 2165253-2167535
NCBI BlastP on this gene
Accession: QEK12818
Location: 2165253-2167535
NCBI BlastP on this gene
FQB35_11040
PTS trehalose transporter subunit IIBC
Accession: QEK12817
Location: 2163660-2165075
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 302
Sequence coverage: 88 %
E-value: 1e-92
NCBI BlastP on this gene
Accession: QEK12817
Location: 2163660-2165075
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 302
Sequence coverage: 88 %
E-value: 1e-92
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QEK12816
Location: 2161918-2163582
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 508
Sequence coverage: 99 %
E-value: 4e-171
NCBI BlastP on this gene
Accession: QEK12816
Location: 2161918-2163582
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 508
Sequence coverage: 99 %
E-value: 4e-171
NCBI BlastP on this gene
treC
treR
Na+/H+ antiporter NhaC family protein
Accession: QEK12814
Location: 2158963-2160669
NCBI BlastP on this gene
Accession: QEK12814
Location: 2158963-2160669
NCBI BlastP on this gene
FQB35_11020
beta-ketoacyl-ACP synthase II
Accession: QEK12813
Location: 2157656-2158906
NCBI BlastP on this gene
Accession: QEK12813
Location: 2157656-2158906
NCBI BlastP on this gene
fabF
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP039703 : Clostridium butyricum strain 29-1 plasmid p-butyl_plas_29-1 Total score: 2.5 Cumulative Blast bit score: 806
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
FBD77_17400
FBD77_17395
FBD77_17390
FBD77_17385
FBD77_17380
PTS trehalose transporter subunit IIBC
Accession: QCJ04253
Location: 73414-74826
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: QCJ04253
Location: 73414-74826
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QCJ04252
Location: 71618-73276
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: QCJ04252
Location: 71618-73276
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
treC
treR
FBD77_17360
FBD77_17355
FBD77_17350
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP016333 : Clostridium butyricum strain TK520 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
BBB49_17735
BBB49_17740
BBB49_17745
BBB49_17750
BBB49_17755
PTS maltose transporter subunit IIBC
Accession: AOR95920
Location: 327993-329405
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: AOR95920
Location: 327993-329405
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
BBB49_17760
alpha,alpha-phosphotrehalase
Accession: AOR95921
Location: 329543-331201
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: AOR95921
Location: 329543-331201
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
BBB49_17765
BBB49_17770
BBB49_17775
BBB49_17780
BBB49_17785
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP014705 : Clostridium butyricum strain TOA chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
AZ909_18265
AZ909_18270
AZ909_18275
AZ909_18280
AZ909_18285
PTS maltose transporter subunit IIBC
Accession: ANF16008
Location: 368762-370174
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: ANF16008
Location: 368762-370174
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
AZ909_18290
alpha,alpha-phosphotrehalase
Accession: ANF16009
Location: 370312-371970
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: ANF16009
Location: 370312-371970
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
AZ909_18295
AZ909_18300
AZ909_18305
AZ909_18310
AZ909_18315
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013489 : Clostridium butyricum strain KNU-L09 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ATN24_19285
ATN24_19295
ATN24_19300
ATN24_19305
PTS maltose transporter subunit IIBC
Accession: ALR90585
Location: 527024-528436
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: ALR90585
Location: 527024-528436
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
ATN24_19310
glucohydrolase
Accession: ALR90586
Location: 528574-530232
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: ALR90586
Location: 528574-530232
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
ATN24_19315
ATN24_19320
ATN24_19325
ATN24_19330
ATN24_19335
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013353 : Clostridium butyricum strain JKY6D1 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ATD26_18235
ATD26_18240
ATD26_18245
ATD26_18250
ATD26_18255
PTS maltose transporter subunit IIBC
Accession: ALS18825
Location: 301171-302583
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: ALS18825
Location: 301171-302583
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
ATD26_18260
glucohydrolase
Accession: ALS18826
Location: 302721-304379
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: ALS18826
Location: 302721-304379
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
ATD26_18265
ATD26_18270
ATD26_18275
ATD26_18280
ATD26_18285
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP017080 : Bacillus muralis strain G25-68 chromosome Total score: 2.5 Cumulative Blast bit score: 805
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ABE28_009835
ABE28_009840
DUF4867 domain-containing protein
Accession: AOH54651
Location: 2007344-2008042
NCBI BlastP on this gene
Accession: AOH54651
Location: 2007344-2008042
NCBI BlastP on this gene
ABE28_009845
ABE28_009850
ABE28_009855
PTS maltose transporter subunit IIBC
Accession: AOH54654
Location: 2009794-2011245
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 85 %
E-value: 3e-90
NCBI BlastP on this gene
Accession: AOH54654
Location: 2009794-2011245
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 85 %
E-value: 3e-90
NCBI BlastP on this gene
ABE28_009860
alpha,alpha-phosphotrehalase
Accession: AOH54655
Location: 2011325-2013004
BlastP hit with treA
Percentage identity: 50 %
BlastP bit score: 509
Sequence coverage: 91 %
E-value: 1e-171
NCBI BlastP on this gene
Accession: AOH54655
Location: 2011325-2013004
BlastP hit with treA
Percentage identity: 50 %
BlastP bit score: 509
Sequence coverage: 91 %
E-value: 1e-171
NCBI BlastP on this gene
ABE28_009865
ABE28_009870
ABE28_009875
quaternary ammonium transporter
Accession: AOH54658
Location: 2014215-2014487
NCBI BlastP on this gene
Accession: AOH54658
Location: 2014215-2014487
NCBI BlastP on this gene
ABE28_009880
DNA polymerase III subunit beta
Accession: AOH54659
Location: 2015049-2016182
NCBI BlastP on this gene
Accession: AOH54659
Location: 2015049-2016182
NCBI BlastP on this gene
ABE28_009885
ABE28_009890
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013238 : Clostridium butyricum strain CDC_51208 plasmid pNPD4_2 Total score: 2.5 Cumulative Blast bit score: 803
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
NPD4_3705
NPD4_3704
NPD4_3703
NPD4_3702
NPD4_3701
PTS system, trehalose-specific IIBC component
Accession: APF21357
Location: 193121-194533
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
Accession: APF21357
Location: 193121-194533
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: APF21430
Location: 191325-192983
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: APF21430
Location: 191325-192983
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
NPD4_3697
NPD4_3696
putative zinc-binding domain protein
Accession: APF21092
Location: 187380-188012
NCBI BlastP on this gene
Accession: APF21092
Location: 187380-188012
NCBI BlastP on this gene
NPD4_3695
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
AP019717 : Clostridium butyricum NBRC 13949 plasmid pCBU1 DNA Total score: 2.5 Cumulative Blast bit score: 803
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
Cbu04g_41250
Cbu04g_41260
Cbu04g_41270
serine/threonine protein kinase
Accession: BBK79120
Location: 655954-656673
NCBI BlastP on this gene
Accession: BBK79120
Location: 655954-656673
NCBI BlastP on this gene
Cbu04g_41280
Cbu04g_41290
PTS maltose transporter subunit IIBC
Accession: BBK79122
Location: 657693-659105
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
Accession: BBK79122
Location: 657693-659105
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: BBK79123
Location: 659243-660901
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: BBK79123
Location: 659243-660901
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treA
treR
FprA family A-type flavoprotein
Accession: BBK79125
Location: 661961-663127
NCBI BlastP on this gene
Accession: BBK79125
Location: 661961-663127
NCBI BlastP on this gene
Cbu04g_41330
dedA
Cbu04g_41350
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP039706 : Clostridium butyricum strain 4-1 plasmid p-butyl_plas_4-1 Total score: 2.5 Cumulative Blast bit score: 801
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
FBD76_20225
FBD76_20230
FBD76_20235
FBD76_20240
FBD76_20245
PTS trehalose transporter subunit IIBC
Accession: QCJ08656
Location: 619277-620689
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: QCJ08656
Location: 619277-620689
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QCJ08657
Location: 620827-622485
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: QCJ08657
Location: 620827-622485
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: QCJ08659
Location: 623545-624711
NCBI BlastP on this gene
Accession: QCJ08659
Location: 623545-624711
NCBI BlastP on this gene
FBD76_20265
FBD76_20270
FBD76_20275
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP033248 : Clostridium butyricum strain CFSA3989 plasmid pCFSA3989 Total score: 2.5 Cumulative Blast bit score: 801
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
EBQ27_01605
EBQ27_01610
EBQ27_01615
EBQ27_01620
EBQ27_01625
PTS trehalose transporter subunit IIBC
Accession: QGH24389
Location: 383227-384639
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: QGH24389
Location: 383227-384639
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QGH24390
Location: 384777-386435
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: QGH24390
Location: 384777-386435
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: QGH24392
Location: 387495-388661
NCBI BlastP on this gene
Accession: QGH24392
Location: 387495-388661
NCBI BlastP on this gene
EBQ27_01645
EBQ27_01650
EBQ27_01655
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP033246 : Clostridium butyricum strain CFSA3987 plasmid pCFSA3987 Total score: 2.5 Cumulative Blast bit score: 801
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
EBL75_01605
EBL75_01610
EBL75_01615
EBL75_01620
EBL75_01625
PTS trehalose transporter subunit IIBC
Accession: QGH20353
Location: 383188-384600
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: QGH20353
Location: 383188-384600
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QGH20354
Location: 384738-386396
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: QGH20354
Location: 384738-386396
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: QGH20356
Location: 387456-388622
NCBI BlastP on this gene
Accession: QGH20356
Location: 387456-388622
NCBI BlastP on this gene
EBL75_01645
EBL75_01650
EBL75_01655
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP030776 : Clostridium butyricum strain S-45-5 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 801
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
DRB99_17735
DRB99_17740
DRB99_17745
DRB99_17750
DRB99_17755
PTS trehalose transporter subunit IIBC
Accession: AXB86768
Location: 108634-110046
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: AXB86768
Location: 108634-110046
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
DRB99_17760
alpha,alpha-phosphotrehalase
Accession: AXB86769
Location: 110184-111842
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: AXB86769
Location: 110184-111842
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: AXB86771
Location: 112902-114068
NCBI BlastP on this gene
Accession: AXB86771
Location: 112902-114068
NCBI BlastP on this gene
DRB99_17775
DRB99_17780
DRB99_17785
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LT630003 : [Clostridium] sphenoides JCM 1415 strain ATCC 19403 genome assembly, chromosome: I. Total score: 2.5 Cumulative Blast bit score: 801
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SAMN02745906_3037
HD-GYP domain, c-di-GMP phosphodiesterase class II (or its inactivated variant)
Accession: SET92268
Location: 3311405-3312736
NCBI BlastP on this gene
Accession: SET92268
Location: 3311405-3312736
NCBI BlastP on this gene
SAMN02745906_3036
GntR family transcriptional regulator, trehalose operon transcriptional repressor
Accession: SET92260
Location: 3310436-3311149
NCBI BlastP on this gene
Accession: SET92260
Location: 3310436-3311149
NCBI BlastP on this gene
SAMN02745906_3035
PTS system, trehalose-specific IIC component
Accession: SET92249
Location: 3308799-3310229
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 300
Sequence coverage: 92 %
E-value: 2e-91
NCBI BlastP on this gene
Accession: SET92249
Location: 3308799-3310229
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 300
Sequence coverage: 92 %
E-value: 2e-91
NCBI BlastP on this gene
SAMN02745906_3034
trehalose-6-phosphate hydrolase
Accession: SET92238
Location: 3307140-3308798
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 501
Sequence coverage: 91 %
E-value: 3e-168
NCBI BlastP on this gene
Accession: SET92238
Location: 3307140-3308798
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 501
Sequence coverage: 91 %
E-value: 3e-168
NCBI BlastP on this gene
SAMN02745906_3033
Putative Mn2+ efflux pump MntP
Accession: SET92227
Location: 3306580-3307143
NCBI BlastP on this gene
Accession: SET92227
Location: 3306580-3307143
NCBI BlastP on this gene
SAMN02745906_3032
Predicted RNA-binding protein containing a PIN domain
Accession: SET92215
Location: 3305999-3306520
NCBI BlastP on this gene
Accession: SET92215
Location: 3305999-3306520
NCBI BlastP on this gene
SAMN02745906_3031
putative efflux protein, MATE family
Accession: SET92203
Location: 3304572-3305942
NCBI BlastP on this gene
Accession: SET92203
Location: 3304572-3305942
NCBI BlastP on this gene
SAMN02745906_3030
nondiscriminating aspartyl-tRNA synthetase
Accession: SET92191
Location: 3302545-3303876
NCBI BlastP on this gene
Accession: SET92191
Location: 3302545-3303876
NCBI BlastP on this gene
SAMN02745906_3029
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP017761 : Marinilactibacillus sp. 15R chromosome Total score: 2.5 Cumulative Blast bit score: 797
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ribonuclease P protein component
Accession: API88312
Location: 592341-592682
NCBI BlastP on this gene
Accession: API88312
Location: 592341-592682
NCBI BlastP on this gene
BKP56_02885
BKP56_02890
BKP56_02895
BKP56_02900
PTS maltose transporter subunit IIBC
Accession: API88315
Location: 596392-597849
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: API88315
Location: 596392-597849
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
BKP56_02905
alpha,alpha-phosphotrehalase
Accession: API88316
Location: 598034-599686
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 489
Sequence coverage: 99 %
E-value: 9e-164
NCBI BlastP on this gene
Accession: API88316
Location: 598034-599686
