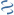MultiGeneBlast hits

Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP016843 : Carnobacterium divergens strain TMW 2.1579 chromosome Total score: 1.0 Cumulative Blast bit score: 478
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
gldA
BFC22_01070
BFC22_01065
dihydroxyacetone kinase transcriptional activator DhaS
Accession: ANZ98767
Location: 233076-233654
NCBI BlastP on this gene
Accession: ANZ98767
Location: 233076-233654
NCBI BlastP on this gene
BFC22_01060
PTS maltose transporter subunit IIBC
Accession: ANZ98766
Location: 231412-232869
NCBI BlastP on this gene
Accession: ANZ98766
Location: 231412-232869
NCBI BlastP on this gene
BFC22_01055
alpha,alpha-phosphotrehalase
Accession: ANZ98765
Location: 229711-231381
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 478
Sequence coverage: 99 %
E-value: 4e-159
NCBI BlastP on this gene
Accession: ANZ98765
Location: 229711-231381
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 478
Sequence coverage: 99 %
E-value: 4e-159
NCBI BlastP on this gene
BFC22_01050
BFC22_01045
BFC22_01040
BFC22_01035
tRNA uridine(34) 5-carboxymethylaminomethyl synthesis GTPase MnmE
Accession: ANZ98761
Location: 225607-227004
NCBI BlastP on this gene
Accession: ANZ98761
Location: 225607-227004
NCBI BlastP on this gene
BFC22_01030
tRNA uridine(34) 5-carboxymethylaminomethyl synthesis enzyme MnmG
Accession: ANZ98760
Location: 223696-225591
NCBI BlastP on this gene
Accession: ANZ98760
Location: 223696-225591
NCBI BlastP on this gene
BFC22_01025
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP024848 : Oceanobacillus sp. 160 chromosome Total score: 1.0 Cumulative Blast bit score: 477
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
typA
CUC15_02980
PTS trehalose transporter subunit IIBC
Accession: AXI11077
Location: 576415-578283
NCBI BlastP on this gene
Accession: AXI11077
Location: 576415-578283
NCBI BlastP on this gene
CUC15_02985
alpha,alpha-phosphotrehalase
Accession: AXI08008
Location: 578519-580183
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 6e-159
NCBI BlastP on this gene
Accession: AXI08008
Location: 578519-580183
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 6e-159
NCBI BlastP on this gene
treC
treR
CUC15_03000
CUC15_03005
CUC15_03010
CUC15_03015
bacitracin ABC transporter ATP-binding protein
Accession: AXI08014
Location: 583655-584578
NCBI BlastP on this gene
Accession: AXI08014
Location: 583655-584578
NCBI BlastP on this gene
CUC15_03020
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP048837 : [Clostridium] innocuum strain LC-LUMC-CI-001 chromosome Total score: 1.0 Cumulative Blast bit score: 476
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
G4D55_00695
G4D55_00690
O-acetyl-ADP-ribose deacetylase
Accession: QIX07678
Location: 144818-145324
NCBI BlastP on this gene
Accession: QIX07678
Location: 144818-145324
NCBI BlastP on this gene
G4D55_00685
treR
PTS system trehalose-specific EIIBC component
Accession: QIX07676
Location: 141967-143850
NCBI BlastP on this gene
Accession: QIX07676
Location: 141967-143850
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: QIX07675
Location: 140308-141957
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 476
Sequence coverage: 100 %
E-value: 1e-158
NCBI BlastP on this gene
Accession: QIX07675
Location: 140308-141957
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 476
Sequence coverage: 100 %
E-value: 1e-158
NCBI BlastP on this gene
treC
DUF1622 domain-containing protein
Accession: QIX07674
Location: 139317-139673
NCBI BlastP on this gene
Accession: QIX07674
Location: 139317-139673
NCBI BlastP on this gene
G4D55_00665
ribose-phosphate pyrophosphokinase
Accession: QIX07673
Location: 138341-139294
NCBI BlastP on this gene
Accession: QIX07673
Location: 138341-139294
NCBI BlastP on this gene
G4D55_00660
bifunctional UDP-N-acetylglucosamine
Accession: QIX07672
Location: 136962-138317
NCBI BlastP on this gene
Accession: QIX07672
Location: 136962-138317
NCBI BlastP on this gene
glmU
Cof-type HAD-IIB family hydrolase
Accession: QIX07671
Location: 135908-136684
NCBI BlastP on this gene
Accession: QIX07671
Location: 135908-136684
NCBI BlastP on this gene
G4D55_00650
HAMP domain-containing histidine kinase
Accession: QIX07670
Location: 134347-135690
NCBI BlastP on this gene
Accession: QIX07670
Location: 134347-135690
NCBI BlastP on this gene
G4D55_00645
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
AP022323 : Aeribacillus pallidus PI8 DNA Total score: 1.0 Cumulative Blast bit score: 474
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
APP_11320
yxxE
APP_11340
PTS system glucose-specific EIIA component
Accession: BBU38843
Location: 1193787-1194272
NCBI BlastP on this gene
Accession: BBU38843
Location: 1193787-1194272
NCBI BlastP on this gene
crr_1
PTS system trehalose-specific EIIBC component
Accession: BBU38844
Location: 1194308-1195723
NCBI BlastP on this gene
Accession: BBU38844
Location: 1194308-1195723
NCBI BlastP on this gene
treP
trehalose-6-phosphate hydrolase
Accession: BBU38845
Location: 1195841-1197526
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 474
Sequence coverage: 101 %
E-value: 1e-157
NCBI BlastP on this gene
Accession: BBU38845
Location: 1195841-1197526
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 474
Sequence coverage: 101 %
E-value: 1e-157
NCBI BlastP on this gene
treA
treR
APP_11390
APP_11400
APP_11410
APP_11420
APP_11430
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP017703 : Aeribacillus pallidus strain KCTC3564 Total score: 1.0 Cumulative Blast bit score: 473
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
AP3564_10570
AP3564_10575
PTS glucose transporter subunit IIA
Accession: ASS90606
Location: 2168983-2169468
NCBI BlastP on this gene
Accession: ASS90606
Location: 2168983-2169468
NCBI BlastP on this gene
AP3564_10580
trehalose permease IIC protein
Accession: ASS90607
Location: 2169503-2170918
NCBI BlastP on this gene
Accession: ASS90607
Location: 2169503-2170918
NCBI BlastP on this gene
AP3564_10585
alpha,alpha-phosphotrehalase
Accession: ASS90608
Location: 2171036-2172721
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 473
Sequence coverage: 101 %
E-value: 3e-157
NCBI BlastP on this gene
Accession: ASS90608
Location: 2171036-2172721
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 473
Sequence coverage: 101 %
E-value: 3e-157
NCBI BlastP on this gene
AP3564_10590
AP3564_10595
group II intron reverse transcriptase/maturase
Accession: ASS92364
Location: 2174544-2175803
NCBI BlastP on this gene
Accession: ASS92364
Location: 2174544-2175803
NCBI BlastP on this gene
AP3564_10600
AP3564_10605
AP3564_10610
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP025518 : Bacillaceae bacterium ZC4 chromosome Total score: 1.0 Cumulative Blast bit score: 471
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CX649_07230
TetR/AcrR family transcriptional regulator
Accession: CX649_07235
Location: 1487090-1487623
NCBI BlastP on this gene
Accession: CX649_07235
Location: 1487090-1487623
NCBI BlastP on this gene
CX649_07235
HXXEE domain-containing protein
Accession: AXI39448
Location: 1487616-1488200
NCBI BlastP on this gene
Accession: AXI39448
Location: 1487616-1488200
NCBI BlastP on this gene
CX649_07240
PTS glucose transporter subunit IIA
Accession: AXI39449
Location: 1488409-1488894
NCBI BlastP on this gene
Accession: AXI39449
Location: 1488409-1488894
NCBI BlastP on this gene
CX649_07245
PTS trehalose transporter subunit IIBC
Accession: AXI39450
Location: 1488930-1490345
NCBI BlastP on this gene
Accession: AXI39450
Location: 1488930-1490345
NCBI BlastP on this gene
CX649_07250
alpha,alpha-phosphotrehalase
Accession: AXI39451
Location: 1490463-1492148
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 471
Sequence coverage: 101 %
E-value: 1e-156
NCBI BlastP on this gene
Accession: AXI39451
Location: 1490463-1492148
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 471
Sequence coverage: 101 %
E-value: 1e-156
NCBI BlastP on this gene
treC
treR
CX649_07265
LacI family transcriptional regulator
Accession: AXI39453
Location: 1493819-1494817
NCBI BlastP on this gene
Accession: AXI39453
Location: 1493819-1494817
NCBI BlastP on this gene
CX649_07270
PTS maltose transporter subunit IIBC
Accession: AXI39454
Location: 1494914-1496356
NCBI BlastP on this gene
Accession: AXI39454
Location: 1494914-1496356
NCBI BlastP on this gene
CX649_07275
sucrose-6-phosphate hydrolase
Accession: AXI39455
Location: 1496436-1497917
NCBI BlastP on this gene
Accession: AXI39455
Location: 1496436-1497917
NCBI BlastP on this gene
CX649_07280
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
FP929037 : Clostridium saccharolyticum-like K10 draft genome. Total score: 1.0 Cumulative Blast bit score: 469
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CLS_00510
trehalose operon repressor, B. subtilis-type
Accession: CBK75955
Location: 45196-45909
NCBI BlastP on this gene
Accession: CBK75955
Location: 45196-45909
NCBI BlastP on this gene
CLS_00550
PTS system, glucose-like IIB component
Accession: CBK75956
Location: 46062-47552
NCBI BlastP on this gene
Accession: CBK75956
Location: 46062-47552
NCBI BlastP on this gene
CLS_00560
Glycosidases
Accession: CBK75957
Location: 47604-49316
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 469
Sequence coverage: 97 %
E-value: 1e-155
NCBI BlastP on this gene
Accession: CBK75957
Location: 47604-49316
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 469
Sequence coverage: 97 %
E-value: 1e-155
NCBI BlastP on this gene
CLS_00570
HAD-superfamily hydrolase, subfamily IIB
Accession: CBK75958
Location: 51295-52143
NCBI BlastP on this gene
Accession: CBK75958
Location: 51295-52143
NCBI BlastP on this gene
CLS_00590
ABC-type sugar transport system, permease component
Accession: CBK75959
Location: 52978-53802
NCBI BlastP on this gene
Accession: CBK75959
Location: 52978-53802
NCBI BlastP on this gene
CLS_00610
ABC-type sugar transport system, periplasmic component
Accession: CBK75960
Location: 53835-55199
NCBI BlastP on this gene
Accession: CBK75960
Location: 53835-55199
NCBI BlastP on this gene
CLS_00620
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022437 : Virgibacillus necropolis strain LMG 19488 chromosome Total score: 1.0 Cumulative Blast bit score: 468
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
alpha,alpha-phosphotrehalase
Accession: ASN07129
Location: 4205715-4207373
BlastP hit with treA
Percentage identity: 42 %
BlastP bit score: 468
Sequence coverage: 98 %
E-value: 1e-155
NCBI BlastP on this gene
Accession: ASN07129
Location: 4205715-4207373
BlastP hit with treA
Percentage identity: 42 %
BlastP bit score: 468
Sequence coverage: 98 %
E-value: 1e-155
NCBI BlastP on this gene
treC
treR
CFK40_20045
CFK40_20040
bacitracin ABC transporter ATP-binding protein
Accession: ASN07125
Location: 4202187-4203107
NCBI BlastP on this gene
Accession: ASN07125
Location: 4202187-4203107
NCBI BlastP on this gene
CFK40_20035
RNA polymerase subunit sigma-70
Accession: ASN07124
Location: 4201230-4201796
NCBI BlastP on this gene
Accession: ASN07124
Location: 4201230-4201796
NCBI BlastP on this gene
CFK40_20030
CFK40_20025
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002394 : Bacillus cellulosilyticus DSM 2522 Total score: 1.0 Cumulative Blast bit score: 464
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ABC transporter related protein
Accession: ADU29191
Location: 1039275-1040126
NCBI BlastP on this gene
Accession: ADU29191
Location: 1039275-1040126
NCBI BlastP on this gene
Bcell_0915
Bcell_0916
Bcell_0917
protein of unknown function DUF711
Accession: ADU29194
Location: 1041101-1042468
NCBI BlastP on this gene
Accession: ADU29194
Location: 1041101-1042468
NCBI BlastP on this gene
Bcell_0918
ACT domain-containing protein
Accession: ADU29195
Location: 1042483-1042758
NCBI BlastP on this gene
Accession: ADU29195
Location: 1042483-1042758
NCBI BlastP on this gene
Bcell_0919
Bcell_0920
alpha,alpha-phosphotrehalase
Accession: ADU29197
Location: 1044253-1045938
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 5e-154
NCBI BlastP on this gene
Accession: ADU29197
Location: 1044253-1045938
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 5e-154
NCBI BlastP on this gene
Bcell_0921
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP001878 : Bacillus pseudofirmus OF4 Total score: 1.0 Cumulative Blast bit score: 462
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
BpOF4_10335
BpOF4_10340
Phosphotransferase system (PTS) trehalose-specific enzyme II, BC component
Accession: ADC50122
Location: 501460-502872
NCBI BlastP on this gene
Accession: ADC50122
Location: 501460-502872
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: ADC50123
Location: 503025-504704
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 462
Sequence coverage: 92 %
E-value: 5e-153
NCBI BlastP on this gene
Accession: ADC50123
Location: 503025-504704
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 462
Sequence coverage: 92 %
E-value: 5e-153
NCBI BlastP on this gene
BpOF4_10350
GntR family transcriptional repressor of trehalose operon
Accession: ADC50124
Location: 504729-505445
NCBI BlastP on this gene
Accession: ADC50124
Location: 504729-505445
NCBI BlastP on this gene
BpOF4_10355
BpOF4_10360
Nitric oxide synthase, oxygenase domain protein
Accession: ADC50126
Location: 506432-507532
NCBI BlastP on this gene
Accession: ADC50126
Location: 506432-507532
NCBI BlastP on this gene
nos
ABC transporter ATP-binding protein
Accession: ADC50127
Location: 507719-509602
NCBI BlastP on this gene
Accession: ADC50127
Location: 507719-509602
NCBI BlastP on this gene
BpOF4_10370
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP040367 : Bacillus flexus isolate 1-2-1 chromosome Total score: 1.0 Cumulative Blast bit score: 461
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
alpha,alpha-phosphotrehalase
Accession: QCS54022
Location: 3014890-3016575
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
Accession: QCS54022
Location: 3014890-3016575
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
treC
treR
pxpB
biotin-dependent carboxyltransferase
Accession: QCS54019
Location: 3012188-3013195
NCBI BlastP on this gene
Accession: QCS54019
Location: 3012188-3013195
NCBI BlastP on this gene
FED53_16180
FED53_16175
divalent metal cation transporter
Accession: FED53_16170
Location: 3011711-3012103
NCBI BlastP on this gene
Accession: FED53_16170
Location: 3011711-3012103
NCBI BlastP on this gene
FED53_16170
FED53_16165
FED53_16160
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP015506 : Bacillus oceanisediminis 2691 chromosome Total score: 1.0 Cumulative Blast bit score: 461
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
A361_08210
NAD-dependent protein deacylase
Accession: AND39100
Location: 1603930-1604643
NCBI BlastP on this gene
Accession: AND39100
Location: 1603930-1604643
NCBI BlastP on this gene
A361_08215
preprotein translocase subunit TatA
Accession: AND39101
Location: 1604761-1604934
NCBI BlastP on this gene
Accession: AND39101
Location: 1604761-1604934
NCBI BlastP on this gene
A361_08220
trehalose permease IIC protein
Accession: AND39102
Location: 1605218-1606624
NCBI BlastP on this gene
Accession: AND39102
Location: 1605218-1606624
NCBI BlastP on this gene
A361_08225
A361_08230
alpha,alpha-phosphotrehalase
Accession: AND39104
Location: 1608429-1610114
BlastP hit with treA
Percentage identity: 43 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
Accession: AND39104
Location: 1608429-1610114
BlastP hit with treA
Percentage identity: 43 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
A361_08235
A361_08240
A361_08245
peptide chain release factor 3
Accession: AND39107
Location: 1612903-1614480
NCBI BlastP on this gene
Accession: AND39107
Location: 1612903-1614480
NCBI BlastP on this gene
A361_08250
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013019 : Clostridium pasteurianum strain M150B Total score: 1.0 Cumulative Blast bit score: 461
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
AQ984_01810
AQ984_01805
5-aminoimidazole-4-carboxamide ribonucleotide transformylase
Accession: AOZ77698
Location: 400206-401381
NCBI BlastP on this gene
Accession: AOZ77698
Location: 400206-401381
NCBI BlastP on this gene
AQ984_01800
PTS beta-glucoside transporter subunit IIABC
Accession: AOZ77697
Location: 397856-399799
NCBI BlastP on this gene
Accession: AOZ77697
Location: 397856-399799
NCBI BlastP on this gene
AQ984_01795
trehalose-6-phosphate hydrolase
Accession: AOZ77696
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AOZ77696
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
AQ984_01790
GntR family transcriptional regulator
Accession: AOZ77695
Location: 395268-395978
NCBI BlastP on this gene
Accession: AOZ77695
Location: 395268-395978
NCBI BlastP on this gene
AQ984_01785
AQ984_01780
HxlR family transcriptional regulator
Accession: AOZ77693
Location: 393710-394018
NCBI BlastP on this gene
Accession: AOZ77693
Location: 393710-394018
NCBI BlastP on this gene
AQ984_01775
AQ984_01770
AQ984_01765
AQ984_01760
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013018 : Clostridium pasteurianum DSM 525 = ATCC 6013 Total score: 1.0 Cumulative Blast bit score: 461
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
AQ983_01810
AQ983_01805
5-aminoimidazole-4-carboxamide ribonucleotide transformylase
Accession: AOZ73901
Location: 400206-401381
NCBI BlastP on this gene
Accession: AOZ73901
Location: 400206-401381
NCBI BlastP on this gene
AQ983_01800
PTS beta-glucoside transporter subunit IIABC
Accession: AOZ73900
Location: 397856-399799
NCBI BlastP on this gene
Accession: AOZ73900
Location: 397856-399799
NCBI BlastP on this gene
AQ983_01795
trehalose-6-phosphate hydrolase
Accession: AOZ73899
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AOZ73899
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
AQ983_01790
GntR family transcriptional regulator