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 489
Sequence coverage: 99 %
E-value: 9e-164
NCBI BlastP on this gene
BKP56_02910
BKP56_02915
BKP56_02920
BKP56_02925
BKP56_02930
BKP56_02935
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022464 : Enterocloster bolteae strain ATCC BAA-613 chromosome Total score: 2.5 Cumulative Blast bit score: 796
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CGC65_03455
CGC65_03460
treR
PTS beta-glucoside transporter subunit EIIBCA
Accession: ASN93805
Location: 716811-718754
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 1e-91
NCBI BlastP on this gene
Accession: ASN93805
Location: 716811-718754
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 1e-91
NCBI BlastP on this gene
CGC65_03470
alpha,alpha-phosphotrehalase
Accession: ASN93806
Location: 718820-720466
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 491
Sequence coverage: 99 %
E-value: 2e-164
NCBI BlastP on this gene
Accession: ASN93806
Location: 718820-720466
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 491
Sequence coverage: 99 %
E-value: 2e-164
NCBI BlastP on this gene
treC
hybrid sensor histidine kinase/response regulator
Accession: ASN93807
Location: 720566-723400
NCBI BlastP on this gene
Accession: ASN93807
Location: 720566-723400
NCBI BlastP on this gene
CGC65_03480
glf
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002109 : [Clostridium] saccharolyticum WM1 chromosome Total score: 2.5 Cumulative Blast bit score: 793
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
Closa_3364
metal dependent phosphohydrolase
Accession: ADL05890
Location: 3626170-3627501
NCBI BlastP on this gene
Accession: ADL05890
Location: 3626170-3627501
NCBI BlastP on this gene
Closa_3363
transcriptional regulator, GntR family
Accession: ADL05889
Location: 3625204-3625917
NCBI BlastP on this gene
Accession: ADL05889
Location: 3625204-3625917
NCBI BlastP on this gene
Closa_3362
PTS system, trehalose-specific IIBC subunit
Accession: ADL05888
Location: 3623600-3625009
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 295
Sequence coverage: 91 %
E-value: 5e-90
NCBI BlastP on this gene
Accession: ADL05888
Location: 3623600-3625009
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 295
Sequence coverage: 91 %
E-value: 5e-90
NCBI BlastP on this gene
Closa_3361
alpha,alpha-phosphotrehalase
Accession: ADL05887
Location: 3621942-3623600
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 498
Sequence coverage: 95 %
E-value: 5e-167
NCBI BlastP on this gene
Accession: ADL05887
Location: 3621942-3623600
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 498
Sequence coverage: 95 %
E-value: 5e-167
NCBI BlastP on this gene
Closa_3360
protein of unknown function DUF204
Accession: ADL05886
Location: 3621382-3621945
NCBI BlastP on this gene
Accession: ADL05886
Location: 3621382-3621945
NCBI BlastP on this gene
Closa_3359
protein of unknown function DUF901
Accession: ADL05885
Location: 3620798-3621319
NCBI BlastP on this gene
Accession: ADL05885
Location: 3620798-3621319
NCBI BlastP on this gene
Closa_3358
Closa_3357
Closa_3356
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
HE999757 : Carnobacterium maltaromaticum LMA28 complete genome. Total score: 2.5 Cumulative Blast bit score: 790
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
BN424_3617
BN424_3616
BN424_3615
bacterial regulatory s, tetR family protein
Accession: CCO13019
Location: 3612368-3612934
NCBI BlastP on this gene
Accession: CCO13019
Location: 3612368-3612934
NCBI BlastP on this gene
BN424_3614
BN424_3613
PTS system, trehalose-specific IIBC component
Accession: CCO13017
Location: 3609697-3611139
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: CCO13017
Location: 3609697-3611139
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 2e-92
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: CCO13016
Location: 3607905-3609563
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
Accession: CCO13016
Location: 3607905-3609563
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
treC
BN424_3610
BN424_3609
ilvD
acetolactate synthase, large subunit,biosynthetic type
Accession: CCO13012
Location: 3603353-3605026
NCBI BlastP on this gene
Accession: CCO13012
Location: 3603353-3605026
NCBI BlastP on this gene
ilvB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP029329 : Clostridium beijerinckii isolate WB53 chromosome Total score: 2.5 Cumulative Blast bit score: 790
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uxuA
DIC82_03555
DIC82_03550
PTS trehalose transporter subunit IIBC
Accession: AWK50203
Location: 878136-879545
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 300
Sequence coverage: 86 %
E-value: 8e-92
NCBI BlastP on this gene
Accession: AWK50203
Location: 878136-879545
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 300
Sequence coverage: 86 %
E-value: 8e-92
NCBI BlastP on this gene
DIC82_03545
alpha,alpha-phosphotrehalase
Accession: AWK50202
Location: 876395-878053
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 490
Sequence coverage: 100 %
E-value: 4e-164
NCBI BlastP on this gene
Accession: AWK50202
Location: 876395-878053
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 490
Sequence coverage: 100 %
E-value: 4e-164
NCBI BlastP on this gene
treC
PTS glucose transporter subunit IIA
Accession: AWK50201
Location: 875817-876317
NCBI BlastP on this gene
Accession: AWK50201
Location: 875817-876317
NCBI BlastP on this gene
DIC82_03535
treR
DIC82_03525
DIC82_03520
tRNA-specific adenosine deaminase
Accession: AWK50197
Location: 872018-872461
NCBI BlastP on this gene
Accession: AWK50197
Location: 872018-872461
NCBI BlastP on this gene
DIC82_03515
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP016844 : Carnobacterium maltaromaticum strain TMW 2.1581 chromosome Total score: 2.5 Cumulative Blast bit score: 790
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
BFC23_13565
DhaKLM operon coactivator DhaQ
Accession: BFC23_13560
Location: 2884066-2885054
NCBI BlastP on this gene
Accession: BFC23_13560
Location: 2884066-2885054
NCBI BlastP on this gene
BFC23_13560
dihydroxyacetone kinase transcriptional activator DhaS
Accession: AOA03467
Location: 2883173-2883739
NCBI BlastP on this gene
Accession: AOA03467
Location: 2883173-2883739
NCBI BlastP on this gene
BFC23_13555
BFC23_13550
PTS maltose transporter subunit IIBC
Accession: AOA03465
Location: 2880501-2881943
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 3e-92
NCBI BlastP on this gene
Accession: AOA03465
Location: 2880501-2881943
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 3e-92
NCBI BlastP on this gene
BFC23_13545
alpha,alpha-phosphotrehalase
Accession: AOA03464
Location: 2878709-2880367
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
Accession: AOA03464
Location: 2878709-2880367
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
BFC23_13540
BFC23_13535
acetolactate synthase, large subunit, biosynthetic type
Accession: AOA03462
Location: 2874169-2875860
NCBI BlastP on this gene
Accession: AOA03462
Location: 2874169-2875860
NCBI BlastP on this gene
BFC23_13530
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP015399 : Lachnoclostridium sp. YL32 Total score: 2.5 Cumulative Blast bit score: 788
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
A4V08_13160
A4V08_13155
A4V08_13150
A4V08_13145
PTS trehalose transporter subunit IIBC
Accession: ANU46605
Location: 2891840-2893231
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 8e-94
NCBI BlastP on this gene
Accession: ANU46605
Location: 2891840-2893231
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 8e-94
NCBI BlastP on this gene
A4V08_13140
alpha,alpha-phosphotrehalase
Accession: ANU46604
Location: 2890156-2891802
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 483
Sequence coverage: 99 %
E-value: 2e-161
NCBI BlastP on this gene
Accession: ANU46604
Location: 2890156-2891802
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 483
Sequence coverage: 99 %
E-value: 2e-161
NCBI BlastP on this gene
A4V08_13135
A4V08_13130
A4V08_13125
A4V08_13120
A4V08_13115
lipopolysaccharide biosynthesis protein
Accession: ANU46599
Location: 2884805-2886256
NCBI BlastP on this gene
Accession: ANU46599
Location: 2884805-2886256
NCBI BlastP on this gene
A4V08_13110
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP050964 : Enterocloster clostridioformis strain FDAARGOS_739 chromosome. Total score: 2.5 Cumulative Blast bit score: 784
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
FOC47_19250
FOC47_19255
treR
PTS system trehalose-specific EIIBC component
Accession: QIX92484
Location: 3854983-3856434
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 301
Sequence coverage: 85 %
E-value: 7e-92
NCBI BlastP on this gene
Accession: QIX92484
Location: 3854983-3856434
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 301
Sequence coverage: 85 %
E-value: 7e-92
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: QIX94111
Location: 3856629-3858266
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 484
Sequence coverage: 99 %
E-value: 5e-162
NCBI BlastP on this gene
Accession: QIX94111
Location: 3856629-3858266
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 484
Sequence coverage: 99 %
E-value: 5e-162
NCBI BlastP on this gene
treC
FOC47_19275
glf
FOC47_19285
glycosyltransferase family 2 protein
Accession: QIX94112
Location: 3860916-3861971
NCBI BlastP on this gene
Accession: QIX94112
Location: 3860916-3861971
NCBI BlastP on this gene
FOC47_19290
lipopolysaccharide biosynthesis protein
Accession: QIX92488
Location: 3862049-3863500
NCBI BlastP on this gene
Accession: QIX92488
Location: 3862049-3863500
NCBI BlastP on this gene
FOC47_19295
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP008855 : Bacillus sp. X1(2014) Total score: 2.5 Cumulative Blast bit score: 783
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
heme ABC transporter ATP-binding protein
Accession: AIM15027
Location: 157302-158855
NCBI BlastP on this gene
Accession: AIM15027
Location: 157302-158855
NCBI BlastP on this gene
HW35_00885
HW35_00880
HW35_00875
FMN-dependent NADH-azoreductase
Accession: AIM15024
Location: 154320-154955
NCBI BlastP on this gene
Accession: AIM15024
Location: 154320-154955
NCBI BlastP on this gene
HW35_00865
PTS maltose transporter subunit IIBC
Accession: AIM15023
Location: 152158-153582
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 8e-93
NCBI BlastP on this gene
Accession: AIM15023
Location: 152158-153582
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 8e-93
NCBI BlastP on this gene
HW35_00860
trehalose-6-phosphate hydrolase
Accession: AIM15022
Location: 150425-152092
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 3e-160
NCBI BlastP on this gene
Accession: AIM15022
Location: 150425-152092
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 3e-160
NCBI BlastP on this gene
HW35_00855
trehalose operon transcriptional repressor
Accession: AIM15021
Location: 149695-150411
NCBI BlastP on this gene
Accession: AIM15021
Location: 149695-150411
NCBI BlastP on this gene
HW35_00850
HW35_00845
HW35_00840
HW35_00835
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP049740 : Jeotgalibaca arthritidis strain CECT 9157 chromosome Total score: 2.5 Cumulative Blast bit score: 782
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
flagellar hook-basal body protein
Accession: QII81985
Location: 1096113-1096775
NCBI BlastP on this gene
Accession: QII81985
Location: 1096113-1096775
NCBI BlastP on this gene
G7057_05650
ATP-binding cassette domain-containing protein
Accession: QII81986
Location: 1096791-1097516
NCBI BlastP on this gene
Accession: QII81986
Location: 1096791-1097516
NCBI BlastP on this gene
G7057_05655
carbamoyl phosphate synthase-like protein
Accession: QII81987
Location: 1097546-1098859
NCBI BlastP on this gene
Accession: QII81987
Location: 1097546-1098859
NCBI BlastP on this gene
G7057_05660
NERD domain-containing protein
Accession: QII81988
Location: 1098978-1099847
NCBI BlastP on this gene
Accession: QII81988
Location: 1098978-1099847
NCBI BlastP on this gene
G7057_05665
PTS system trehalose-specific EIIBC component
Location: 1100100-1101614
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 317
Sequence coverage: 95 %
E-value: 5e-96
Location: 1100100-1101614
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 317
Sequence coverage: 95 %
E-value: 5e-96
treP
alpha,alpha-phosphotrehalase
Accession: QII81989
Location: 1101689-1103341
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 2e-154
NCBI BlastP on this gene
Accession: QII81989
Location: 1101689-1103341
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 2e-154
NCBI BlastP on this gene
treC
G7057_05680
G7057_05685
GerMN domain-containing protein
Accession: QII81992
Location: 1104621-1105727
NCBI BlastP on this gene
Accession: QII81992
Location: 1104621-1105727
NCBI BlastP on this gene
G7057_05690
YeiH family putative sulfate export transporter
Accession: QII81993
Location: 1105921-1106940
NCBI BlastP on this gene
Accession: QII81993
Location: 1105921-1106940
NCBI BlastP on this gene
G7057_05695
LysR family transcriptional regulator
Accession: QII81994
Location: 1107083-1107952
NCBI BlastP on this gene
Accession: QII81994
Location: 1107083-1107952
NCBI BlastP on this gene
G7057_05700
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP039727 : Bacillus sp. S3 chromosome Total score: 2.5 Cumulative Blast bit score: 776
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
inositol phosphorylceramide synthase
Accession: FAY30_12760
Location: 2665098-2665745
NCBI BlastP on this gene
Accession: FAY30_12760
Location: 2665098-2665745
NCBI BlastP on this gene
FAY30_12760
FAY30_12755
thiol-disulfide oxidoreductase DCC family protein
Accession: QCJ42708
Location: 2663323-2663715
NCBI BlastP on this gene
Accession: QCJ42708
Location: 2663323-2663715
NCBI BlastP on this gene
FAY30_12750
LysM peptidoglycan-binding domain-containing protein
Accession: QCJ42707
Location: 2662057-2663247
NCBI BlastP on this gene
Accession: QCJ42707
Location: 2662057-2663247
NCBI BlastP on this gene
FAY30_12745
PTS trehalose transporter subunit IIBC
Accession: QCJ42706
Location: 2659999-2661423
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 7e-93
NCBI BlastP on this gene
Accession: QCJ42706
Location: 2659999-2661423
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 7e-93
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QCJ42705
Location: 2658250-2659917
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 473
Sequence coverage: 100 %
E-value: 3e-157
NCBI BlastP on this gene
Accession: QCJ42705
Location: 2658250-2659917
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 473
Sequence coverage: 100 %
E-value: 3e-157
NCBI BlastP on this gene
treC
treR
SOS response-associated peptidase
Accession: QCJ45201
Location: 2656722-2657399
NCBI BlastP on this gene
Accession: QCJ45201
Location: 2656722-2657399
NCBI BlastP on this gene
FAY30_12725
FAY30_12720
FAY30_12715
FAY30_12710
FAY30_12705
FAY30_12700