Accession: AOZ73898
Location: 395268-395978
NCBI BlastP on this gene
Accession: AOZ73898
Location: 395268-395978
NCBI BlastP on this gene
AQ983_01785
AQ983_01780
HxlR family transcriptional regulator
Accession: AOZ73896
Location: 393710-394018
NCBI BlastP on this gene
Accession: AOZ73896
Location: 393710-394018
NCBI BlastP on this gene
AQ983_01775
AQ983_01770
AQ983_01765
AQ983_01760
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP009268 : Clostridium pasteurianum DSM 525 = ATCC 6013 Total score: 1.0 Cumulative Blast bit score: 461
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CLPA_c03730
CLPA_c03720
bifunctional purine biosynthesis protein PurH
Accession: AJA50459
Location: 400208-401383
NCBI BlastP on this gene
Accession: AJA50459
Location: 400208-401383
NCBI BlastP on this gene
purH1
PTS system trehalose-specific EIIBC component
Accession: AJA50458
Location: 397858-399801
NCBI BlastP on this gene
Accession: AJA50458
Location: 397858-399801
NCBI BlastP on this gene
treP
trehalose-6-phosphate hydrolase
Accession: AJA50457
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AJA50457
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
treA
trehalose operon transcriptional repressor
Accession: AJA50456
Location: 395270-395980
NCBI BlastP on this gene
Accession: AJA50456
Location: 395270-395980
NCBI BlastP on this gene
treR
CLPA_c03670
transcriptional regulator, HxlR family
Accession: AJA50454
Location: 393712-394020
NCBI BlastP on this gene
Accession: AJA50454
Location: 393712-394020
NCBI BlastP on this gene
CLPA_c03660
CLPA_c03650
CLPA_c03640
transcriptional regulator, TetR family
Accession: AJA50451
Location: 391286-391915
NCBI BlastP on this gene
Accession: AJA50451
Location: 391286-391915
NCBI BlastP on this gene
CLPA_c03630
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP009267 : Clostridium pasteurianum DSM 525 = ATCC 6013 Total score: 1.0 Cumulative Blast bit score: 461
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CPAST_c03730
CPAST_c03720
bifunctional purine biosynthesis protein PurH
Accession: AJA46471
Location: 400208-401383
NCBI BlastP on this gene
Accession: AJA46471
Location: 400208-401383
NCBI BlastP on this gene
purH1
PTS system trehalose-specific EIIBC component
Accession: AJA46470
Location: 397858-399801
NCBI BlastP on this gene
Accession: AJA46470
Location: 397858-399801
NCBI BlastP on this gene
treP
trehalose-6-phosphate hydrolase
Accession: AJA46469
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AJA46469
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
treA
trehalose operon transcriptional repressor
Accession: AJA46468
Location: 395270-395980
NCBI BlastP on this gene
Accession: AJA46468
Location: 395270-395980
NCBI BlastP on this gene
treR
CPAST_c03670
transcriptional regulator, HxlR family
Accession: AJA46466
Location: 393712-394020
NCBI BlastP on this gene
Accession: AJA46466
Location: 393712-394020
NCBI BlastP on this gene
CPAST_c03660
CPAST_c03650
CPAST_c03640
transcriptional regulator, TetR family
Accession: AJA46463
Location: 391286-391915
NCBI BlastP on this gene
Accession: AJA46463
Location: 391286-391915
NCBI BlastP on this gene
CPAST_c03630
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022535 : Spiroplasma corruscae strain EC-1 chromosome Total score: 1.0 Cumulative Blast bit score: 430
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system, trehalose-specific IIBC component
Accession: ASP28428
Location: 765801-767363
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 431
Sequence coverage: 93 %
E-value: 9e-142
NCBI BlastP on this gene
Accession: ASP28428
Location: 765801-767363
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 431
Sequence coverage: 93 %
E-value: 9e-142
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: ASP28427
Location: 764166-765788
NCBI BlastP on this gene
Accession: ASP28427
Location: 764166-765788
NCBI BlastP on this gene
treC
SCORR_v1c06540
SCORR_v1c06530
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP031376 : Spiroplasma alleghenense strain PLHS-1 chromosome Total score: 1.0 Cumulative Blast bit score: 430
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system, trehalose-specific IIBC component
Accession: AXK51694
Location: 1177891-1179567
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 430
Sequence coverage: 97 %
E-value: 6e-141
NCBI BlastP on this gene
Accession: AXK51694
Location: 1177891-1179567
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 430
Sequence coverage: 97 %
E-value: 6e-141
NCBI BlastP on this gene
treB
SALLE_v1c10230
SALLE_v1c10220
SALLE_v1c10210
SALLE_v1c10200
SALLE_v1c10190
SALLE_v1c10180
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP025257 : Mesoplasma syrphidae strain YJS chromosome Total score: 1.0 Cumulative Blast bit score: 426
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS sugar transporter subunit IIA
Accession: AUF83741
Location: 692942-694519
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 426
Sequence coverage: 95 %
E-value: 6e-140
NCBI BlastP on this gene
Accession: AUF83741
Location: 692942-694519
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 426
Sequence coverage: 95 %
E-value: 6e-140
NCBI BlastP on this gene
CXP39_02940
CXP39_02935
CXP39_02930
phosphoenolpyruvate--protein phosphotransferase
Accession: AUF83738
Location: 689137-690858
NCBI BlastP on this gene
Accession: AUF83738
Location: 689137-690858
NCBI BlastP on this gene
ptsP
PTS glucose transporter subunit IIA
Accession: AUF83737
Location: 688567-689043
NCBI BlastP on this gene
Accession: AUF83737
Location: 688567-689043
NCBI BlastP on this gene
CXP39_02920
CXP39_02915
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP043026 : Spiroplasma chinense strain CCH chromosome Total score: 1.0 Cumulative Blast bit score: 422
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
methionyl-tRNA formyltransferase
Accession: QEH61694
Location: 554037-554987
NCBI BlastP on this gene
Accession: QEH61694
Location: 554037-554987
NCBI BlastP on this gene
fmt
SCHIN_v1c04980
efp
SCHIN_v1c05000
pldB
SCHIN_v1c05020
SCHIN_v1c05030
PTS system, trehalose-specific IIB component
Accession: QEH61701
Location: 559595-561274
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 422
Sequence coverage: 97 %
E-value: 9e-138
NCBI BlastP on this gene
Accession: QEH61701
Location: 559595-561274
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 422
Sequence coverage: 97 %
E-value: 9e-138
NCBI BlastP on this gene
treB
SCHIN_v1c05050
mapA
pgmB
SCHIN_v1c05080
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP024870 : Spiroplasma clarkii strain CN-5 chromosome Total score: 1.0 Cumulative Blast bit score: 419
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SCLAR_v1c11980
GntR family transcriptional regulator
Accession: ATX71499
Location: 1329155-1329832
NCBI BlastP on this gene
Accession: ATX71499
Location: 1329155-1329832
NCBI BlastP on this gene
SCLAR_v1c11990
SCLAR_v1c12000
SCLAR_v1c12010
PTS system, trehalose-specific IIB component
Accession: ATX71502
Location: 1332100-1333767
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 9e-137
NCBI BlastP on this gene
Accession: ATX71502
Location: 1332100-1333767
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 9e-137
NCBI BlastP on this gene
treB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013860 : Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 412
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system, trehalose-specific IIBC component
Accession: ALX70904
Location: 766378-767934
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
Accession: ALX70904
Location: 766378-767934
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: ALX70903
Location: 764747-766363
NCBI BlastP on this gene
Accession: ALX70903
Location: 764747-766363
NCBI BlastP on this gene
treC
STURO_v1c06430
STURO_v1c06420
STURO_v1c06410
ldhA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP012328 : Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 412
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system, trehalose-specific IIB component PTS system, trehalose-specific IIC component
Accession: AKU79893
Location: 766480-768036
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
Accession: AKU79893
Location: 766480-768036
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
STURON_00647
trehalose-6-phosphate hydrolase
Accession: AKU79892
Location: 764849-766465
NCBI BlastP on this gene
Accession: AKU79892
Location: 764849-766465
NCBI BlastP on this gene
treA
STURON_00645
STURON_00644
STURON_00643
ldhA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP006681 : Spiroplasma culicicola AES-1 Total score: 1.0 Cumulative Blast bit score: 408
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SCULI_v1c04520
SCULI_v1c04510
SCULI_v1c04500
SCULI_v1c04490
SCULI_v1c04480
SCULI_v1c04470
PTS system trehalose-specific IIBC component
Accession: AHI52787
Location: 492197-493849
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 408
Sequence coverage: 97 %
E-value: 1e-132
NCBI BlastP on this gene
Accession: AHI52787
Location: 492197-493849
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 408
Sequence coverage: 97 %
E-value: 1e-132
NCBI BlastP on this gene
treB
SCULI_v1c04450
pgmB
mapA
SCULI_v1c04420
SCULI_v1c04410
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP025420 : Streptococcus parauberis strain SPOF3K chromosome Total score: 1.0 Cumulative Blast bit score: 313
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SPSF3K_00109
SPSF3K_00110
[Formate-C-acetyltransferase]-activating enzyme
Accession: AUT04853
Location: 96866-97480
NCBI BlastP on this gene
Accession: AUT04853
Location: 96866-97480
NCBI BlastP on this gene
nrdG
SPSF3K_00112
catE
Streptomycin 3''-adenylyltransferase
Accession: AUT04856
Location: 99438-99764
NCBI BlastP on this gene
Accession: AUT04856
Location: 99438-99764
NCBI BlastP on this gene
aadA
HTH-type transcriptional regulator GmuR
Accession: AUT04857
Location: 99788-100522
NCBI BlastP on this gene
Accession: AUT04857
Location: 99788-100522
NCBI BlastP on this gene
SPSF3K_00115
Protein-N(pi)-phosphohistidine--sugar phosphotransferase
Accession: AUT04858
Location: 100723-102732
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: AUT04858
Location: 100723-102732
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
treP
treC
SPSF3K_00118
Protein-N(pi)-phosphohistidine--sugar phosphotransferase
Accession: AUT04861
Location: 106686-106970
NCBI BlastP on this gene
Accession: AUT04861
Location: 106686-106970
NCBI BlastP on this gene
ulaB
Ascorbate-specific permease IIC component UlaA
Accession: AUT04862
Location: 106983-108323
NCBI BlastP on this gene
Accession: AUT04862
Location: 106983-108323
NCBI BlastP on this gene
SPSF3K_00120
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002471 : Streptococcus parauberis KCTC 11537 Total score: 1.0 Cumulative Blast bit score: 313
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
sugar phosphotransferase system (PTS), IIABC component
Accession: AEF26225
Location: 2042784-2044769
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 8e-95
NCBI BlastP on this gene
Accession: AEF26225
Location: 2042784-2044769
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 8e-95
NCBI BlastP on this gene
STP_1777
trehalose-6-phosphate hydrolase
Accession: AEF26224
Location: 2041091-2042716
NCBI BlastP on this gene
Accession: AEF26224
Location: 2041091-2042716
NCBI BlastP on this gene
treA
STP_1775
sugar-specific permease, SgaT/UlaA family
Accession: AEF26222
Location: 2037194-2038534
NCBI BlastP on this gene
Accession: AEF26222
Location: 2037194-2038534
NCBI BlastP on this gene
STP_1774
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR595858 : Streptococcus sp. NCTC 10228 strain NCTC10228 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 311
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system sugar transporter subunit IIABC
Accession: VUD00898
Location: 2040139-2042133
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
Accession: VUD00898
Location: 2040139-2042133
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: VUD00895
Location: 2038442-2040073
NCBI BlastP on this gene
Accession: VUD00895
Location: 2038442-2040073
NCBI BlastP on this gene
treA
pepO
NCTC10228_02064
tsf
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR134341 : Streptococcus pseudoporcinus strain NCTC13786 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 311
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
anaerobic ribonucleoside triphosphate reductase
Accession: VEF93911
Location: 1303615-1305810
NCBI BlastP on this gene
Accession: VEF93911
Location: 1303615-1305810
NCBI BlastP on this gene
nrdD_2
NCTC13786_01278
mviM
GNAT family acetyltransferase
Accession: VEF93914
Location: 1307036-1307539
NCBI BlastP on this gene
Accession: VEF93914
Location: 1307036-1307539
NCBI BlastP on this gene
NCTC13786_01280
anaerobic ribonucleoside-triphosphate reductase activating protein
Accession: VEF93915
Location: 1307546-1308160
NCBI BlastP on this gene
Accession: VEF93915
Location: 1307546-1308160
NCBI BlastP on this gene
pflA_1
hypothetical cytosolic protein
Accession: VEF93916
Location: 1308281-1309012
NCBI BlastP on this gene
Accession: VEF93916
Location: 1308281-1309012
NCBI BlastP on this gene
yaaA
GntR family transcriptional regulator
Accession: VEF93917
Location: 1309145-1309858
NCBI BlastP on this gene
Accession: VEF93917
Location: 1309145-1309858
NCBI BlastP on this gene
treR
PTS system sugar transporter subunit IIABC
Accession: VEF93918
Location: 1310075-1312069
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
Accession: VEF93918
Location: 1310075-1312069
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: VEF93919
Location: 1312135-1313766
NCBI BlastP on this gene
Accession: VEF93919
Location: 1312135-1313766
NCBI BlastP on this gene
treA
pepO
NCTC13786_01287
tsf
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483383 : Streptococcus cristatus strain NCTC12479 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 310
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
phosphoenolpyruvate carboxykinase (ATP)
Accession: SQG31908
Location: 561604-563250
NCBI BlastP on this gene
Accession: SQG31908
Location: 561604-563250
NCBI BlastP on this gene
NCTC12479_00557
NCTC12479_00558
GntR family transcriptional regulator
Accession: SQG31910
Location: 565036-565749
NCBI BlastP on this gene
Accession: SQG31910
Location: 565036-565749
NCBI BlastP on this gene
treR
trehalose PTS system, IIABC components
Accession: SQG31911
Location: 565936-567903
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 310
Sequence coverage: 95 %
E-value: 2e-93
NCBI BlastP on this gene
Accession: SQG31911
Location: 565936-567903
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 310
Sequence coverage: 95 %
E-value: 2e-93
NCBI BlastP on this gene
treP
dexS
pyruvate formate-lyase-activating enzyme
Accession: SQG31913
Location: 569968-570765
NCBI BlastP on this gene
Accession: SQG31913
Location: 569968-570765
NCBI BlastP on this gene
act
glycosyl transferase family protein
Accession: SQG31914
Location: 571269-572492
NCBI BlastP on this gene
Accession: SQG31914
Location: 571269-572492
NCBI BlastP on this gene
gspA
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP050133 : Streptococcus cristatus ATCC 51100 chromosome Total score: 1.0 Cumulative Blast bit score: 308
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system trehalose-specific EIIBC component
Accession: QIP49760
Location: 1439035-1441002
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 95 %
E-value: 6e-93
NCBI BlastP on this gene
Accession: QIP49760
Location: 1439035-1441002
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 95 %
E-value: 6e-93
NCBI BlastP on this gene
treP
treC
pyruvate formate lyase-activating protein
Accession: QIP49758
Location: 1436172-1436969
NCBI BlastP on this gene
Accession: QIP49758
Location: 1436172-1436969
NCBI BlastP on this gene
pflA
HBA50_07140
HBA50_07135
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002394 : Bacillus cellulosilyticus DSM 2522 Total score: 1.0 Cumulative Blast bit score: 308
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SUF system FeS assembly protein, NifU family
Accession: ADU29113
Location: 959073-959522
NCBI BlastP on this gene
Accession: ADU29113
Location: 959073-959522
NCBI BlastP on this gene
Bcell_0834
Bcell_0836
GCN5-related N-acetyltransferase
Accession: ADU29115
Location: 961158-961712
NCBI BlastP on this gene
Accession: ADU29115
Location: 961158-961712
NCBI BlastP on this gene
Bcell_0837
Bcell_0838
Bcell_0839
Bcell_0840
PTS system, trehalose-specific IIBC subunit
Accession: ADU29119
Location: 964229-965665
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 308
Sequence coverage: 86 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: ADU29119
Location: 964229-965665
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 308
Sequence coverage: 86 %
E-value: 1e-94
NCBI BlastP on this gene
Bcell_0841
DNA-(apurinic or apyrimidinic site) lyase
Accession: ADU29120
Location: 965819-966613
NCBI BlastP on this gene
Accession: ADU29120
Location: 965819-966613
NCBI BlastP on this gene
Bcell_0842
Bcell_0843
RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: ADU29122
Location: 967481-968005
NCBI BlastP on this gene
Accession: ADU29122
Location: 967481-968005
NCBI BlastP on this gene
Bcell_0844
cell envelope-related transcriptional attenuator
Accession: ADU29123
Location: 967998-968957
NCBI BlastP on this gene
Accession: ADU29123
Location: 967998-968957
NCBI BlastP on this gene
Bcell_0845
small acid-soluble spore protein alpha/beta type
Accession: ADU29124
Location: 969096-969296
NCBI BlastP on this gene
Accession: ADU29124
Location: 969096-969296
NCBI BlastP on this gene
Bcell_0846
PAS/PAC sensor signal transduction histidine kinase
Accession: ADU29125
Location: 969533-971368
NCBI BlastP on this gene
Accession: ADU29125