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483476 : Bacillus lentus strain NCTC4824 genome assembly, chromosome: 1. Total score: 2.5 Cumulative Blast bit score: 772
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
L-seryl-tRNA(Sec) selenium transferase
Accession: SQI51388
Location: 197648-198751
NCBI BlastP on this gene
Accession: SQI51388
Location: 197648-198751
NCBI BlastP on this gene
selA
4-hydroxy-2-ketovalerate aldolase
Accession: SQI51390
Location: 198752-199507
NCBI BlastP on this gene
Accession: SQI51390
Location: 198752-199507
NCBI BlastP on this gene
NCTC4824_00220
iolC_1
UDP-N-acetylenolpyruvoylglucosamine reductase
Accession: SQI51393
Location: 200757-201674
NCBI BlastP on this gene
Accession: SQI51393
Location: 200757-201674
NCBI BlastP on this gene
murB
PTS system, trehalose-specific IIBC subunit
Accession: SQI51395
Location: 201902-203338
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 302
Sequence coverage: 86 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: SQI51395
Location: 201902-203338
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 302
Sequence coverage: 86 %
E-value: 2e-92
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: SQI51396
Location: 203483-205153
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 470
Sequence coverage: 101 %
E-value: 3e-156
NCBI BlastP on this gene
Accession: SQI51396
Location: 203483-205153
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 470
Sequence coverage: 101 %
E-value: 3e-156
NCBI BlastP on this gene
treA
GntR family transcriptional regulator
Accession: SQI51398
Location: 205168-205881
NCBI BlastP on this gene
Accession: SQI51398
Location: 205168-205881
NCBI BlastP on this gene
treR
methyl-accepting chemotaxis sensory transducer with Cache sensor
Accession: SQI51400
Location: 206072-207763
NCBI BlastP on this gene
Accession: SQI51400
Location: 206072-207763
NCBI BlastP on this gene
mcpA_1
NCTC4824_00227
pyruvate formate-lyase-activating enzyme
Accession: SQI51403
Location: 208153-208929
NCBI BlastP on this gene
Accession: SQI51403
Location: 208153-208929
NCBI BlastP on this gene
pflA_1
pflB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP051128 : Bacillus megaterium strain S2 chromosome Total score: 2.5 Cumulative Blast bit score: 762
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
phosphatase PAP2 family protein
Accession: QIZ08862
Location: 4321141-4321782
NCBI BlastP on this gene
Accession: QIZ08862
Location: 4321141-4321782
NCBI BlastP on this gene
HFZ78_20910
thiol-disulfide oxidoreductase DCC family protein
Accession: QIZ11053
Location: 4322113-4322505
NCBI BlastP on this gene
Accession: QIZ11053
Location: 4322113-4322505
NCBI BlastP on this gene
HFZ78_20915
LysM peptidoglycan-binding domain-containing protein
Accession: QIZ08863
Location: 4322628-4323818
NCBI BlastP on this gene
Accession: QIZ08863
Location: 4322628-4323818
NCBI BlastP on this gene
HFZ78_20920
HFZ78_20925
GNAT family N-acetyltransferase
Accession: QIZ08865
Location: 4324350-4324922
NCBI BlastP on this gene
Accession: QIZ08865
Location: 4324350-4324922
NCBI BlastP on this gene
HFZ78_20930
PTS system trehalose-specific EIIBC component
Accession: QIZ08866
Location: 4325339-4326763
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 301
Sequence coverage: 87 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: QIZ08866
Location: 4325339-4326763
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 301
Sequence coverage: 87 %
E-value: 2e-92
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: QIZ08867
Location: 4326849-4328516
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 99 %
E-value: 7e-153
NCBI BlastP on this gene
Accession: QIZ08867
Location: 4326849-4328516
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 99 %
E-value: 7e-153
NCBI BlastP on this gene
treC
treR
SOS response-associated peptidase
Accession: QIZ08869
Location: 4329384-4330061
NCBI BlastP on this gene
Accession: QIZ08869
Location: 4329384-4330061
NCBI BlastP on this gene
HFZ78_20950
HFZ78_20955
HFZ78_20960
GNAT family N-acetyltransferase
Accession: QIZ08872
Location: 4331312-4331749
NCBI BlastP on this gene
Accession: QIZ08872
Location: 4331312-4331749
NCBI BlastP on this gene
HFZ78_20965
yyaC
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP010796 : Carnobacterium sp. CP1 Total score: 2.5 Cumulative Blast bit score: 759
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
NY10_1745
Alpha-glycerophosphate oxidase
Accession: ALV22342
Location: 1800352-1802187
NCBI BlastP on this gene
Accession: ALV22342
Location: 1800352-1802187
NCBI BlastP on this gene
NY10_1744
NY10_1743
NY10_1742
PTS system, trehalose-specific IIB component
Accession: ALV22339
Location: 1797069-1798529
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 299
Sequence coverage: 96 %
E-value: 3e-91
NCBI BlastP on this gene
Accession: ALV22339
Location: 1797069-1798529
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 299
Sequence coverage: 96 %
E-value: 3e-91
NCBI BlastP on this gene
NY10_1741
Trehalose-6-phosphate hydrolase
Accession: ALV22338
Location: 1795390-1797045
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 99 %
E-value: 3e-152
NCBI BlastP on this gene
Accession: ALV22338
Location: 1795390-1797045
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 99 %
E-value: 3e-152
NCBI BlastP on this gene
NY10_1740
Trehalose operon transcriptional repressor
Accession: ALV22337
Location: 1794021-1794749
NCBI BlastP on this gene
Accession: ALV22337
Location: 1794021-1794749
NCBI BlastP on this gene
NY10_1739
NY10_1738
GTPase and tRNA-U34 5-formylation enzyme TrmE
Accession: ALV22335
Location: 1790645-1792033
NCBI BlastP on this gene
Accession: ALV22335
Location: 1790645-1792033
NCBI BlastP on this gene
NY10_1737
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP045915 : Gracilibacillus sp. SCU50 chromosome Total score: 2.5 Cumulative Blast bit score: 758
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
NADP-specific glutamate dehydrogenase
Accession: QGH36608
Location: 4750566-4751927
NCBI BlastP on this gene
Accession: QGH36608
Location: 4750566-4751927
NCBI BlastP on this gene
GI584_22250
GI584_22245
GI584_22240
PTS trehalose transporter subunit IIBC
Accession: QGH36605
Location: 4746780-4748222
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 298
Sequence coverage: 86 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QGH36605
Location: 4746780-4748222
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 298
Sequence coverage: 86 %
E-value: 6e-91
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QGH36604
Location: 4744676-4746355
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 101 %
E-value: 3e-152
NCBI BlastP on this gene
Accession: QGH36604
Location: 4744676-4746355
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 101 %
E-value: 3e-152
NCBI BlastP on this gene
treC
treR
GI584_22220
GI584_22215
methyltransferase domain-containing protein
Accession: QGH36600
Location: 4741875-4742597
NCBI BlastP on this gene
Accession: QGH36600
Location: 4741875-4742597
NCBI BlastP on this gene
GI584_22210
Lrp/AsnC family transcriptional regulator
Accession: QGH36599
Location: 4741108-4741605
NCBI BlastP on this gene
Accession: QGH36599
Location: 4741108-4741605
NCBI BlastP on this gene
GI584_22205
GI584_22200
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022512 : Mesoplasma florum strain MQ3 chromosome Total score: 2.5 Cumulative Blast bit score: 536
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG006_02770
CG006_02765
CG006_02760
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN63883
Location: 599875-601473
NCBI BlastP on this gene
Accession: AVN63883
Location: 599875-601473
NCBI BlastP on this gene
CG006_02755
hypothetical protein
Accession: AVN63882
Location: 598839-599786
BlastP hit with treR
Percentage identity: 33 %
BlastP bit score: 104
Sequence coverage: 85 %
E-value: 3e-22
NCBI BlastP on this gene
Accession: AVN63882
Location: 598839-599786
BlastP hit with treR
Percentage identity: 33 %
BlastP bit score: 104
Sequence coverage: 85 %
E-value: 3e-22
NCBI BlastP on this gene
CG006_02750
PTS sugar transporter subunit IIA
Accession: AVN63881
Location: 597217-598776
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AVN63881
Location: 597217-598776
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
CG006_02745
CG006_02740
CG006_02735
CG006_02730
magnesium-translocating P-type ATPase
Accession: AVN63877
Location: 591349-594009
NCBI BlastP on this gene
Accession: AVN63877
Location: 591349-594009
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022511 : Mesoplasma coleopterae strain BARC 781 chromosome. Total score: 2.5 Cumulative Blast bit score: 535
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG000_02735
CG000_02730
CG000_02725
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN63196
Location: 607126-608724
NCBI BlastP on this gene
Accession: AVN63196
Location: 607126-608724
NCBI BlastP on this gene
CG000_02720
hypothetical protein
Accession: AVN63195
Location: 606090-607037
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVN63195
Location: 606090-607037
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
CG000_02715
PTS sugar transporter subunit IIA
Accession: AVN63194
Location: 604468-606027
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AVN63194
Location: 604468-606027
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 6e-144
NCBI BlastP on this gene
CG000_02710
CG000_02705
CG000_02700
CG000_02695
magnesium-translocating P-type ATPase
Accession: AVN63190
Location: 598589-601249
NCBI BlastP on this gene
Accession: AVN63190
Location: 598589-601249
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011855 : Spiroplasma atrichopogonis strain GNAT3597 Total score: 2.5 Cumulative Blast bit score: 535
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SATRI_v1c09280
SATRI_v1c09210
SATRI_v1c09200
SATRI_v1c09190
RNA methyltransferase, TrmH family
Accession: AKM53313
Location: 830808-831599
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
Accession: AKM53313
Location: 830808-831599
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
spoU
GTP-binding protein YsxC
Accession: AKM53312
Location: 830212-830805
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
Accession: AKM53312
Location: 830212-830805
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
engB
SATRI_v1c09160
SATRI_v1c09090
SATRI_v1c09080
prolipoprotein diacylglyceryl transferase
Accession: AKM53308
Location: 824134-825657
NCBI BlastP on this gene
Accession: AKM53308
Location: 824134-825657
NCBI BlastP on this gene
lgt
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP006720 : Spiroplasma mirum ATCC 29335 strain SMCA Total score: 2.5 Cumulative Blast bit score: 535
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
P344_05140
P344_05135
P344_05130
P344_05125
P344_05120
P344_05115
P344_05110
P344_05105
P344_05100
P344_05095
P344_05090
hypothetical protein
Accession: AHI58339
Location: 803561-804352
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
Accession: AHI58339
Location: 803561-804352
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
P344_05085
hypothetical protein
Accession: AHI58338
Location: 802965-803558
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
Accession: AHI58338
Location: 802965-803558
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
P344_05080
P344_05075
P344_05070
P344_05065
P344_05060
P344_05055
P344_05050
P344_05045
P344_05040
P344_05035
P344_05030
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
AE017263 : Mesoplasma florum L1 complete genome. Total score: 2.5 Cumulative Blast bit score: 535
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
Mfl505
Mfl504
cof-like hydrolase, HAD-superfamily
Accession: AAT75861
Location: 590775-591596
NCBI BlastP on this gene
Accession: AAT75861
Location: 590775-591596
NCBI BlastP on this gene
Mfl503
Mfl502
trehalose operon repressor, repressor of treA,B,C
Accession: AAT75859
Location: 588125-589072
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 103
Sequence coverage: 85 %
E-value: 8e-22
NCBI BlastP on this gene
Accession: AAT75859
Location: 588125-589072
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 103
Sequence coverage: 85 %
E-value: 8e-22
NCBI BlastP on this gene
Mfl501
trehalose PTS system IIABC component
Accession: AAT75858
Location: 586503-588062
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AAT75858
Location: 586503-588062
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Mfl500
trehalose-6-phosphate hydrolase
Accession: AAT75857
Location: 584838-586457
NCBI BlastP on this gene
Accession: AAT75857
Location: 584838-586457
NCBI BlastP on this gene
Mfl499
Mfl498
Mfl497
Mfl496
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022514 : Mesoplasma florum strain BARC 787 chromosome Total score: 2.5 Cumulative Blast bit score: 534
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG002_02570
CG002_02565
CG002_02560
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN65229
Location: 547905-549503
NCBI BlastP on this gene
Accession: AVN65229
Location: 547905-549503
NCBI BlastP on this gene
CG002_02555
hypothetical protein
Accession: AVN65228
Location: 546869-547816
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AVN65228
Location: 546869-547816
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
CG002_02550
PTS sugar transporter subunit IIA
Accession: AVN65227
Location: 545247-546806
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AVN65227
Location: 545247-546806
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
CG002_02545
CG002_02540
CG002_02535
CG002_02530
magnesium-translocating P-type ATPase
Accession: AVN65223
Location: 539381-542038
NCBI BlastP on this gene
Accession: AVN65223
Location: 539381-542038
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022510 : Mesoplasma coleopterae strain BARC 786 chromosome. Total score: 2.5 Cumulative Blast bit score: 534
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG001_02635
CG001_02630
CG001_02625
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN62520
Location: 573794-575392
NCBI BlastP on this gene
Accession: AVN62520
Location: 573794-575392
NCBI BlastP on this gene
CG001_02620
hypothetical protein
Accession: AVN62519
Location: 572758-573705
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVN62519
Location: 572758-573705
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
CG001_02615
PTS sugar transporter subunit IIA
Accession: AVN62518
Location: 571136-572695
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 435
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AVN62518
Location: 571136-572695
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 435
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
CG001_02610
CG001_02605
CG001_02600
CG001_02595
magnesium-translocating P-type ATPase
Accession: AVN62514
Location: 565257-567917
NCBI BlastP on this gene
Accession: AVN62514
Location: 565257-567917
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022505 : Mesoplasma florum strain W23 chromosome Total score: 2.5 Cumulative Blast bit score: 534