Location: 969533-971368
NCBI BlastP on this gene
Bcell_0847
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483408 : Streptococcus uberis strain NCTC4674 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 307
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system sugar transporter subunit IIABC
Accession: SQG84040
Location: 1923576-1925561
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: SQG84040
Location: 1923576-1925561
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: SQG84039
Location: 1921877-1923508
NCBI BlastP on this gene
Accession: SQG84039
Location: 1921877-1923508
NCBI BlastP on this gene
treA
ulaC_2
PTS system ascorbate-specific transporter subunits IICB
Accession: SQG84037
Location: 1919314-1919598
NCBI BlastP on this gene
Accession: SQG84037
Location: 1919314-1919598
NCBI BlastP on this gene
NCTC4674_02014
SgaT/UlaA family sugar-specific permease
Accession: SQG84036
Location: 1917958-1919301
NCBI BlastP on this gene
Accession: SQG84036
Location: 1917958-1919301
NCBI BlastP on this gene
ulaA_2
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022435 : Streptococcus uberis strain NZ01 chromosome Total score: 1.0 Cumulative Blast bit score: 307
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS beta-glucoside transporter subunit EIIBCA
Accession: AUC25774
Location: 1774797-1776782
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: AUC25774
Location: 1774797-1776782
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
CGZ53_09000
treC
transcription antiterminator BglG
Accession: AUC25772
Location: 1770823-1772865
NCBI BlastP on this gene
Accession: AUC25772
Location: 1770823-1772865
NCBI BlastP on this gene
CGZ53_08990
PTS lactose transporter subunit IIB
Accession: AUC25771
Location: 1770534-1770818
NCBI BlastP on this gene
Accession: AUC25771
Location: 1770534-1770818
NCBI BlastP on this gene
CGZ53_08985
PTS ascorbate transporter subunit IIC
Accession: AUC25770
Location: 1769178-1770521
NCBI BlastP on this gene
Accession: AUC25770
Location: 1769178-1770521
NCBI BlastP on this gene
CGZ53_08980
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
AM946015 : Streptococcus uberis 0140J complete genome. Total score: 1.0 Cumulative Blast bit score: 307
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
sugar phosphotransferase system (PTS), IIABC component
Accession: CAR43740
Location: 1750789-1752774
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: CAR43740
Location: 1750789-1752774
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
SUB1763
putative trehalose-6-phosphate hydrolase
Accession: CAR43735
Location: 1749090-1750721
NCBI BlastP on this gene
Accession: CAR43735
Location: 1749090-1750721
NCBI BlastP on this gene
treA
putative PTS multi-domain regulator
Accession: CAR43734
Location: 1746815-1748857
NCBI BlastP on this gene
Accession: CAR43734
Location: 1746815-1748857
NCBI BlastP on this gene
SUB1761
sugar phosphotransferase system (PTS),
Accession: CAR43732
Location: 1746526-1746810
NCBI BlastP on this gene
Accession: CAR43732
Location: 1746526-1746810
NCBI BlastP on this gene
SUB1760
putative sugar-specific permease, SgaT/UlaA family
Accession: CAR43730
Location: 1745170-1746513
NCBI BlastP on this gene
Accession: CAR43730
Location: 1745170-1746513
NCBI BlastP on this gene
SUB1759
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483397 : Streptococcus uberis strain NCTC3858 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 305
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system sugar transporter subunit IIABC
Accession: SQG47165
Location: 1886561-1888546
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 96 %
E-value: 1e-91
NCBI BlastP on this gene
Accession: SQG47165
Location: 1886561-1888546
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 96 %
E-value: 1e-91
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: SQG47164
Location: 1884862-1886493
NCBI BlastP on this gene
Accession: SQG47164
Location: 1884862-1886493
NCBI BlastP on this gene
treA
ulaC_3
PTS system ascorbate-specific transporter subunits IICB
Accession: SQG47162
Location: 1882298-1882582
NCBI BlastP on this gene
Accession: SQG47162
Location: 1882298-1882582
NCBI BlastP on this gene
NCTC3858_01941
SgaT/UlaA family sugar-specific permease
Accession: SQG47161
Location: 1880942-1882285
NCBI BlastP on this gene
Accession: SQG47161
Location: 1880942-1882285
NCBI BlastP on this gene
ulaA_3
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP028415 : Streptococcus mitis strain SK637 chromosome Total score: 1.0 Cumulative Blast bit score: 305
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SK637_00328
SK637_00329
ABC transporter substrate binding family protein
Accession: QBZ12892
Location: 320443-321252
NCBI BlastP on this gene
Accession: QBZ12892
Location: 320443-321252
NCBI BlastP on this gene
SK637_00330
branched-chain amino acid transport system / permease component family protein
Accession: QBZ12893
Location: 321263-322156
NCBI BlastP on this gene
Accession: QBZ12893
Location: 321263-322156
NCBI BlastP on this gene
SK637_00331
SK637_00332
SK637_00333
PTS system trehalose-specific IIBC component
Accession: QBZ12896
Location: 323900-325867
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 93 %
E-value: 1e-91
NCBI BlastP on this gene
Accession: QBZ12896
Location: 323900-325867
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 93 %
E-value: 1e-91
NCBI BlastP on this gene
SK637_00334
SK637_00335
SK637_00336
SK637_00337
non-canonical purine NTP pyrophosphatase RdgB
Accession: QBZ12900
Location: 328942-329913
NCBI BlastP on this gene
Accession: QBZ12900
Location: 328942-329913
NCBI BlastP on this gene
SK637_00338
SK637_00339
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP018187 : Streptococcus salivarius strain ICDC2 Total score: 1.0 Cumulative Blast bit score: 305
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS beta-glucoside transporter subunit EIIBCA
Accession: QGU81410
Location: 2140805-2142784
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
Accession: QGU81410
Location: 2140805-2142784
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
BSR19_09965
BSR19_09960
BSR19_09955
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: QGU81407
Location: 2134411-2136999
NCBI BlastP on this gene
Accession: QGU81407
Location: 2134411-2136999
NCBI BlastP on this gene
BSR19_09950
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002888 : Streptococcus salivarius 57.I Total score: 1.0 Cumulative Blast bit score: 305
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
Ssal_00112
fructose-1,6-bisphosphate aldolase, class II
Accession: AEJ52537
Location: 98328-99209
NCBI BlastP on this gene
Accession: AEJ52537
Location: 98328-99209
NCBI BlastP on this gene
fbaA
Ssal_00115
Ssal_00116
Ssal_00117
treR
pts system, trehalose-specific iibc component
Accession: AEJ52542
Location: 101771-103816
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 4e-91
NCBI BlastP on this gene
Accession: AEJ52542
Location: 101771-103816
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 4e-91
NCBI BlastP on this gene
treB
treC
pepO
aldehyde-alcohol dehydrogenase 2
Accession: AEJ52545
Location: 107614-110211
NCBI BlastP on this gene
Accession: AEJ52545
Location: 107614-110211
NCBI BlastP on this gene
adhE
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP015282 : Streptococcus salivarius strain ATCC 27945 Total score: 1.0 Cumulative Blast bit score: 304
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
gamma-glutamylcysteine synthetase
Accession: ARI58245
Location: 1616084-1617379
NCBI BlastP on this gene
Accession: ARI58245
Location: 1616084-1617379
NCBI BlastP on this gene
NX99_07305
fructose-bisphosphate aldolase
Accession: ARI58246
Location: 1617803-1618684
NCBI BlastP on this gene
Accession: ARI58246
Location: 1617803-1618684
NCBI BlastP on this gene
NX99_07315
NX99_07320
NX99_07325
NX99_07330
PTS beta-glucoside transporter subunit EIIBCA
Accession: ARI58250
Location: 1621210-1623189
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
Accession: ARI58250
Location: 1621210-1623189
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
NX99_07335
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
AP018338 : Streptococcus oralis subsp. tigurinus DNA Total score: 1.0 Cumulative Blast bit score: 304
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system trehalose-specific IIBC component
Accession: BBA09167
Location: 1594737-1596704
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 94 %
E-value: 4e-91
NCBI BlastP on this gene
Accession: BBA09167
Location: 1594737-1596704
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 94 %
E-value: 4e-91
NCBI BlastP on this gene
treP
treC
uncharacterised protein family UPF0154
Accession: BBA09165
Location: 1592658-1592903
NCBI BlastP on this gene
Accession: BBA09165
Location: 1592658-1592903
NCBI BlastP on this gene
STO1_015610
murI
Non-canonical purine NTP pyrophosphatase