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG009_02710
CG009_02705
CG009_02700
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AVN59115
Location: 573220-574818
NCBI BlastP on this gene
Accession: AVN59115
Location: 573220-574818
NCBI BlastP on this gene
CG009_02695
hypothetical protein
Accession: AVN59114
Location: 572184-573131
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AVN59114
Location: 572184-573131
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CG009_02690
PTS sugar transporter subunit IIA
Accession: AVN59113
Location: 570562-572121
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AVN59113
Location: 570562-572121
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
CG009_02685
CG009_02680
CG009_02675
CG009_02670
CG009_02665
PTS sugar transporter subunit IIA
Accession: AVN59108
Location: 564863-566590
NCBI BlastP on this gene
Accession: AVN59108
Location: 564863-566590
NCBI BlastP on this gene
CG009_02660
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP023736 : Mesoplasma florum strain PPA1 chromosome Total score: 2.5 Cumulative Blast bit score: 533
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CQZ70_02780
CQZ70_02775
CQZ70_02770
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: ATI74154
Location: 613071-614669
NCBI BlastP on this gene
Accession: ATI74154
Location: 613071-614669
NCBI BlastP on this gene
CQZ70_02765
hypothetical protein
Accession: ATI74153
Location: 612035-612982
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: ATI74153
Location: 612035-612982
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CQZ70_02760
PTS sugar transporter subunit IIA
Accession: ATI74152
Location: 610413-611972
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: ATI74152
Location: 610413-611972
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
CQZ70_02755
CQZ70_02750
CQZ70_02745
CQZ70_02740
magnesium-translocating P-type ATPase
Accession: ATI74148
Location: 604546-607203
NCBI BlastP on this gene
Accession: ATI74148
Location: 604546-607203
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022513 : Mesoplasma florum strain CnuA-2 chromosome Total score: 2.5 Cumulative Blast bit score: 533
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG003_02765
CG003_02760
CG003_02755
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AVN64567
Location: 608119-609717
NCBI BlastP on this gene
Accession: AVN64567
Location: 608119-609717
NCBI BlastP on this gene
CG003_02750
LacI family transcriptional regulator
Accession: AVN64566
Location: 607083-608030
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 101
Sequence coverage: 85 %
E-value: 3e-21
NCBI BlastP on this gene
Accession: AVN64566
Location: 607083-608030
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 101
Sequence coverage: 85 %
E-value: 3e-21
NCBI BlastP on this gene
CG003_02745
PTS sugar transporter subunit IIA
Accession: AVN64565
Location: 605461-607020
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 4e-142
NCBI BlastP on this gene
Accession: AVN64565
Location: 605461-607020
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 4e-142
NCBI BlastP on this gene
CG003_02740
CG003_02735
CG003_02730
CG003_02725
magnesium-translocating P-type ATPase
Accession: AVN64561
Location: 599592-602252
NCBI BlastP on this gene
Accession: AVN64561
Location: 599592-602252
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022508 : Mesoplasma florum strain MouA-2 chromosome Total score: 2.5 Cumulative Blast bit score: 533
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG005_02640
CG005_02635
CG005_02630
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN61167
Location: 578742-580340
NCBI BlastP on this gene
Accession: AVN61167
Location: 578742-580340
NCBI BlastP on this gene
CG005_02625
hypothetical protein
Accession: AVN61166
Location: 577706-578653
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AVN61166
Location: 577706-578653
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CG005_02620
PTS sugar transporter subunit IIA
Accession: AVN61165
Location: 576084-577643
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AVN61165
Location: 576084-577643
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
CG005_02615
CG005_02610
CG005_02605
CG005_02600
magnesium-translocating P-type ATPase
Accession: AVN61161
Location: 570219-572876
NCBI BlastP on this gene
Accession: AVN61161
Location: 570219-572876
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022506 : Mesoplasma florum strain W20 chromosome. Total score: 2.5 Cumulative Blast bit score: 533
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG008_02805
CG008_02800
CG008_02795
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN59810
Location: 604263-605861
NCBI BlastP on this gene
Accession: AVN59810
Location: 604263-605861
NCBI BlastP on this gene
CG008_02790
LacI family transcriptional regulator
Accession: AVN59809
Location: 603227-604174
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AVN59809
Location: 603227-604174
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
CG008_02785
PTS sugar transporter subunit IIA
Accession: AVN59808
Location: 601605-603164
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AVN59808
Location: 601605-603164
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
CG008_02780
CG008_02775
CG008_02770
CG008_02765
magnesium-translocating P-type ATPase
Accession: AVN59804
Location: 595739-598396
NCBI BlastP on this gene
Accession: AVN59804
Location: 595739-598396
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022432 : Mesoplasma florum strain W12 chromosome. Total score: 2.5 Cumulative Blast bit score: 533
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MflW12_5340
MflW12_5330
MflW12_5320
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AVN65936
Location: 602812-604410
NCBI BlastP on this gene
Accession: AVN65936
Location: 602812-604410
NCBI BlastP on this gene
MflW12_5310
Sucrose operon repressor ScrR, LacI family
Accession: AVN65935
Location: 601776-602723
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AVN65935
Location: 601776-602723
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
MflW12_5300
PTS system trehalose-specific IIB component, PTS system trehalose-specific IIC component
Accession: AVN65934
Location: 600154-601713
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AVN65934
Location: 600154-601713
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
MflW12_5290
Trehalose-6-phosphate hydrolase
Accession: AVN65933
Location: 598500-600119
NCBI BlastP on this gene
Accession: AVN65933
Location: 598500-600119
NCBI BlastP on this gene
MflW12_5280
MflW12_5270
Mg(2+) transport ATPase, P-type
Accession: AVN65931
Location: 594288-596945
NCBI BlastP on this gene
Accession: AVN65931
Location: 594288-596945
NCBI BlastP on this gene
MflW12_5260
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP006778 : Mesoplasma florum W37 Total score: 2.5 Cumulative Blast bit score: 533
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
mflW37_5340
mflW37_5330
cof-like hydrolase, HAD-superfamily
Accession: AGY41599
Location: 601139-601960
NCBI BlastP on this gene
Accession: AGY41599
Location: 601139-601960
NCBI BlastP on this gene
mflW37_5320
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AGY41598
Location: 599525-601123
NCBI BlastP on this gene
Accession: AGY41598
Location: 599525-601123
NCBI BlastP on this gene
mflW37_5310
trehalose operon repressor, repressor of treA,B,C
Accession: AGY41597
Location: 598489-599436
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AGY41597
Location: 598489-599436
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
mflW37_5300
PTS system, trehalose-specific IIB component PTS system, trehalose-specific IIC component
Accession: AGY41596
Location: 596867-598426
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AGY41596
Location: 596867-598426
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
mflW37_5290
Trehalose-6-phosphate hydrolase
Accession: AGY41595
Location: 595213-596832
NCBI BlastP on this gene
Accession: AGY41595
Location: 595213-596832
NCBI BlastP on this gene
mflW37_5280
mflW37_5270
Mg(2+) transport ATPase, P-type
Accession: AGY41593
Location: 591001-593658
NCBI BlastP on this gene
Accession: AGY41593
Location: 591001-593658
NCBI BlastP on this gene
mflW37_5260
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP023735 : Mesoplasma florum strain GF1 chromosome Total score: 2.5 Cumulative Blast bit score: 530
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CQZ69_02785
CQZ69_02780
CQZ69_02775
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: ATI73468
Location: 612720-614318
NCBI BlastP on this gene
Accession: ATI73468
Location: 612720-614318
NCBI BlastP on this gene
CQZ69_02770
hypothetical protein
Accession: ATI73467
Location: 611684-612631
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: ATI73467
Location: 611684-612631
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CQZ69_02765
PTS sugar transporter subunit IIA
Accession: ATI73466
Location: 610062-611621
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
Accession: ATI73466
Location: 610062-611621
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
CQZ69_02760
CQZ69_02755
CQZ69_02750
CQZ69_02745
CQZ69_02740
magnesium-translocating P-type ATPase
Accession: ATI73461
Location: 603704-606364
NCBI BlastP on this gene
Accession: ATI73461
Location: 603704-606364
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022509 : Mesoplasma florum strain GF chromosome. Total score: 2.5 Cumulative Blast bit score: 530
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CG004_02770
CG004_02765
CG004_02760
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN61861
Location: 597872-599470
NCBI BlastP on this gene
Accession: AVN61861
Location: 597872-599470
NCBI BlastP on this gene
CG004_02755
hypothetical protein
Accession: AVN61860
Location: 596836-597783
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AVN61860
Location: 596836-597783
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CG004_02750
PTS sugar transporter subunit IIA
Accession: AVN61859
Location: 595214-596773
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
Accession: AVN61859
Location: 595214-596773
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
CG004_02745
CG004_02740
CG004_02735
CG004_02730
CG004_02725
magnesium-translocating P-type ATPase
Accession: AVN61854
Location: 588856-591516
NCBI BlastP on this gene
Accession: AVN61854
Location: 588856-591516
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011856 : Spiroplasma eriocheiris strain DSM 21848 Total score: 2.5 Cumulative Blast bit score: 530
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
RpiR family transcriptional regulator
Accession: AKM54451
Location: 970715-971524
NCBI BlastP on this gene
Accession: AKM54451
Location: 970715-971524
NCBI BlastP on this gene
SERIO_v1c08910
patB
pepA
SERIO_v1c08880
SERIO_v1c08870
SERIO_v1c08860
RNA methyltransferase, TrmH family
Accession: AKM54445
Location: 965424-966215
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
Accession: AKM54445
Location: 965424-966215
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
spoU
GTP-binding protein YsxC
Accession: AKM54444
Location: 964828-965421
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
Accession: AKM54444
Location: 964828-965421
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
engB
focA
pflD
SERIO_v1c08810
prolipoprotein diacylglyceryl transferase
Accession: AKM54440
Location: 958610-960130
NCBI BlastP on this gene
Accession: AKM54440
Location: 958610-960130
NCBI BlastP on this gene
lgt
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP001973 : Spiroplasma eriocheiris CCTCC M 207170 Total score: 2.5 Cumulative Blast bit score: 530
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
putative phosphosugar-binding transcriptional regulator
Accession: AHF58009
Location: 969776-970585
NCBI BlastP on this gene
Accession: AHF58009
Location: 969776-970585
NCBI BlastP on this gene
SPE_0889
SPE_0888
pepB1
SPE_0886
SPE_0885
SPE_0884
rRNA methylase
Accession: AHF58003
Location: 964485-965276
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
Accession: AHF58003
Location: 964485-965276
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
spoU
putative GTP binding protein
Accession: AHF58002
Location: 963889-964482
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
Accession: AHF58002
Location: 963889-964482
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
engB
focA
pflB
SPE_0879
prolipoprotein diacylglyceryl transferase
Accession: AHF57998
Location: 957671-959191
NCBI BlastP on this gene
Accession: AHF57998
Location: 957671-959191
NCBI BlastP on this gene
lgt1
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP024969 : Mesoplasma tabanidae strain BARC 857 chromosome Total score: 2.5 Cumulative Blast bit score: 525
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MTABA_v1c05510
tpiA
MTABA_v1c05490
gpm
trehalose operon repressor, repressor of treA,B,C
Accession: ATZ21741
Location: 633782-634726
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 84 %
E-value: 1e-18
NCBI BlastP on this gene
Accession: ATZ21741
Location: 633782-634726
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 84 %
E-value: 1e-18
NCBI BlastP on this gene
MTABA_v1c05470
PTS system, trehalose-specific IIBC component
Accession: ATZ21740
Location: 632160-633719
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 5e-142
NCBI BlastP on this gene
Accession: ATZ21740
Location: 632160-633719
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 5e-142
NCBI BlastP on this gene
treB
MTABA_v1c05450
trehalose-6-phosphate hydrolase
Accession: ATZ21738
Location: 630022-631641
NCBI BlastP on this gene
Accession: ATZ21738
Location: 630022-631641
NCBI BlastP on this gene
treC
MTABA_v1c05430
glk
Mg(2+) transport ATPase, P-type
Accession: ATZ21735
Location: 625788-628448
NCBI BlastP on this gene
Accession: ATZ21735
Location: 625788-628448
NCBI BlastP on this gene
mgtA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP005077 : Spiroplasma chrysopicola DF-1 Total score: 2.5 Cumulative Blast bit score: 502
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SCHRY_v1c03080
hemN
ytjP
SCHRY_v1c03110
RNA methyltransferase, TrmH family
Accession: AGM24897
Location: 339535-340335
BlastP hit with AAP55640.1
Percentage identity: 49 %
BlastP bit score: 214
Sequence coverage: 100 %
E-value: 2e-64
NCBI BlastP on this gene
Accession: AGM24897
Location: 339535-340335
BlastP hit with AAP55640.1
Percentage identity: 49 %
BlastP bit score: 214
Sequence coverage: 100 %
E-value: 2e-64
NCBI BlastP on this gene
spoU
GTP-binding protein YsxC
Accession: AGM24898
Location: 340328-340924
BlastP hit with AAP55641.1
Percentage identity: 70 %
BlastP bit score: 288
Sequence coverage: 98 %
E-value: 2e-95
NCBI BlastP on this gene
Accession: AGM24898
Location: 340328-340924
BlastP hit with AAP55641.1
Percentage identity: 70 %
BlastP bit score: 288
Sequence coverage: 98 %
E-value: 2e-95