Accession: BBA09163
Location: 1590691-1591662
NCBI BlastP on this gene
Accession: BBA09163
Location: 1590691-1591662
NCBI BlastP on this gene
rdgB
STO1_015580
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR134336 : Streptococcus oralis strain NCTC11427 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
trehalose PTS system, IIABC components
Accession: VEF79619
Location: 1614753-1616720
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 7e-91
NCBI BlastP on this gene
Accession: VEF79619
Location: 1614753-1616720
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 7e-91
NCBI BlastP on this gene
treP
treC
NCTC11427_01597
murI
deoxyribonucleotide triphosphate
Accession: VEF79615
Location: 1610705-1611676
NCBI BlastP on this gene
Accession: VEF79615
Location: 1610705-1611676
NCBI BlastP on this gene
rdgB
NCTC11427_01594
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR134323 : Streptococcus urinalis strain NCTC13766 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
mviM
NCTC13766_00060
acetyltransferase (GNAT) family protein
Accession: VEF30997
Location: 59832-60329
NCBI BlastP on this gene
Accession: VEF30997
Location: 59832-60329
NCBI BlastP on this gene
NCTC13766_00061
anaerobic ribonucleoside-triphosphate reductase activating protein
Accession: VEF30998
Location: 60347-60955
NCBI BlastP on this gene
Accession: VEF30998
Location: 60347-60955
NCBI BlastP on this gene
pflA
yaaA
catE
trehalose operon transcriptional repressor
Accession: VEF31001
Location: 62803-63516
NCBI BlastP on this gene
Accession: VEF31001
Location: 62803-63516
NCBI BlastP on this gene
treR
PTS system transporter subunit IIABC
Accession: VEF31002
Location: 63735-65717
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: VEF31002
Location: 63735-65717
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 6e-91
NCBI BlastP on this gene
treB
treA
NADP or NAD utilizing oxidoreductase
Accession: VEF31004
Location: 67440-68420
NCBI BlastP on this gene
Accession: VEF31004
Location: 67440-68420
NCBI BlastP on this gene
afr
pepO
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR134274 : Streptococcus salivarius strain NCTC8618 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system trehalose-specific transporter subunit IIBC
Accession: VED90220
Location: 2101140-2103116
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
Accession: VED90220
Location: 2101140-2103116
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
treB_1
putative trehalose-6-phosphate hydrolase
Accession: VED90219
Location: 2099451-2101076
NCBI BlastP on this gene
Accession: VED90219
Location: 2099451-2101076
NCBI BlastP on this gene
treC
pepO
aldehyde-alcohol dehydrogenase 2
Accession: VED90217
Location: 2094746-2097334
NCBI BlastP on this gene
Accession: VED90217
Location: 2094746-2097334
NCBI BlastP on this gene
adhE_4
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP040804 : Streptococcus salivarius strain LAB813 chromosome Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
IS30-like element IS1139 family transposase
Accession: QEM31807
Location: 250403-251428
NCBI BlastP on this gene
Accession: QEM31807
Location: 250403-251428
NCBI BlastP on this gene
FHI56_02420
FHI56_02430
FHI56_02435
FHI56_02440
FHI56_02445
treR
PTS trehalose transporter subunit IIBC
Location: 255186-257165
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 93 %
E-value: 7e-91
Location: 255186-257165
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 93 %
E-value: 7e-91
treB
treC
FHI56_02465
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: QEM31814
Location: 260963-263560
NCBI BlastP on this gene
Accession: QEM31814
Location: 260963-263560
NCBI BlastP on this gene
adhE
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP023507 : Streptococcus oralis strain FDAARGOS_367 chromosome Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CO686_00930
treR
PTS trehalose transporter subunit IIBC
Accession: ATF57782
Location: 180261-182378
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 2e-90
NCBI BlastP on this gene
Accession: ATF57782
Location: 180261-182378
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 2e-90
NCBI BlastP on this gene
CO686_00940
treC
CO686_00950
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP016786 : Clostridium isatidis strain DSM 15098 chromosome Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
BEN51_09190
BEN51_09185
energy-coupled thiamine transporter ThiT
Accession: ASW43645
Location: 1943664-1944287
NCBI BlastP on this gene
Accession: ASW43645
Location: 1943664-1944287
NCBI BlastP on this gene
BEN51_09180
BEN51_09175
BEN51_09170
BEN51_09165
PTS maltose transporter subunit IIBC
Accession: ASW43641
Location: 1939146-1940606
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 96 %
E-value: 9e-93
NCBI BlastP on this gene
Accession: ASW43641
Location: 1939146-1940606
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 96 %
E-value: 9e-93
NCBI BlastP on this gene
BEN51_09160
BEN51_09155
BEN51_09145
BEN51_09140
BEN51_09135
BEN51_09130
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP016207 : Streptococcus sp. oral taxon 064 strain W10853 sequence. Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS beta-glucoside transporter subunit EIIBCA
Accession: ANR74721
Location: 343050-345017
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: ANR74721
Location: 343050-345017
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 6e-91
NCBI BlastP on this gene
AXF18_01655
AXF18_01650
AXF18_01645
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP014144 : Streptococcus salivarius strain JF Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
gamma-glutamylcysteine synthetase
Accession: AMB83681
Location: 2117640-2118935
NCBI BlastP on this gene
Accession: AMB83681
Location: 2117640-2118935
NCBI BlastP on this gene
AWB63_09820
fructose-bisphosphate aldolase
Accession: AMB83682
Location: 2119357-2120238
NCBI BlastP on this gene
Accession: AMB83682
Location: 2119357-2120238
NCBI BlastP on this gene
AWB63_09830
AWB63_09835
AWB63_09840
AWB63_09845
PTS beta-glucoside transporter subunit EIIBCA
Accession: AMB83686
Location: 2122723-2124699
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
Accession: AMB83686
Location: 2122723-2124699
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
AWB63_09850
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP009913 : Streptococcus salivarius strain NCTC 8618 Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system trehalose-specific transporter subunits IIBCA
Accession: AIY21959
Location: 2084011-2085987
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
Accession: AIY21959
Location: 2084011-2085987
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
SSAL8618_09625
SSAL8618_09620
SSAL8618_09615
SSAL8618_09610
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP007628 : Streptococcus sp. VT 162 Total score: 1.0 Cumulative Blast bit score: 303
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
GntR family transcriptional regulator
Accession: AHZ47129
Location: 305639-306352
NCBI BlastP on this gene
Accession: AHZ47129
Location: 305639-306352
NCBI BlastP on this gene
V470_01515
PTS system trehalose-specific transporter subunits IIBCA
Accession: AHZ47130
Location: 306537-308504
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 5e-91
NCBI BlastP on this gene
Accession: AHZ47130
Location: 306537-308504
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 5e-91
NCBI BlastP on this gene
V470_01520
V470_01525
V470_01530
V470_01535
deoxyribonucleotide triphosphate pyrophosphatase
Accession: AHZ47134
Location: 311583-312554
NCBI BlastP on this gene
Accession: AHZ47134
Location: 311583-312554
NCBI BlastP on this gene
V470_01540
V470_01545
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
151. : CP016843 Carnobacterium divergens strain TMW 2.1579 chromosome Total score: 1.0 Cumulative Blast bit score: 478
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
gldA
BFC22_01070
BFC22_01065
dihydroxyacetone kinase transcriptional activator DhaS
Accession: ANZ98767
Location: 233076-233654
NCBI BlastP on this gene
Accession: ANZ98767
Location: 233076-233654
NCBI BlastP on this gene
BFC22_01060
PTS maltose transporter subunit IIBC
Accession: ANZ98766
Location: 231412-232869
NCBI BlastP on this gene
Accession: ANZ98766
Location: 231412-232869
NCBI BlastP on this gene
BFC22_01055
alpha,alpha-phosphotrehalase
Accession: ANZ98765
Location: 229711-231381
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 478
Sequence coverage: 99 %
E-value: 4e-159
NCBI BlastP on this gene
Accession: ANZ98765
Location: 229711-231381
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 478
Sequence coverage: 99 %
E-value: 4e-159
NCBI BlastP on this gene
BFC22_01050
BFC22_01045
BFC22_01040
BFC22_01035
tRNA uridine(34) 5-carboxymethylaminomethyl synthesis GTPase MnmE
Accession: ANZ98761
Location: 225607-227004
NCBI BlastP on this gene
Accession: ANZ98761
Location: 225607-227004
NCBI BlastP on this gene