NCBI BlastP on this gene
engB
prolipoprotein diacylglyceryl transferase
Accession: AGM24899
Location: 341007-342851
NCBI BlastP on this gene
Accession: AGM24899
Location: 341007-342851
NCBI BlastP on this gene
SCHRY_v1c03140
SCHRY_v1c03150
SCHRY_v1c03160
SCHRY_v1c03170
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
51. : CP002582 Clostridium lentocellum DSM 5427 Total score: 2.5 Cumulative Blast bit score: 812
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
metal-dependent phosphohydrolase HD sub domain
Accession: ADZ85143
Location: 3781070-3781555
NCBI BlastP on this gene
Accession: ADZ85143
Location: 3781070-3781555
NCBI BlastP on this gene
Clole_3456
NADPH-dependent FMN reductase
Accession: ADZ85142
Location: 3780549-3781031
NCBI BlastP on this gene
Accession: ADZ85142
Location: 3780549-3781031
NCBI BlastP on this gene
Clole_3455
cation diffusion facilitator family transporter
Accession: ADZ85141
Location: 3779386-3780273
NCBI BlastP on this gene
Accession: ADZ85141
Location: 3779386-3780273
NCBI BlastP on this gene
Clole_3454
phenazine biosynthesis protein PhzF family
Accession: ADZ85140
Location: 3778411-3779289
NCBI BlastP on this gene
Accession: ADZ85140
Location: 3778411-3779289
NCBI BlastP on this gene
Clole_3453
transcriptional regulator, GntR family with UTRA sensor domain
Accession: ADZ85139
Location: 3777431-3778168
NCBI BlastP on this gene
Accession: ADZ85139
Location: 3777431-3778168
NCBI BlastP on this gene
Clole_3452
PTS system, trehalose-specific IIBC subunit
Accession: ADZ85138
Location: 3775822-3777243
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 311
Sequence coverage: 85 %
E-value: 6e-96
NCBI BlastP on this gene
Accession: ADZ85138
Location: 3775822-3777243
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 311
Sequence coverage: 85 %
E-value: 6e-96
NCBI BlastP on this gene
Clole_3451
alpha,alpha-phosphotrehalase
Accession: ADZ85137
Location: 3774083-3775720
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 501
Sequence coverage: 98 %
E-value: 3e-168
NCBI BlastP on this gene
Accession: ADZ85137
Location: 3774083-3775720
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 501
Sequence coverage: 98 %
E-value: 3e-168
NCBI BlastP on this gene
Clole_3450
Clole_3449
Clole_3448
Clole_3447
52. : CP042243 Crassaminicella sp. SY095 chromosome Total score: 2.5 Cumulative Blast bit score: 810
LL-diaminopimelate aminotransferase
Accession: QEK12819
Location: 2167749-2168981
NCBI BlastP on this gene
Accession: QEK12819
Location: 2167749-2168981
NCBI BlastP on this gene
FQB35_11045
ATP-dependent Clp protease ATP-binding subunit
Accession: QEK12818
Location: 2165253-2167535
NCBI BlastP on this gene
Accession: QEK12818
Location: 2165253-2167535
NCBI BlastP on this gene
FQB35_11040
PTS trehalose transporter subunit IIBC
Accession: QEK12817
Location: 2163660-2165075
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 302
Sequence coverage: 88 %
E-value: 1e-92
NCBI BlastP on this gene
Accession: QEK12817
Location: 2163660-2165075
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 302
Sequence coverage: 88 %
E-value: 1e-92
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QEK12816
Location: 2161918-2163582
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 508
Sequence coverage: 99 %
E-value: 4e-171
NCBI BlastP on this gene
Accession: QEK12816
Location: 2161918-2163582
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 508
Sequence coverage: 99 %
E-value: 4e-171
NCBI BlastP on this gene
treC
treR
Na+/H+ antiporter NhaC family protein
Accession: QEK12814
Location: 2158963-2160669
NCBI BlastP on this gene
Accession: QEK12814
Location: 2158963-2160669
NCBI BlastP on this gene
FQB35_11020
beta-ketoacyl-ACP synthase II
Accession: QEK12813
Location: 2157656-2158906
NCBI BlastP on this gene
Accession: QEK12813
Location: 2157656-2158906
NCBI BlastP on this gene
fabF
53. : CP039703 Clostridium butyricum strain 29-1 plasmid p-butyl_plas_29-1 Total score: 2.5 Cumulative Blast bit score: 806
FBD77_17400
FBD77_17395
FBD77_17390
FBD77_17385
FBD77_17380
PTS trehalose transporter subunit IIBC
Accession: QCJ04253
Location: 73414-74826
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: QCJ04253
Location: 73414-74826
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QCJ04252
Location: 71618-73276
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: QCJ04252
Location: 71618-73276
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
treC
treR
FBD77_17360
FBD77_17355
FBD77_17350
54. : CP016333 Clostridium butyricum strain TK520 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
BBB49_17735
BBB49_17740
BBB49_17745
BBB49_17750
BBB49_17755
PTS maltose transporter subunit IIBC
Accession: AOR95920
Location: 327993-329405
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: AOR95920
Location: 327993-329405
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
BBB49_17760
alpha,alpha-phosphotrehalase
Accession: AOR95921
Location: 329543-331201
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: AOR95921
Location: 329543-331201
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
BBB49_17765
BBB49_17770
BBB49_17775
BBB49_17780
BBB49_17785
55. : CP014705 Clostridium butyricum strain TOA chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
AZ909_18265
AZ909_18270
AZ909_18275
AZ909_18280
AZ909_18285
PTS maltose transporter subunit IIBC
Accession: ANF16008
Location: 368762-370174
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: ANF16008
Location: 368762-370174
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
AZ909_18290
alpha,alpha-phosphotrehalase
Accession: ANF16009
Location: 370312-371970
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: ANF16009
Location: 370312-371970
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
AZ909_18295
AZ909_18300
AZ909_18305
AZ909_18310
AZ909_18315
56. : CP013489 Clostridium butyricum strain KNU-L09 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
ATN24_19285
ATN24_19295
ATN24_19300
ATN24_19305
PTS maltose transporter subunit IIBC
Accession: ALR90585
Location: 527024-528436
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: ALR90585
Location: 527024-528436
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
ATN24_19310
glucohydrolase
Accession: ALR90586
Location: 528574-530232
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: ALR90586
Location: 528574-530232
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
ATN24_19315
ATN24_19320
ATN24_19325
ATN24_19330
ATN24_19335
57. : CP013353 Clostridium butyricum strain JKY6D1 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 806
ATD26_18235
ATD26_18240
ATD26_18245
ATD26_18250
ATD26_18255
PTS maltose transporter subunit IIBC
Accession: ALS18825
Location: 301171-302583
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
Accession: ALS18825
Location: 301171-302583
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 4e-90
NCBI BlastP on this gene
ATD26_18260
glucohydrolase
Accession: ALS18826
Location: 302721-304379
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
Accession: ALS18826
Location: 302721-304379
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 510
Sequence coverage: 99 %
E-value: 6e-172
NCBI BlastP on this gene
ATD26_18265
ATD26_18270
ATD26_18275
ATD26_18280
ATD26_18285
58. : CP017080 Bacillus muralis strain G25-68 chromosome Total score: 2.5 Cumulative Blast bit score: 805
ABE28_009835
ABE28_009840
DUF4867 domain-containing protein
Accession: AOH54651
Location: 2007344-2008042
NCBI BlastP on this gene
Accession: AOH54651
Location: 2007344-2008042
NCBI BlastP on this gene
ABE28_009845
ABE28_009850
ABE28_009855
PTS maltose transporter subunit IIBC
Accession: AOH54654
Location: 2009794-2011245
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 85 %
E-value: 3e-90
NCBI BlastP on this gene
Accession: AOH54654
Location: 2009794-2011245
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 85 %
E-value: 3e-90
NCBI BlastP on this gene
ABE28_009860
alpha,alpha-phosphotrehalase
Accession: AOH54655
Location: 2011325-2013004
BlastP hit with treA
Percentage identity: 50 %
BlastP bit score: 509
Sequence coverage: 91 %
E-value: 1e-171
NCBI BlastP on this gene
Accession: AOH54655
Location: 2011325-2013004
BlastP hit with treA
Percentage identity: 50 %
BlastP bit score: 509
Sequence coverage: 91 %
E-value: 1e-171
NCBI BlastP on this gene
ABE28_009865
ABE28_009870
ABE28_009875
quaternary ammonium transporter
Accession: AOH54658
Location: 2014215-2014487
NCBI BlastP on this gene
Accession: AOH54658
Location: 2014215-2014487
NCBI BlastP on this gene
ABE28_009880
DNA polymerase III subunit beta
Accession: AOH54659
Location: 2015049-2016182
NCBI BlastP on this gene
Accession: AOH54659
Location: 2015049-2016182
NCBI BlastP on this gene
ABE28_009885
ABE28_009890
59. : CP013238 Clostridium butyricum strain CDC_51208 plasmid pNPD4_2 Total score: 2.5 Cumulative Blast bit score: 803
NPD4_3705
NPD4_3704
NPD4_3703
NPD4_3702
NPD4_3701
PTS system, trehalose-specific IIBC component
Accession: APF21357
Location: 193121-194533
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
Accession: APF21357
Location: 193121-194533
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: APF21430
Location: 191325-192983
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: APF21430
Location: 191325-192983
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
NPD4_3697
NPD4_3696
putative zinc-binding domain protein
Accession: APF21092
Location: 187380-188012
NCBI BlastP on this gene
Accession: APF21092
Location: 187380-188012
NCBI BlastP on this gene
NPD4_3695
60. : AP019717 Clostridium butyricum NBRC 13949 plasmid pCBU1 DNA Total score: 2.5 Cumulative Blast bit score: 803
Cbu04g_41250
Cbu04g_41260
Cbu04g_41270
serine/threonine protein kinase
Accession: BBK79120
Location: 655954-656673
NCBI BlastP on this gene
Accession: BBK79120
Location: 655954-656673
NCBI BlastP on this gene
Cbu04g_41280
Cbu04g_41290
PTS maltose transporter subunit IIBC
Accession: BBK79122
Location: 657693-659105
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
Accession: BBK79122
Location: 657693-659105
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 296
Sequence coverage: 86 %
E-value: 3e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: BBK79123
Location: 659243-660901
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: BBK79123
Location: 659243-660901
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treA
treR
FprA family A-type flavoprotein
Accession: BBK79125
Location: 661961-663127
NCBI BlastP on this gene
Accession: BBK79125
Location: 661961-663127
NCBI BlastP on this gene
Cbu04g_41330
dedA
Cbu04g_41350
61. : CP039706 Clostridium butyricum strain 4-1 plasmid p-butyl_plas_4-1 Total score: 2.5 Cumulative Blast bit score: 801
FBD76_20225
FBD76_20230
FBD76_20235
FBD76_20240
FBD76_20245
PTS trehalose transporter subunit IIBC
Accession: QCJ08656
Location: 619277-620689
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: QCJ08656
Location: 619277-620689
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QCJ08657
Location: 620827-622485
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: QCJ08657
Location: 620827-622485
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: QCJ08659
Location: 623545-624711
NCBI BlastP on this gene
Accession: QCJ08659
Location: 623545-624711
NCBI BlastP on this gene
FBD76_20265
FBD76_20270
FBD76_20275
62. : CP033248 Clostridium butyricum strain CFSA3989 plasmid pCFSA3989 Total score: 2.5 Cumulative Blast bit score: 801
EBQ27_01605
EBQ27_01610
EBQ27_01615
EBQ27_01620
EBQ27_01625
PTS trehalose transporter subunit IIBC
Accession: QGH24389
Location: 383227-384639
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: QGH24389
Location: 383227-384639
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QGH24390
Location: 384777-386435
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: QGH24390
Location: 384777-386435
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: QGH24392
Location: 387495-388661
NCBI BlastP on this gene
Accession: QGH24392
Location: 387495-388661
NCBI BlastP on this gene
EBQ27_01645
EBQ27_01650
EBQ27_01655
63. : CP033246 Clostridium butyricum strain CFSA3987 plasmid pCFSA3987 Total score: 2.5 Cumulative Blast bit score: 801
EBL75_01605
EBL75_01610
EBL75_01615
EBL75_01620
EBL75_01625
PTS trehalose transporter subunit IIBC
Accession: QGH20353
Location: 383188-384600
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: QGH20353
Location: 383188-384600
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QGH20354
Location: 384738-386396
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: QGH20354
Location: 384738-386396
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: QGH20356
Location: 387456-388622
NCBI BlastP on this gene
Accession: QGH20356
Location: 387456-388622
NCBI BlastP on this gene
EBL75_01645
EBL75_01650
EBL75_01655
64. : CP030776 Clostridium butyricum strain S-45-5 chromosome 2 Total score: 2.5 Cumulative Blast bit score: 801
DRB99_17735
DRB99_17740
DRB99_17745
DRB99_17750
DRB99_17755
PTS trehalose transporter subunit IIBC
Accession: AXB86768
Location: 108634-110046
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
Accession: AXB86768
Location: 108634-110046
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 295
Sequence coverage: 86 %
E-value: 9e-90
NCBI BlastP on this gene
DRB99_17760
alpha,alpha-phosphotrehalase
Accession: AXB86769
Location: 110184-111842
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
Accession: AXB86769
Location: 110184-111842
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 507
Sequence coverage: 99 %
E-value: 9e-171
NCBI BlastP on this gene
treC
treR
FprA family A-type flavoprotein
Accession: AXB86771
Location: 112902-114068
NCBI BlastP on this gene
Accession: AXB86771
Location: 112902-114068
NCBI BlastP on this gene
DRB99_17775
DRB99_17780
DRB99_17785
65. : LT630003 [Clostridium] sphenoides JCM 1415 strain ATCC 19403 genome assembly, chromosome: I. Total score: 2.5 Cumulative Blast bit score: 801
SAMN02745906_3037
HD-GYP domain, c-di-GMP phosphodiesterase class II (or its inactivated variant)
Accession: SET92268
Location: 3311405-3312736
NCBI BlastP on this gene
Accession: SET92268
Location: 3311405-3312736
NCBI BlastP on this gene
SAMN02745906_3036
GntR family transcriptional regulator, trehalose operon transcriptional repressor
Accession: SET92260
Location: 3310436-3311149
NCBI BlastP on this gene
Accession: SET92260
Location: 3310436-3311149
NCBI BlastP on this gene
SAMN02745906_3035
PTS system, trehalose-specific IIC component
Accession: SET92249
Location: 3308799-3310229
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 300
Sequence coverage: 92 %
E-value: 2e-91
NCBI BlastP on this gene
Accession: SET92249
Location: 3308799-3310229
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 300
Sequence coverage: 92 %
E-value: 2e-91
NCBI BlastP on this gene
SAMN02745906_3034
trehalose-6-phosphate hydrolase
Accession: SET92238
Location: 3307140-3308798
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 501
Sequence coverage: 91 %
E-value: 3e-168
NCBI BlastP on this gene
Accession: SET92238
Location: 3307140-3308798
BlastP hit with treA
Percentage identity: 48 %
BlastP bit score: 501
Sequence coverage: 91 %
E-value: 3e-168
NCBI BlastP on this gene
SAMN02745906_3033
Putative Mn2+ efflux pump MntP
Accession: SET92227
Location: 3306580-3307143
NCBI BlastP on this gene
Accession: SET92227
Location: 3306580-3307143
NCBI BlastP on this gene
SAMN02745906_3032
Predicted RNA-binding protein containing a PIN domain
Accession: SET92215
Location: 3305999-3306520
NCBI BlastP on this gene
Accession: SET92215
Location: 3305999-3306520
NCBI BlastP on this gene
SAMN02745906_3031
putative efflux protein, MATE family
Accession: SET92203
Location: 3304572-3305942
NCBI BlastP on this gene
Accession: SET92203
Location: 3304572-3305942
NCBI BlastP on this gene
SAMN02745906_3030
nondiscriminating aspartyl-tRNA synthetase
Accession: SET92191
Location: 3302545-3303876
NCBI BlastP on this gene
Accession: SET92191
Location: 3302545-3303876
NCBI BlastP on this gene
SAMN02745906_3029
66. : CP017761 Marinilactibacillus sp. 15R chromosome Total score: 2.5 Cumulative Blast bit score: 797
ribonuclease P protein component
Accession: API88312
Location: 592341-592682
NCBI BlastP on this gene
Accession: API88312
Location: 592341-592682
NCBI BlastP on this gene
BKP56_02885
BKP56_02890
BKP56_02895
BKP56_02900
PTS maltose transporter subunit IIBC
Accession: API88315
Location: 596392-597849
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: API88315
Location: 596392-597849
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
BKP56_02905
alpha,alpha-phosphotrehalase
Accession: API88316
Location: 598034-599686
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 489
Sequence coverage: 99 %
E-value: 9e-164
NCBI BlastP on this gene
Accession: API88316
Location: 598034-599686
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 489
Sequence coverage: 99 %
E-value: 9e-164
NCBI BlastP on this gene
BKP56_02910
BKP56_02915
BKP56_02920
BKP56_02925
BKP56_02930
BKP56_02935
67. : CP022464 Enterocloster bolteae strain ATCC BAA-613 chromosome Total score: 2.5 Cumulative Blast bit score: 796
CGC65_03455
CGC65_03460
treR
PTS beta-glucoside transporter subunit EIIBCA
Accession: ASN93805
Location: 716811-718754
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 1e-91
NCBI BlastP on this gene
Accession: ASN93805
Location: 716811-718754
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 1e-91
NCBI BlastP on this gene
CGC65_03470
alpha,alpha-phosphotrehalase
Accession: ASN93806
Location: 718820-720466
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 491
Sequence coverage: 99 %
E-value: 2e-164
NCBI BlastP on this gene
Accession: ASN93806
Location: 718820-720466
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 491
Sequence coverage: 99 %
E-value: 2e-164
NCBI BlastP on this gene
treC
hybrid sensor histidine kinase/response regulator
Accession: ASN93807
Location: 720566-723400
NCBI BlastP on this gene
Accession: ASN93807
Location: 720566-723400
NCBI BlastP on this gene
CGC65_03480
glf
68. : CP002109 [Clostridium] saccharolyticum WM1 chromosome Total score: 2.5 Cumulative Blast bit score: 793
Closa_3364
metal dependent phosphohydrolase
Accession: ADL05890
Location: 3626170-3627501
NCBI BlastP on this gene
Accession: ADL05890
Location: 3626170-3627501
NCBI BlastP on this gene
Closa_3363
transcriptional regulator, GntR family
Accession: ADL05889
Location: 3625204-3625917
NCBI BlastP on this gene
Accession: ADL05889
Location: 3625204-3625917
NCBI BlastP on this gene
Closa_3362
PTS system, trehalose-specific IIBC subunit
Accession: ADL05888
Location: 3623600-3625009
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 295
Sequence coverage: 91 %
E-value: 5e-90
NCBI BlastP on this gene
Accession: ADL05888
Location: 3623600-3625009
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 295
Sequence coverage: 91 %
E-value: 5e-90
NCBI BlastP on this gene
Closa_3361
alpha,alpha-phosphotrehalase
Accession: ADL05887
Location: 3621942-3623600
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 498
Sequence coverage: 95 %
E-value: 5e-167
NCBI BlastP on this gene
Accession: ADL05887
Location: 3621942-3623600
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 498
Sequence coverage: 95 %
E-value: 5e-167
NCBI BlastP on this gene
Closa_3360
protein of unknown function DUF204
Accession: ADL05886
Location: 3621382-3621945
NCBI BlastP on this gene
Accession: ADL05886
Location: 3621382-3621945
NCBI BlastP on this gene
Closa_3359
protein of unknown function DUF901
Accession: ADL05885
Location: 3620798-3621319
NCBI BlastP on this gene
Accession: ADL05885
Location: 3620798-3621319
NCBI BlastP on this gene
Closa_3358
Closa_3357
Closa_3356
69. : HE999757 Carnobacterium maltaromaticum LMA28 complete genome. Total score: 2.5 Cumulative Blast bit score: 790
BN424_3617
BN424_3616
BN424_3615
bacterial regulatory s, tetR family protein
Accession: CCO13019
Location: 3612368-3612934
NCBI BlastP on this gene
Accession: CCO13019
Location: 3612368-3612934
NCBI BlastP on this gene
BN424_3614
BN424_3613
PTS system, trehalose-specific IIBC component
Accession: CCO13017
Location: 3609697-3611139
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: CCO13017
Location: 3609697-3611139
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 2e-92
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: CCO13016
Location: 3607905-3609563
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
Accession: CCO13016
Location: 3607905-3609563
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
treC
BN424_3610
BN424_3609
ilvD
acetolactate synthase, large subunit,biosynthetic type
Accession: CCO13012
Location: 3603353-3605026
NCBI BlastP on this gene
Accession: CCO13012
Location: 3603353-3605026
NCBI BlastP on this gene
ilvB
70. : CP029329 Clostridium beijerinckii isolate WB53 chromosome Total score: 2.5 Cumulative Blast bit score: 790
uxuA
DIC82_03555
DIC82_03550
PTS trehalose transporter subunit IIBC
Accession: AWK50203
Location: 878136-879545
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 300
Sequence coverage: 86 %
E-value: 8e-92
NCBI BlastP on this gene
Accession: AWK50203
Location: 878136-879545
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 300
Sequence coverage: 86 %
E-value: 8e-92
NCBI BlastP on this gene
DIC82_03545
alpha,alpha-phosphotrehalase
Accession: AWK50202
Location: 876395-878053
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 490
Sequence coverage: 100 %
E-value: 4e-164
NCBI BlastP on this gene
Accession: AWK50202
Location: 876395-878053
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 490
Sequence coverage: 100 %
E-value: 4e-164
NCBI BlastP on this gene
treC
PTS glucose transporter subunit IIA
Accession: AWK50201
Location: 875817-876317
NCBI BlastP on this gene
Accession: AWK50201
Location: 875817-876317
NCBI BlastP on this gene
DIC82_03535
treR
DIC82_03525
DIC82_03520
tRNA-specific adenosine deaminase
Accession: AWK50197
Location: 872018-872461
NCBI BlastP on this gene
Accession: AWK50197
Location: 872018-872461
NCBI BlastP on this gene
DIC82_03515
71. : CP016844 Carnobacterium maltaromaticum strain TMW 2.1581 chromosome Total score: 2.5 Cumulative Blast bit score: 790
BFC23_13565
DhaKLM operon coactivator DhaQ
Accession: BFC23_13560
Location: 2884066-2885054
NCBI BlastP on this gene
Accession: BFC23_13560
Location: 2884066-2885054
NCBI BlastP on this gene
BFC23_13560
dihydroxyacetone kinase transcriptional activator DhaS
Accession: AOA03467
Location: 2883173-2883739
NCBI BlastP on this gene
Accession: AOA03467
Location: 2883173-2883739
NCBI BlastP on this gene
BFC23_13555
BFC23_13550
PTS maltose transporter subunit IIBC
Accession: AOA03465
Location: 2880501-2881943
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 3e-92
NCBI BlastP on this gene
Accession: AOA03465
Location: 2880501-2881943
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 302
Sequence coverage: 95 %
E-value: 3e-92
NCBI BlastP on this gene
BFC23_13545
alpha,alpha-phosphotrehalase
Accession: AOA03464
Location: 2878709-2880367
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
Accession: AOA03464
Location: 2878709-2880367
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 488
Sequence coverage: 99 %
E-value: 2e-163
NCBI BlastP on this gene
BFC23_13540
BFC23_13535
acetolactate synthase, large subunit, biosynthetic type
Accession: AOA03462
Location: 2874169-2875860
NCBI BlastP on this gene
Accession: AOA03462
Location: 2874169-2875860
NCBI BlastP on this gene
BFC23_13530
72. : CP015399 Lachnoclostridium sp. YL32 Total score: 2.5 Cumulative Blast bit score: 788
A4V08_13160
A4V08_13155
A4V08_13150
A4V08_13145
PTS trehalose transporter subunit IIBC
Accession: ANU46605
Location: 2891840-2893231
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 8e-94
NCBI BlastP on this gene
Accession: ANU46605
Location: 2891840-2893231
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 85 %
E-value: 8e-94
NCBI BlastP on this gene
A4V08_13140
alpha,alpha-phosphotrehalase
Accession: ANU46604
Location: 2890156-2891802
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 483
Sequence coverage: 99 %
E-value: 2e-161
NCBI BlastP on this gene
Accession: ANU46604
Location: 2890156-2891802
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 483
Sequence coverage: 99 %
E-value: 2e-161
NCBI BlastP on this gene
A4V08_13135
A4V08_13130
A4V08_13125
A4V08_13120
A4V08_13115
lipopolysaccharide biosynthesis protein
Accession: ANU46599
Location: 2884805-2886256
NCBI BlastP on this gene
Accession: ANU46599
Location: 2884805-2886256
NCBI BlastP on this gene
A4V08_13110
73. : CP050964 Enterocloster clostridioformis strain FDAARGOS_739 chromosome. Total score: 2.5 Cumulative Blast bit score: 784
FOC47_19250
FOC47_19255
treR
PTS system trehalose-specific EIIBC component
Accession: QIX92484
Location: 3854983-3856434
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 301
Sequence coverage: 85 %
E-value: 7e-92
NCBI BlastP on this gene
Accession: QIX92484
Location: 3854983-3856434
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 301
Sequence coverage: 85 %
E-value: 7e-92
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: QIX94111
Location: 3856629-3858266
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 484
Sequence coverage: 99 %
E-value: 5e-162
NCBI BlastP on this gene
Accession: QIX94111
Location: 3856629-3858266
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 484
Sequence coverage: 99 %
E-value: 5e-162
NCBI BlastP on this gene
treC
FOC47_19275
glf
FOC47_19285
glycosyltransferase family 2 protein
Accession: QIX94112
Location: 3860916-3861971
NCBI BlastP on this gene
Accession: QIX94112
Location: 3860916-3861971
NCBI BlastP on this gene
FOC47_19290
lipopolysaccharide biosynthesis protein
Accession: QIX92488
Location: 3862049-3863500
NCBI BlastP on this gene
Accession: QIX92488
Location: 3862049-3863500
NCBI BlastP on this gene
FOC47_19295
74. : CP008855 Bacillus sp. X1(2014) Total score: 2.5 Cumulative Blast bit score: 783
heme ABC transporter ATP-binding protein
Accession: AIM15027
Location: 157302-158855
NCBI BlastP on this gene
Accession: AIM15027
Location: 157302-158855
NCBI BlastP on this gene
HW35_00885
HW35_00880
HW35_00875
FMN-dependent NADH-azoreductase
Accession: AIM15024
Location: 154320-154955
NCBI BlastP on this gene
Accession: AIM15024
Location: 154320-154955
NCBI BlastP on this gene
HW35_00865
PTS maltose transporter subunit IIBC
Accession: AIM15023
Location: 152158-153582
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 8e-93
NCBI BlastP on this gene
Accession: AIM15023
Location: 152158-153582
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 8e-93
NCBI BlastP on this gene
HW35_00860
trehalose-6-phosphate hydrolase
Accession: AIM15022
Location: 150425-152092
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 3e-160
NCBI BlastP on this gene
Accession: AIM15022
Location: 150425-152092
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 3e-160
NCBI BlastP on this gene
HW35_00855
trehalose operon transcriptional repressor
Accession: AIM15021
Location: 149695-150411
NCBI BlastP on this gene
Accession: AIM15021
Location: 149695-150411
NCBI BlastP on this gene
HW35_00850
HW35_00845
HW35_00840
HW35_00835
75. : CP049740 Jeotgalibaca arthritidis strain CECT 9157 chromosome Total score: 2.5 Cumulative Blast bit score: 782
flagellar hook-basal body protein
Accession: QII81985
Location: 1096113-1096775
NCBI BlastP on this gene
Accession: QII81985
Location: 1096113-1096775
NCBI BlastP on this gene
G7057_05650
ATP-binding cassette domain-containing protein
Accession: QII81986
Location: 1096791-1097516
NCBI BlastP on this gene
Accession: QII81986
Location: 1096791-1097516
NCBI BlastP on this gene
G7057_05655
carbamoyl phosphate synthase-like protein
Accession: QII81987
Location: 1097546-1098859
NCBI BlastP on this gene
Accession: QII81987
Location: 1097546-1098859
NCBI BlastP on this gene
G7057_05660
NERD domain-containing protein
Accession: QII81988
Location: 1098978-1099847
NCBI BlastP on this gene
Accession: QII81988
Location: 1098978-1099847
NCBI BlastP on this gene
G7057_05665
PTS system trehalose-specific EIIBC component
Location: 1100100-1101614
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 317
Sequence coverage: 95 %
E-value: 5e-96
Location: 1100100-1101614
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 317
Sequence coverage: 95 %
E-value: 5e-96
treP
alpha,alpha-phosphotrehalase
Accession: QII81989
Location: 1101689-1103341
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 2e-154
NCBI BlastP on this gene
Accession: QII81989
Location: 1101689-1103341
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 465
Sequence coverage: 99 %
E-value: 2e-154
NCBI BlastP on this gene
treC
G7057_05680
G7057_05685
GerMN domain-containing protein
Accession: QII81992
Location: 1104621-1105727
NCBI BlastP on this gene
Accession: QII81992
Location: 1104621-1105727
NCBI BlastP on this gene
G7057_05690
YeiH family putative sulfate export transporter
Accession: QII81993
Location: 1105921-1106940
NCBI BlastP on this gene
Accession: QII81993
Location: 1105921-1106940
NCBI BlastP on this gene
G7057_05695
LysR family transcriptional regulator
Accession: QII81994
Location: 1107083-1107952
NCBI BlastP on this gene
Accession: QII81994
Location: 1107083-1107952
NCBI BlastP on this gene
G7057_05700
76. : CP039727 Bacillus sp. S3 chromosome Total score: 2.5 Cumulative Blast bit score: 776
inositol phosphorylceramide synthase
Accession: FAY30_12760
Location: 2665098-2665745
NCBI BlastP on this gene
Accession: FAY30_12760
Location: 2665098-2665745
NCBI BlastP on this gene
FAY30_12760
FAY30_12755
thiol-disulfide oxidoreductase DCC family protein
Accession: QCJ42708
Location: 2663323-2663715
NCBI BlastP on this gene
Accession: QCJ42708
Location: 2663323-2663715
NCBI BlastP on this gene
FAY30_12750
LysM peptidoglycan-binding domain-containing protein
Accession: QCJ42707
Location: 2662057-2663247
NCBI BlastP on this gene
Accession: QCJ42707
Location: 2662057-2663247