BFC22_01030
tRNA uridine(34) 5-carboxymethylaminomethyl synthesis enzyme MnmG
Accession: ANZ98760
Location: 223696-225591
NCBI BlastP on this gene
Accession: ANZ98760
Location: 223696-225591
NCBI BlastP on this gene
BFC22_01025
152. : CP024848 Oceanobacillus sp. 160 chromosome Total score: 1.0 Cumulative Blast bit score: 477
typA
CUC15_02980
PTS trehalose transporter subunit IIBC
Accession: AXI11077
Location: 576415-578283
NCBI BlastP on this gene
Accession: AXI11077
Location: 576415-578283
NCBI BlastP on this gene
CUC15_02985
alpha,alpha-phosphotrehalase
Accession: AXI08008
Location: 578519-580183
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 6e-159
NCBI BlastP on this gene
Accession: AXI08008
Location: 578519-580183
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 477
Sequence coverage: 100 %
E-value: 6e-159
NCBI BlastP on this gene
treC
treR
CUC15_03000
CUC15_03005
CUC15_03010
CUC15_03015
bacitracin ABC transporter ATP-binding protein
Accession: AXI08014
Location: 583655-584578
NCBI BlastP on this gene
Accession: AXI08014
Location: 583655-584578
NCBI BlastP on this gene
CUC15_03020
153. : CP048837 [Clostridium] innocuum strain LC-LUMC-CI-001 chromosome Total score: 1.0 Cumulative Blast bit score: 476
G4D55_00695
G4D55_00690
O-acetyl-ADP-ribose deacetylase
Accession: QIX07678
Location: 144818-145324
NCBI BlastP on this gene
Accession: QIX07678
Location: 144818-145324
NCBI BlastP on this gene
G4D55_00685
treR
PTS system trehalose-specific EIIBC component
Accession: QIX07676
Location: 141967-143850
NCBI BlastP on this gene
Accession: QIX07676
Location: 141967-143850
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: QIX07675
Location: 140308-141957
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 476
Sequence coverage: 100 %
E-value: 1e-158
NCBI BlastP on this gene
Accession: QIX07675
Location: 140308-141957
BlastP hit with treA
Percentage identity: 46 %
BlastP bit score: 476
Sequence coverage: 100 %
E-value: 1e-158
NCBI BlastP on this gene
treC
DUF1622 domain-containing protein
Accession: QIX07674
Location: 139317-139673
NCBI BlastP on this gene
Accession: QIX07674
Location: 139317-139673
NCBI BlastP on this gene
G4D55_00665
ribose-phosphate pyrophosphokinase
Accession: QIX07673
Location: 138341-139294
NCBI BlastP on this gene
Accession: QIX07673
Location: 138341-139294
NCBI BlastP on this gene
G4D55_00660
bifunctional UDP-N-acetylglucosamine
Accession: QIX07672
Location: 136962-138317
NCBI BlastP on this gene
Accession: QIX07672
Location: 136962-138317
NCBI BlastP on this gene
glmU
Cof-type HAD-IIB family hydrolase
Accession: QIX07671
Location: 135908-136684
NCBI BlastP on this gene
Accession: QIX07671
Location: 135908-136684
NCBI BlastP on this gene
G4D55_00650
HAMP domain-containing histidine kinase
Accession: QIX07670
Location: 134347-135690
NCBI BlastP on this gene
Accession: QIX07670
Location: 134347-135690
NCBI BlastP on this gene
G4D55_00645
154. : AP022323 Aeribacillus pallidus PI8 DNA Total score: 1.0 Cumulative Blast bit score: 474
APP_11320
yxxE
APP_11340
PTS system glucose-specific EIIA component
Accession: BBU38843
Location: 1193787-1194272
NCBI BlastP on this gene
Accession: BBU38843
Location: 1193787-1194272
NCBI BlastP on this gene
crr_1
PTS system trehalose-specific EIIBC component
Accession: BBU38844
Location: 1194308-1195723
NCBI BlastP on this gene
Accession: BBU38844
Location: 1194308-1195723
NCBI BlastP on this gene
treP
trehalose-6-phosphate hydrolase
Accession: BBU38845
Location: 1195841-1197526
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 474
Sequence coverage: 101 %
E-value: 1e-157
NCBI BlastP on this gene
Accession: BBU38845
Location: 1195841-1197526
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 474
Sequence coverage: 101 %
E-value: 1e-157
NCBI BlastP on this gene
treA
treR
APP_11390
APP_11400
APP_11410
APP_11420
APP_11430
155. : CP017703 Aeribacillus pallidus strain KCTC3564 Total score: 1.0 Cumulative Blast bit score: 473
AP3564_10570
AP3564_10575
PTS glucose transporter subunit IIA
Accession: ASS90606
Location: 2168983-2169468
NCBI BlastP on this gene
Accession: ASS90606
Location: 2168983-2169468
NCBI BlastP on this gene
AP3564_10580
trehalose permease IIC protein
Accession: ASS90607
Location: 2169503-2170918
NCBI BlastP on this gene
Accession: ASS90607
Location: 2169503-2170918
NCBI BlastP on this gene
AP3564_10585
alpha,alpha-phosphotrehalase
Accession: ASS90608
Location: 2171036-2172721
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 473
Sequence coverage: 101 %
E-value: 3e-157
NCBI BlastP on this gene
Accession: ASS90608
Location: 2171036-2172721
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 473
Sequence coverage: 101 %
E-value: 3e-157
NCBI BlastP on this gene
AP3564_10590
AP3564_10595
group II intron reverse transcriptase/maturase
Accession: ASS92364
Location: 2174544-2175803
NCBI BlastP on this gene
Accession: ASS92364
Location: 2174544-2175803
NCBI BlastP on this gene
AP3564_10600
AP3564_10605
AP3564_10610
156. : CP025518 Bacillaceae bacterium ZC4 chromosome Total score: 1.0 Cumulative Blast bit score: 471
CX649_07230
TetR/AcrR family transcriptional regulator
Accession: CX649_07235
Location: 1487090-1487623
NCBI BlastP on this gene
Accession: CX649_07235
Location: 1487090-1487623
NCBI BlastP on this gene
CX649_07235
HXXEE domain-containing protein
Accession: AXI39448
Location: 1487616-1488200
NCBI BlastP on this gene
Accession: AXI39448
Location: 1487616-1488200
NCBI BlastP on this gene
CX649_07240
PTS glucose transporter subunit IIA
Accession: AXI39449
Location: 1488409-1488894
NCBI BlastP on this gene
Accession: AXI39449
Location: 1488409-1488894
NCBI BlastP on this gene
CX649_07245
PTS trehalose transporter subunit IIBC
Accession: AXI39450
Location: 1488930-1490345
NCBI BlastP on this gene
Accession: AXI39450
Location: 1488930-1490345
NCBI BlastP on this gene
CX649_07250
alpha,alpha-phosphotrehalase
Accession: AXI39451
Location: 1490463-1492148
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 471
Sequence coverage: 101 %
E-value: 1e-156
NCBI BlastP on this gene
Accession: AXI39451
Location: 1490463-1492148
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 471
Sequence coverage: 101 %
E-value: 1e-156
NCBI BlastP on this gene
treC
treR
CX649_07265
LacI family transcriptional regulator
Accession: AXI39453
Location: 1493819-1494817
NCBI BlastP on this gene
Accession: AXI39453
Location: 1493819-1494817
NCBI BlastP on this gene
CX649_07270
PTS maltose transporter subunit IIBC
Accession: AXI39454
Location: 1494914-1496356
NCBI BlastP on this gene
Accession: AXI39454
Location: 1494914-1496356
NCBI BlastP on this gene
CX649_07275
sucrose-6-phosphate hydrolase
Accession: AXI39455
Location: 1496436-1497917
NCBI BlastP on this gene
Accession: AXI39455
Location: 1496436-1497917
NCBI BlastP on this gene
CX649_07280
157. : FP929037 Clostridium saccharolyticum-like K10 draft genome. Total score: 1.0 Cumulative Blast bit score: 469
CLS_00510
trehalose operon repressor, B. subtilis-type
Accession: CBK75955
Location: 45196-45909
NCBI BlastP on this gene
Accession: CBK75955
Location: 45196-45909
NCBI BlastP on this gene
CLS_00550
PTS system, glucose-like IIB component
Accession: CBK75956
Location: 46062-47552
NCBI BlastP on this gene
Accession: CBK75956
Location: 46062-47552
NCBI BlastP on this gene
CLS_00560
Glycosidases
Accession: CBK75957
Location: 47604-49316
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 469
Sequence coverage: 97 %
E-value: 1e-155
NCBI BlastP on this gene
Accession: CBK75957
Location: 47604-49316
BlastP hit with treA
Percentage identity: 45 %
BlastP bit score: 469
Sequence coverage: 97 %
E-value: 1e-155
NCBI BlastP on this gene
CLS_00570
HAD-superfamily hydrolase, subfamily IIB
Accession: CBK75958
Location: 51295-52143
NCBI BlastP on this gene
Accession: CBK75958
Location: 51295-52143
NCBI BlastP on this gene
CLS_00590
ABC-type sugar transport system, permease component
Accession: CBK75959
Location: 52978-53802
NCBI BlastP on this gene
Accession: CBK75959
Location: 52978-53802
NCBI BlastP on this gene
CLS_00610
ABC-type sugar transport system, periplasmic component
Accession: CBK75960
Location: 53835-55199
NCBI BlastP on this gene
Accession: CBK75960
Location: 53835-55199
NCBI BlastP on this gene
CLS_00620
158. : CP022437 Virgibacillus necropolis strain LMG 19488 chromosome Total score: 1.0 Cumulative Blast bit score: 468
alpha,alpha-phosphotrehalase
Accession: ASN07129
Location: 4205715-4207373
BlastP hit with treA
Percentage identity: 42 %
BlastP bit score: 468
Sequence coverage: 98 %
E-value: 1e-155
NCBI BlastP on this gene
Accession: ASN07129
Location: 4205715-4207373
BlastP hit with treA
Percentage identity: 42 %
BlastP bit score: 468
Sequence coverage: 98 %
E-value: 1e-155
NCBI BlastP on this gene
treC
treR
CFK40_20045
CFK40_20040
bacitracin ABC transporter ATP-binding protein
Accession: ASN07125
Location: 4202187-4203107
NCBI BlastP on this gene
Accession: ASN07125