NCBI BlastP on this gene
FAY30_12745
PTS trehalose transporter subunit IIBC
Accession: QCJ42706
Location: 2659999-2661423
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 7e-93
NCBI BlastP on this gene
Accession: QCJ42706
Location: 2659999-2661423
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 303
Sequence coverage: 86 %
E-value: 7e-93
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QCJ42705
Location: 2658250-2659917
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 473
Sequence coverage: 100 %
E-value: 3e-157
NCBI BlastP on this gene
Accession: QCJ42705
Location: 2658250-2659917
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 473
Sequence coverage: 100 %
E-value: 3e-157
NCBI BlastP on this gene
treC
treR
SOS response-associated peptidase
Accession: QCJ45201
Location: 2656722-2657399
NCBI BlastP on this gene
Accession: QCJ45201
Location: 2656722-2657399
NCBI BlastP on this gene
FAY30_12725
FAY30_12720
FAY30_12715
FAY30_12710
FAY30_12705
FAY30_12700
77. : LS483476 Bacillus lentus strain NCTC4824 genome assembly, chromosome: 1. Total score: 2.5 Cumulative Blast bit score: 772
L-seryl-tRNA(Sec) selenium transferase
Accession: SQI51388
Location: 197648-198751
NCBI BlastP on this gene
Accession: SQI51388
Location: 197648-198751
NCBI BlastP on this gene
selA
4-hydroxy-2-ketovalerate aldolase
Accession: SQI51390
Location: 198752-199507
NCBI BlastP on this gene
Accession: SQI51390
Location: 198752-199507
NCBI BlastP on this gene
NCTC4824_00220
iolC_1
UDP-N-acetylenolpyruvoylglucosamine reductase
Accession: SQI51393
Location: 200757-201674
NCBI BlastP on this gene
Accession: SQI51393
Location: 200757-201674
NCBI BlastP on this gene
murB
PTS system, trehalose-specific IIBC subunit
Accession: SQI51395
Location: 201902-203338
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 302
Sequence coverage: 86 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: SQI51395
Location: 201902-203338
BlastP hit with treP
Percentage identity: 39 %
BlastP bit score: 302
Sequence coverage: 86 %
E-value: 2e-92
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: SQI51396
Location: 203483-205153
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 470
Sequence coverage: 101 %
E-value: 3e-156
NCBI BlastP on this gene
Accession: SQI51396
Location: 203483-205153
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 470
Sequence coverage: 101 %
E-value: 3e-156
NCBI BlastP on this gene
treA
GntR family transcriptional regulator
Accession: SQI51398
Location: 205168-205881
NCBI BlastP on this gene
Accession: SQI51398
Location: 205168-205881
NCBI BlastP on this gene
treR
methyl-accepting chemotaxis sensory transducer with Cache sensor
Accession: SQI51400
Location: 206072-207763
NCBI BlastP on this gene
Accession: SQI51400
Location: 206072-207763
NCBI BlastP on this gene
mcpA_1
NCTC4824_00227
pyruvate formate-lyase-activating enzyme
Accession: SQI51403
Location: 208153-208929
NCBI BlastP on this gene
Accession: SQI51403
Location: 208153-208929
NCBI BlastP on this gene
pflA_1
pflB
78. : CP051128 Bacillus megaterium strain S2 chromosome Total score: 2.5 Cumulative Blast bit score: 762
phosphatase PAP2 family protein
Accession: QIZ08862
Location: 4321141-4321782
NCBI BlastP on this gene
Accession: QIZ08862
Location: 4321141-4321782
NCBI BlastP on this gene
HFZ78_20910
thiol-disulfide oxidoreductase DCC family protein
Accession: QIZ11053
Location: 4322113-4322505
NCBI BlastP on this gene
Accession: QIZ11053
Location: 4322113-4322505
NCBI BlastP on this gene
HFZ78_20915
LysM peptidoglycan-binding domain-containing protein
Accession: QIZ08863
Location: 4322628-4323818
NCBI BlastP on this gene
Accession: QIZ08863
Location: 4322628-4323818
NCBI BlastP on this gene
HFZ78_20920
HFZ78_20925
GNAT family N-acetyltransferase
Accession: QIZ08865
Location: 4324350-4324922
NCBI BlastP on this gene
Accession: QIZ08865
Location: 4324350-4324922
NCBI BlastP on this gene
HFZ78_20930
PTS system trehalose-specific EIIBC component
Accession: QIZ08866
Location: 4325339-4326763
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 301
Sequence coverage: 87 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: QIZ08866
Location: 4325339-4326763
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 301
Sequence coverage: 87 %
E-value: 2e-92
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: QIZ08867
Location: 4326849-4328516
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 99 %
E-value: 7e-153
NCBI BlastP on this gene
Accession: QIZ08867
Location: 4326849-4328516
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 99 %
E-value: 7e-153
NCBI BlastP on this gene
treC
treR
SOS response-associated peptidase
Accession: QIZ08869
Location: 4329384-4330061
NCBI BlastP on this gene
Accession: QIZ08869
Location: 4329384-4330061
NCBI BlastP on this gene
HFZ78_20950
HFZ78_20955
HFZ78_20960
GNAT family N-acetyltransferase
Accession: QIZ08872
Location: 4331312-4331749
NCBI BlastP on this gene
Accession: QIZ08872
Location: 4331312-4331749
NCBI BlastP on this gene
HFZ78_20965
yyaC
79. : CP010796 Carnobacterium sp. CP1 Total score: 2.5 Cumulative Blast bit score: 759
NY10_1745
Alpha-glycerophosphate oxidase
Accession: ALV22342
Location: 1800352-1802187
NCBI BlastP on this gene
Accession: ALV22342
Location: 1800352-1802187
NCBI BlastP on this gene
NY10_1744
NY10_1743
NY10_1742
PTS system, trehalose-specific IIB component
Accession: ALV22339
Location: 1797069-1798529
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 299
Sequence coverage: 96 %
E-value: 3e-91
NCBI BlastP on this gene
Accession: ALV22339
Location: 1797069-1798529
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 299
Sequence coverage: 96 %
E-value: 3e-91
NCBI BlastP on this gene
NY10_1741
Trehalose-6-phosphate hydrolase
Accession: ALV22338
Location: 1795390-1797045
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 99 %
E-value: 3e-152
NCBI BlastP on this gene
Accession: ALV22338
Location: 1795390-1797045
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 99 %
E-value: 3e-152
NCBI BlastP on this gene
NY10_1740
Trehalose operon transcriptional repressor
Accession: ALV22337
Location: 1794021-1794749
NCBI BlastP on this gene
Accession: ALV22337
Location: 1794021-1794749
NCBI BlastP on this gene
NY10_1739
NY10_1738
GTPase and tRNA-U34 5-formylation enzyme TrmE
Accession: ALV22335
Location: 1790645-1792033
NCBI BlastP on this gene
Accession: ALV22335
Location: 1790645-1792033
NCBI BlastP on this gene
NY10_1737
80. : CP045915 Gracilibacillus sp. SCU50 chromosome Total score: 2.5 Cumulative Blast bit score: 758
NADP-specific glutamate dehydrogenase
Accession: QGH36608
Location: 4750566-4751927
NCBI BlastP on this gene
Accession: QGH36608
Location: 4750566-4751927
NCBI BlastP on this gene
GI584_22250
GI584_22245
GI584_22240
PTS trehalose transporter subunit IIBC
Accession: QGH36605
Location: 4746780-4748222
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 298
Sequence coverage: 86 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QGH36605
Location: 4746780-4748222
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 298
Sequence coverage: 86 %
E-value: 6e-91
NCBI BlastP on this gene
treB
alpha,alpha-phosphotrehalase
Accession: QGH36604
Location: 4744676-4746355
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 101 %
E-value: 3e-152
NCBI BlastP on this gene
Accession: QGH36604
Location: 4744676-4746355
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 460
Sequence coverage: 101 %
E-value: 3e-152
NCBI BlastP on this gene
treC
treR
GI584_22220
GI584_22215
methyltransferase domain-containing protein
Accession: QGH36600
Location: 4741875-4742597
NCBI BlastP on this gene
Accession: QGH36600
Location: 4741875-4742597
NCBI BlastP on this gene
GI584_22210
Lrp/AsnC family transcriptional regulator
Accession: QGH36599
Location: 4741108-4741605
NCBI BlastP on this gene
Accession: QGH36599
Location: 4741108-4741605
NCBI BlastP on this gene
GI584_22205
GI584_22200
81. : CP022512 Mesoplasma florum strain MQ3 chromosome Total score: 2.5 Cumulative Blast bit score: 536
CG006_02770
CG006_02765
CG006_02760
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN63883
Location: 599875-601473
NCBI BlastP on this gene
Accession: AVN63883
Location: 599875-601473
NCBI BlastP on this gene
CG006_02755
hypothetical protein
Accession: AVN63882
Location: 598839-599786
BlastP hit with treR
Percentage identity: 33 %
BlastP bit score: 104
Sequence coverage: 85 %
E-value: 3e-22
NCBI BlastP on this gene
Accession: AVN63882
Location: 598839-599786
BlastP hit with treR
Percentage identity: 33 %
BlastP bit score: 104
Sequence coverage: 85 %
E-value: 3e-22
NCBI BlastP on this gene
CG006_02750
PTS sugar transporter subunit IIA
Accession: AVN63881
Location: 597217-598776
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AVN63881
Location: 597217-598776
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
CG006_02745
CG006_02740
CG006_02735
CG006_02730
magnesium-translocating P-type ATPase
Accession: AVN63877
Location: 591349-594009
NCBI BlastP on this gene
Accession: AVN63877
Location: 591349-594009
NCBI BlastP on this gene
mgtA
82. : CP022511 Mesoplasma coleopterae strain BARC 781 chromosome. Total score: 2.5 Cumulative Blast bit score: 535
CG000_02735
CG000_02730
CG000_02725
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN63196
Location: 607126-608724
NCBI BlastP on this gene
Accession: AVN63196
Location: 607126-608724
NCBI BlastP on this gene
CG000_02720
hypothetical protein
Accession: AVN63195
Location: 606090-607037
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVN63195
Location: 606090-607037
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
CG000_02715
PTS sugar transporter subunit IIA
Accession: AVN63194
Location: 604468-606027
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AVN63194
Location: 604468-606027
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 6e-144
NCBI BlastP on this gene
CG000_02710
CG000_02705
CG000_02700
CG000_02695
magnesium-translocating P-type ATPase
Accession: AVN63190
Location: 598589-601249
NCBI BlastP on this gene
Accession: AVN63190
Location: 598589-601249
NCBI BlastP on this gene
mgtA
83. : CP011855 Spiroplasma atrichopogonis strain GNAT3597 Total score: 2.5 Cumulative Blast bit score: 535
SATRI_v1c09280
SATRI_v1c09210
SATRI_v1c09200
SATRI_v1c09190
RNA methyltransferase, TrmH family
Accession: AKM53313
Location: 830808-831599
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
Accession: AKM53313
Location: 830808-831599
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
spoU
GTP-binding protein YsxC
Accession: AKM53312
Location: 830212-830805
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
Accession: AKM53312
Location: 830212-830805
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
engB
SATRI_v1c09160
SATRI_v1c09090
SATRI_v1c09080
prolipoprotein diacylglyceryl transferase
Accession: AKM53308
Location: 824134-825657
NCBI BlastP on this gene
Accession: AKM53308
Location: 824134-825657
NCBI BlastP on this gene
lgt
84. : CP006720 Spiroplasma mirum ATCC 29335 strain SMCA Total score: 2.5 Cumulative Blast bit score: 535
P344_05140
P344_05135
P344_05130
P344_05125
P344_05120
P344_05115
P344_05110
P344_05105
P344_05100
P344_05095
P344_05090
hypothetical protein
Accession: AHI58339
Location: 803561-804352
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
Accession: AHI58339
Location: 803561-804352
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 243
Sequence coverage: 99 %
E-value: 6e-76
NCBI BlastP on this gene
P344_05085
hypothetical protein
Accession: AHI58338
Location: 802965-803558
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
Accession: AHI58338
Location: 802965-803558
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 292
Sequence coverage: 98 %
E-value: 5e-97
NCBI BlastP on this gene
P344_05080
P344_05075
P344_05070
P344_05065
P344_05060
P344_05055
P344_05050
P344_05045
P344_05040
P344_05035
P344_05030
85. : AE017263 Mesoplasma florum L1 complete genome. Total score: 2.5 Cumulative Blast bit score: 535
Mfl505
Mfl504
cof-like hydrolase, HAD-superfamily
Accession: AAT75861
Location: 590775-591596
NCBI BlastP on this gene
Accession: AAT75861
Location: 590775-591596
NCBI BlastP on this gene
Mfl503
Mfl502
trehalose operon repressor, repressor of treA,B,C
Accession: AAT75859
Location: 588125-589072
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 103
Sequence coverage: 85 %
E-value: 8e-22
NCBI BlastP on this gene
Accession: AAT75859
Location: 588125-589072
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 103
Sequence coverage: 85 %
E-value: 8e-22
NCBI BlastP on this gene
Mfl501
trehalose PTS system IIABC component
Accession: AAT75858
Location: 586503-588062
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AAT75858
Location: 586503-588062
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Mfl500
trehalose-6-phosphate hydrolase
Accession: AAT75857
Location: 584838-586457
NCBI BlastP on this gene
Accession: AAT75857
Location: 584838-586457
NCBI BlastP on this gene
Mfl499
Mfl498
Mfl497
Mfl496
86. : CP022514 Mesoplasma florum strain BARC 787 chromosome Total score: 2.5 Cumulative Blast bit score: 534
CG002_02570
CG002_02565
CG002_02560
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN65229
Location: 547905-549503
NCBI BlastP on this gene
Accession: AVN65229
Location: 547905-549503
NCBI BlastP on this gene
CG002_02555
hypothetical protein
Accession: AVN65228
Location: 546869-547816
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AVN65228
Location: 546869-547816
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
CG002_02550
PTS sugar transporter subunit IIA
Accession: AVN65227
Location: 545247-546806
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AVN65227
Location: 545247-546806
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
CG002_02545
CG002_02540
CG002_02535
CG002_02530
magnesium-translocating P-type ATPase
Accession: AVN65223
Location: 539381-542038
NCBI BlastP on this gene
Accession: AVN65223
Location: 539381-542038
NCBI BlastP on this gene
mgtA
87. : CP022510 Mesoplasma coleopterae strain BARC 786 chromosome. Total score: 2.5 Cumulative Blast bit score: 534
CG001_02635
CG001_02630
CG001_02625
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN62520
Location: 573794-575392
NCBI BlastP on this gene
Accession: AVN62520
Location: 573794-575392
NCBI BlastP on this gene
CG001_02620
hypothetical protein
Accession: AVN62519
Location: 572758-573705
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
Accession: AVN62519
Location: 572758-573705
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 99
Sequence coverage: 85 %
E-value: 1e-20
NCBI BlastP on this gene
CG001_02615
PTS sugar transporter subunit IIA
Accession: AVN62518
Location: 571136-572695
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 435
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AVN62518
Location: 571136-572695
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 435
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
CG001_02610
CG001_02605
CG001_02600
CG001_02595
magnesium-translocating P-type ATPase
Accession: AVN62514
Location: 565257-567917
NCBI BlastP on this gene
Accession: AVN62514
Location: 565257-567917
NCBI BlastP on this gene
mgtA
88. : CP022505 Mesoplasma florum strain W23 chromosome Total score: 2.5 Cumulative Blast bit score: 534
CG009_02710
CG009_02705
CG009_02700
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AVN59115
Location: 573220-574818
NCBI BlastP on this gene
Accession: AVN59115
Location: 573220-574818
NCBI BlastP on this gene
CG009_02695
hypothetical protein
Accession: AVN59114
Location: 572184-573131
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AVN59114
Location: 572184-573131
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CG009_02690
PTS sugar transporter subunit IIA
Accession: AVN59113
Location: 570562-572121
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
Accession: AVN59113
Location: 570562-572121
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 2e-142
NCBI BlastP on this gene
CG009_02685
CG009_02680
CG009_02675
CG009_02670
CG009_02665
PTS sugar transporter subunit IIA
Accession: AVN59108
Location: 564863-566590
NCBI BlastP on this gene
Accession: AVN59108
Location: 564863-566590
NCBI BlastP on this gene
CG009_02660
89. : CP023736 Mesoplasma florum strain PPA1 chromosome Total score: 2.5 Cumulative Blast bit score: 533
CQZ70_02780
CQZ70_02775
CQZ70_02770
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: ATI74154
Location: 613071-614669
NCBI BlastP on this gene
Accession: ATI74154
Location: 613071-614669
NCBI BlastP on this gene
CQZ70_02765
hypothetical protein
Accession: ATI74153
Location: 612035-612982
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: ATI74153
Location: 612035-612982
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CQZ70_02760
PTS sugar transporter subunit IIA
Accession: ATI74152
Location: 610413-611972
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: ATI74152
Location: 610413-611972
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
CQZ70_02755
CQZ70_02750
CQZ70_02745
CQZ70_02740
magnesium-translocating P-type ATPase
Accession: ATI74148
Location: 604546-607203
NCBI BlastP on this gene
Accession: ATI74148
Location: 604546-607203
NCBI BlastP on this gene
mgtA
90. : CP022513 Mesoplasma florum strain CnuA-2 chromosome Total score: 2.5 Cumulative Blast bit score: 533
CG003_02765
CG003_02760
CG003_02755
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AVN64567
Location: 608119-609717
NCBI BlastP on this gene
Accession: AVN64567
Location: 608119-609717
NCBI BlastP on this gene
CG003_02750
LacI family transcriptional regulator
Accession: AVN64566
Location: 607083-608030
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 101
Sequence coverage: 85 %
E-value: 3e-21
NCBI BlastP on this gene
Accession: AVN64566
Location: 607083-608030
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 101
Sequence coverage: 85 %
E-value: 3e-21
NCBI BlastP on this gene
CG003_02745
PTS sugar transporter subunit IIA
Accession: AVN64565
Location: 605461-607020
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 4e-142
NCBI BlastP on this gene
Accession: AVN64565
Location: 605461-607020
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 4e-142
NCBI BlastP on this gene
CG003_02740
CG003_02735
CG003_02730
CG003_02725
magnesium-translocating P-type ATPase
Accession: AVN64561
Location: 599592-602252
NCBI BlastP on this gene
Accession: AVN64561
Location: 599592-602252
NCBI BlastP on this gene
mgtA
91. : CP022508 Mesoplasma florum strain MouA-2 chromosome Total score: 2.5 Cumulative Blast bit score: 533
CG005_02640
CG005_02635
CG005_02630
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN61167
Location: 578742-580340
NCBI BlastP on this gene
Accession: AVN61167
Location: 578742-580340
NCBI BlastP on this gene
CG005_02625
hypothetical protein
Accession: AVN61166
Location: 577706-578653
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AVN61166
Location: 577706-578653
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CG005_02620
PTS sugar transporter subunit IIA
Accession: AVN61165
Location: 576084-577643
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AVN61165
Location: 576084-577643
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
CG005_02615
CG005_02610
CG005_02605
CG005_02600
magnesium-translocating P-type ATPase
Accession: AVN61161
Location: 570219-572876
NCBI BlastP on this gene
Accession: AVN61161
Location: 570219-572876
NCBI BlastP on this gene
mgtA
92. : CP022506 Mesoplasma florum strain W20 chromosome. Total score: 2.5 Cumulative Blast bit score: 533
CG008_02805
CG008_02800
CG008_02795
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN59810
Location: 604263-605861
NCBI BlastP on this gene
Accession: AVN59810
Location: 604263-605861
NCBI BlastP on this gene
CG008_02790
LacI family transcriptional regulator
Accession: AVN59809
Location: 603227-604174
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AVN59809
Location: 603227-604174
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
CG008_02785
PTS sugar transporter subunit IIA
Accession: AVN59808
Location: 601605-603164
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AVN59808
Location: 601605-603164
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
CG008_02780
CG008_02775
CG008_02770
CG008_02765
magnesium-translocating P-type ATPase
Accession: AVN59804
Location: 595739-598396
NCBI BlastP on this gene
Accession: AVN59804
Location: 595739-598396
NCBI BlastP on this gene
mgtA
93. : CP022432 Mesoplasma florum strain W12 chromosome. Total score: 2.5 Cumulative Blast bit score: 533
MflW12_5340
MflW12_5330
MflW12_5320
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AVN65936
Location: 602812-604410
NCBI BlastP on this gene
Accession: AVN65936
Location: 602812-604410
NCBI BlastP on this gene
MflW12_5310
Sucrose operon repressor ScrR, LacI family
Accession: AVN65935
Location: 601776-602723
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AVN65935
Location: 601776-602723
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
MflW12_5300
PTS system trehalose-specific IIB component, PTS system trehalose-specific IIC component
Accession: AVN65934
Location: 600154-601713
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AVN65934
Location: 600154-601713
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
MflW12_5290
Trehalose-6-phosphate hydrolase
Accession: AVN65933
Location: 598500-600119
NCBI BlastP on this gene
Accession: AVN65933
Location: 598500-600119
NCBI BlastP on this gene
MflW12_5280
MflW12_5270
Mg(2+) transport ATPase, P-type
Accession: AVN65931
Location: 594288-596945
NCBI BlastP on this gene
Accession: AVN65931
Location: 594288-596945
NCBI BlastP on this gene
MflW12_5260
94. : CP006778 Mesoplasma florum W37 Total score: 2.5 Cumulative Blast bit score: 533
mflW37_5340
mflW37_5330
cof-like hydrolase, HAD-superfamily
Accession: AGY41599
Location: 601139-601960
NCBI BlastP on this gene
Accession: AGY41599
Location: 601139-601960
NCBI BlastP on this gene
mflW37_5320
2,3-bisphosphoglycerate-independent phosphoglycerate mutase
Accession: AGY41598
Location: 599525-601123
NCBI BlastP on this gene
Accession: AGY41598
Location: 599525-601123
NCBI BlastP on this gene
mflW37_5310
trehalose operon repressor, repressor of treA,B,C
Accession: AGY41597
Location: 598489-599436
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
Accession: AGY41597
Location: 598489-599436
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 1e-21
NCBI BlastP on this gene
mflW37_5300
PTS system, trehalose-specific IIB component PTS system, trehalose-specific IIC component
Accession: AGY41596
Location: 596867-598426
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
Accession: AGY41596
Location: 596867-598426
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 6e-142
NCBI BlastP on this gene
mflW37_5290
Trehalose-6-phosphate hydrolase
Accession: AGY41595
Location: 595213-596832
NCBI BlastP on this gene
Accession: AGY41595
Location: 595213-596832
NCBI BlastP on this gene
mflW37_5280
mflW37_5270
Mg(2+) transport ATPase, P-type
Accession: AGY41593
Location: 591001-593658
NCBI BlastP on this gene
Accession: AGY41593
Location: 591001-593658
NCBI BlastP on this gene
mflW37_5260
95. : CP023735 Mesoplasma florum strain GF1 chromosome Total score: 2.5 Cumulative Blast bit score: 530
CQZ69_02785
CQZ69_02780
CQZ69_02775
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: ATI73468
Location: 612720-614318
NCBI BlastP on this gene
Accession: ATI73468
Location: 612720-614318
NCBI BlastP on this gene
CQZ69_02770
hypothetical protein
Accession: ATI73467
Location: 611684-612631
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: ATI73467
Location: 611684-612631
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CQZ69_02765
PTS sugar transporter subunit IIA
Accession: ATI73466
Location: 610062-611621
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
Accession: ATI73466
Location: 610062-611621
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
CQZ69_02760
CQZ69_02755
CQZ69_02750
CQZ69_02745
CQZ69_02740
magnesium-translocating P-type ATPase
Accession: ATI73461
Location: 603704-606364
NCBI BlastP on this gene
Accession: ATI73461
Location: 603704-606364
NCBI BlastP on this gene
mgtA
96. : CP022509 Mesoplasma florum strain GF chromosome. Total score: 2.5 Cumulative Blast bit score: 530
CG004_02770
CG004_02765
CG004_02760
phosphoglycerate mutase (2,3-diphosphoglycerate-independent)
Accession: AVN61861
Location: 597872-599470
NCBI BlastP on this gene
Accession: AVN61861
Location: 597872-599470
NCBI BlastP on this gene
CG004_02755
hypothetical protein
Accession: AVN61860
Location: 596836-597783
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
Accession: AVN61860
Location: 596836-597783
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 102
Sequence coverage: 85 %
E-value: 2e-21
NCBI BlastP on this gene
CG004_02750
PTS sugar transporter subunit IIA
Accession: AVN61859
Location: 595214-596773
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
Accession: AVN61859
Location: 595214-596773
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-140
NCBI BlastP on this gene
CG004_02745
CG004_02740
CG004_02735
CG004_02730
CG004_02725
magnesium-translocating P-type ATPase
Accession: AVN61854
Location: 588856-591516
NCBI BlastP on this gene
Accession: AVN61854
Location: 588856-591516
NCBI BlastP on this gene
mgtA
97. : CP011856 Spiroplasma eriocheiris strain DSM 21848 Total score: 2.5 Cumulative Blast bit score: 530
RpiR family transcriptional regulator
Accession: AKM54451
Location: 970715-971524
NCBI BlastP on this gene
Accession: AKM54451
Location: 970715-971524
NCBI BlastP on this gene
SERIO_v1c08910
patB
pepA
SERIO_v1c08880
SERIO_v1c08870
SERIO_v1c08860
RNA methyltransferase, TrmH family
Accession: AKM54445
Location: 965424-966215
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
Accession: AKM54445
Location: 965424-966215
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
spoU
GTP-binding protein YsxC
Accession: AKM54444
Location: 964828-965421
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
Accession: AKM54444
Location: 964828-965421
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
engB
focA
pflD
SERIO_v1c08810
prolipoprotein diacylglyceryl transferase
Accession: AKM54440
Location: 958610-960130
NCBI BlastP on this gene
Accession: AKM54440
Location: 958610-960130
NCBI BlastP on this gene
lgt
98. : CP001973 Spiroplasma eriocheiris CCTCC M 207170 Total score: 2.5 Cumulative Blast bit score: 530
putative phosphosugar-binding transcriptional regulator
Accession: AHF58009
Location: 969776-970585
NCBI BlastP on this gene
Accession: AHF58009
Location: 969776-970585
NCBI BlastP on this gene
SPE_0889
SPE_0888
pepB1
SPE_0886
SPE_0885
SPE_0884
rRNA methylase
Accession: AHF58003
Location: 964485-965276
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
Accession: AHF58003
Location: 964485-965276
BlastP hit with AAP55640.1
Percentage identity: 51 %
BlastP bit score: 240
Sequence coverage: 100 %
E-value: 7e-75
NCBI BlastP on this gene
spoU
putative GTP binding protein
Accession: AHF58002
Location: 963889-964482
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
Accession: AHF58002
Location: 963889-964482
BlastP hit with AAP55641.1
Percentage identity: 72 %
BlastP bit score: 290
Sequence coverage: 98 %
E-value: 2e-96
NCBI BlastP on this gene
engB
focA
pflB
SPE_0879
prolipoprotein diacylglyceryl transferase
Accession: AHF57998
Location: 957671-959191
NCBI BlastP on this gene
Accession: AHF57998
Location: 957671-959191
NCBI BlastP on this gene
lgt1
99. : CP024969 Mesoplasma tabanidae strain BARC 857 chromosome Total score: 2.5 Cumulative Blast bit score: 525
MTABA_v1c05510
tpiA
MTABA_v1c05490
gpm
trehalose operon repressor, repressor of treA,B,C
Accession: ATZ21741
Location: 633782-634726
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 84 %
E-value: 1e-18
NCBI BlastP on this gene
Accession: ATZ21741
Location: 633782-634726
BlastP hit with treR
Percentage identity: 32 %
BlastP bit score: 94
Sequence coverage: 84 %
E-value: 1e-18
NCBI BlastP on this gene
MTABA_v1c05470
PTS system, trehalose-specific IIBC component
Accession: ATZ21740
Location: 632160-633719
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 5e-142
NCBI BlastP on this gene
Accession: ATZ21740
Location: 632160-633719
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 5e-142
NCBI BlastP on this gene
treB
MTABA_v1c05450
trehalose-6-phosphate hydrolase
Accession: ATZ21738
Location: 630022-631641
NCBI BlastP on this gene
Accession: ATZ21738
Location: 630022-631641
NCBI BlastP on this gene
treC
MTABA_v1c05430
glk
Mg(2+) transport ATPase, P-type
Accession: ATZ21735
Location: 625788-628448
NCBI BlastP on this gene
Accession: ATZ21735
Location: 625788-628448
NCBI BlastP on this gene
mgtA
100. : CP005077 Spiroplasma chrysopicola DF-1 Total score: 2.5 Cumulative Blast bit score: 502
SCHRY_v1c03080
hemN
ytjP
SCHRY_v1c03110
RNA methyltransferase, TrmH family
Accession: AGM24897
Location: 339535-340335
BlastP hit with AAP55640.1
Percentage identity: 49 %
BlastP bit score: 214
Sequence coverage: 100 %
E-value: 2e-64
NCBI BlastP on this gene
Accession: AGM24897
Location: 339535-340335
BlastP hit with AAP55640.1
Percentage identity: 49 %
BlastP bit score: 214
Sequence coverage: 100 %
E-value: 2e-64
NCBI BlastP on this gene
spoU
GTP-binding protein YsxC
Accession: AGM24898
Location: 340328-340924
BlastP hit with AAP55641.1
Percentage identity: 70 %
BlastP bit score: 288
Sequence coverage: 98 %
E-value: 2e-95
NCBI BlastP on this gene
Accession: AGM24898
Location: 340328-340924
BlastP hit with AAP55641.1
Percentage identity: 70 %
BlastP bit score: 288
Sequence coverage: 98 %
E-value: 2e-95
NCBI BlastP on this gene
engB
prolipoprotein diacylglyceryl transferase
Accession: AGM24899
Location: 341007-342851
NCBI BlastP on this gene
Accession: AGM24899
Location: 341007-342851
NCBI BlastP on this gene
SCHRY_v1c03140
SCHRY_v1c03150
SCHRY_v1c03160
SCHRY_v1c03170


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.