Location: 4202187-4203107
NCBI BlastP on this gene
CFK40_20035
RNA polymerase subunit sigma-70
Accession: ASN07124
Location: 4201230-4201796
NCBI BlastP on this gene
Accession: ASN07124
Location: 4201230-4201796
NCBI BlastP on this gene
CFK40_20030
CFK40_20025
159. : CP002394 Bacillus cellulosilyticus DSM 2522 Total score: 1.0 Cumulative Blast bit score: 464
ABC transporter related protein
Accession: ADU29191
Location: 1039275-1040126
NCBI BlastP on this gene
Accession: ADU29191
Location: 1039275-1040126
NCBI BlastP on this gene
Bcell_0915
Bcell_0916
Bcell_0917
protein of unknown function DUF711
Accession: ADU29194
Location: 1041101-1042468
NCBI BlastP on this gene
Accession: ADU29194
Location: 1041101-1042468
NCBI BlastP on this gene
Bcell_0918
ACT domain-containing protein
Accession: ADU29195
Location: 1042483-1042758
NCBI BlastP on this gene
Accession: ADU29195
Location: 1042483-1042758
NCBI BlastP on this gene
Bcell_0919
Bcell_0920
alpha,alpha-phosphotrehalase
Accession: ADU29197
Location: 1044253-1045938
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 5e-154
NCBI BlastP on this gene
Accession: ADU29197
Location: 1044253-1045938
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 464
Sequence coverage: 100 %
E-value: 5e-154
NCBI BlastP on this gene
Bcell_0921
160. : CP001878 Bacillus pseudofirmus OF4 Total score: 1.0 Cumulative Blast bit score: 462
BpOF4_10335
BpOF4_10340
Phosphotransferase system (PTS) trehalose-specific enzyme II, BC component
Accession: ADC50122
Location: 501460-502872
NCBI BlastP on this gene
Accession: ADC50122
Location: 501460-502872
NCBI BlastP on this gene
treP
alpha,alpha-phosphotrehalase
Accession: ADC50123
Location: 503025-504704
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 462
Sequence coverage: 92 %
E-value: 5e-153
NCBI BlastP on this gene
Accession: ADC50123
Location: 503025-504704
BlastP hit with treA
Percentage identity: 47 %
BlastP bit score: 462
Sequence coverage: 92 %
E-value: 5e-153
NCBI BlastP on this gene
BpOF4_10350
GntR family transcriptional repressor of trehalose operon
Accession: ADC50124
Location: 504729-505445
NCBI BlastP on this gene
Accession: ADC50124
Location: 504729-505445
NCBI BlastP on this gene
BpOF4_10355
BpOF4_10360
Nitric oxide synthase, oxygenase domain protein
Accession: ADC50126
Location: 506432-507532
NCBI BlastP on this gene
Accession: ADC50126
Location: 506432-507532
NCBI BlastP on this gene
nos
ABC transporter ATP-binding protein
Accession: ADC50127
Location: 507719-509602
NCBI BlastP on this gene
Accession: ADC50127
Location: 507719-509602
NCBI BlastP on this gene
BpOF4_10370
161. : CP040367 Bacillus flexus isolate 1-2-1 chromosome Total score: 1.0 Cumulative Blast bit score: 461
alpha,alpha-phosphotrehalase
Accession: QCS54022
Location: 3014890-3016575
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
Accession: QCS54022
Location: 3014890-3016575
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
treC
treR
pxpB
biotin-dependent carboxyltransferase
Accession: QCS54019
Location: 3012188-3013195
NCBI BlastP on this gene
Accession: QCS54019
Location: 3012188-3013195
NCBI BlastP on this gene
FED53_16180
FED53_16175
divalent metal cation transporter
Accession: FED53_16170
Location: 3011711-3012103
NCBI BlastP on this gene
Accession: FED53_16170
Location: 3011711-3012103
NCBI BlastP on this gene
FED53_16170
FED53_16165
FED53_16160
162. : CP015506 Bacillus oceanisediminis 2691 chromosome Total score: 1.0 Cumulative Blast bit score: 461
A361_08210
NAD-dependent protein deacylase
Accession: AND39100
Location: 1603930-1604643
NCBI BlastP on this gene
Accession: AND39100
Location: 1603930-1604643
NCBI BlastP on this gene
A361_08215
preprotein translocase subunit TatA
Accession: AND39101
Location: 1604761-1604934
NCBI BlastP on this gene
Accession: AND39101
Location: 1604761-1604934
NCBI BlastP on this gene
A361_08220
trehalose permease IIC protein
Accession: AND39102
Location: 1605218-1606624
NCBI BlastP on this gene
Accession: AND39102
Location: 1605218-1606624
NCBI BlastP on this gene
A361_08225
A361_08230
alpha,alpha-phosphotrehalase
Accession: AND39104
Location: 1608429-1610114
BlastP hit with treA
Percentage identity: 43 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
Accession: AND39104
Location: 1608429-1610114
BlastP hit with treA
Percentage identity: 43 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 2e-152
NCBI BlastP on this gene
A361_08235
A361_08240
A361_08245
peptide chain release factor 3
Accession: AND39107
Location: 1612903-1614480
NCBI BlastP on this gene
Accession: AND39107
Location: 1612903-1614480
NCBI BlastP on this gene
A361_08250
163. : CP013019 Clostridium pasteurianum strain M150B Total score: 1.0 Cumulative Blast bit score: 461
AQ984_01810
AQ984_01805
5-aminoimidazole-4-carboxamide ribonucleotide transformylase
Accession: AOZ77698
Location: 400206-401381
NCBI BlastP on this gene
Accession: AOZ77698
Location: 400206-401381
NCBI BlastP on this gene
AQ984_01800
PTS beta-glucoside transporter subunit IIABC
Accession: AOZ77697
Location: 397856-399799
NCBI BlastP on this gene
Accession: AOZ77697
Location: 397856-399799
NCBI BlastP on this gene
AQ984_01795
trehalose-6-phosphate hydrolase
Accession: AOZ77696
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AOZ77696
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
AQ984_01790
GntR family transcriptional regulator
Accession: AOZ77695
Location: 395268-395978
NCBI BlastP on this gene
Accession: AOZ77695
Location: 395268-395978
NCBI BlastP on this gene
AQ984_01785
AQ984_01780
HxlR family transcriptional regulator
Accession: AOZ77693
Location: 393710-394018
NCBI BlastP on this gene
Accession: AOZ77693
Location: 393710-394018
NCBI BlastP on this gene
AQ984_01775
AQ984_01770
AQ984_01765
AQ984_01760
164. : CP013018 Clostridium pasteurianum DSM 525 = ATCC 6013 Total score: 1.0 Cumulative Blast bit score: 461
AQ983_01810
AQ983_01805
5-aminoimidazole-4-carboxamide ribonucleotide transformylase
Accession: AOZ73901
Location: 400206-401381
NCBI BlastP on this gene
Accession: AOZ73901
Location: 400206-401381
NCBI BlastP on this gene
AQ983_01800
PTS beta-glucoside transporter subunit IIABC
Accession: AOZ73900
Location: 397856-399799
NCBI BlastP on this gene
Accession: AOZ73900
Location: 397856-399799
NCBI BlastP on this gene
AQ983_01795
trehalose-6-phosphate hydrolase
Accession: AOZ73899
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AOZ73899
Location: 396067-397746
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
AQ983_01790
GntR family transcriptional regulator
Accession: AOZ73898
Location: 395268-395978
NCBI BlastP on this gene
Accession: AOZ73898
Location: 395268-395978
NCBI BlastP on this gene
AQ983_01785
AQ983_01780
HxlR family transcriptional regulator
Accession: AOZ73896
Location: 393710-394018
NCBI BlastP on this gene
Accession: AOZ73896
Location: 393710-394018
NCBI BlastP on this gene
AQ983_01775
AQ983_01770
AQ983_01765
AQ983_01760
165. : CP009268 Clostridium pasteurianum DSM 525 = ATCC 6013 Total score: 1.0 Cumulative Blast bit score: 461
CLPA_c03730
CLPA_c03720
bifunctional purine biosynthesis protein PurH
Accession: AJA50459
Location: 400208-401383
NCBI BlastP on this gene
Accession: AJA50459
Location: 400208-401383
NCBI BlastP on this gene
purH1
PTS system trehalose-specific EIIBC component
Accession: AJA50458
Location: 397858-399801
NCBI BlastP on this gene
Accession: AJA50458
Location: 397858-399801
NCBI BlastP on this gene
treP
trehalose-6-phosphate hydrolase
Accession: AJA50457
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AJA50457
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
treA
trehalose operon transcriptional repressor
Accession: AJA50456
Location: 395270-395980
NCBI BlastP on this gene
Accession: AJA50456
Location: 395270-395980
NCBI BlastP on this gene
treR
CLPA_c03670
transcriptional regulator, HxlR family
Accession: AJA50454
Location: 393712-394020
NCBI BlastP on this gene
Accession: AJA50454
Location: 393712-394020
NCBI BlastP on this gene
CLPA_c03660
CLPA_c03650
CLPA_c03640
transcriptional regulator, TetR family
Accession: AJA50451
Location: 391286-391915
NCBI BlastP on this gene
Accession: AJA50451
Location: 391286-391915
NCBI BlastP on this gene
CLPA_c03630
166. : CP009267 Clostridium pasteurianum DSM 525 = ATCC 6013 Total score: 1.0 Cumulative Blast bit score: 461
CPAST_c03730
CPAST_c03720
bifunctional purine biosynthesis protein PurH
Accession: AJA46471
Location: 400208-401383
NCBI BlastP on this gene
Accession: AJA46471
Location: 400208-401383
NCBI BlastP on this gene
purH1
PTS system trehalose-specific EIIBC component
Accession: AJA46470
Location: 397858-399801
NCBI BlastP on this gene
Accession: AJA46470
Location: 397858-399801
NCBI BlastP on this gene
treP
trehalose-6-phosphate hydrolase
Accession: AJA46469
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
Accession: AJA46469
Location: 396069-397748
BlastP hit with treA
Percentage identity: 44 %
BlastP bit score: 461
Sequence coverage: 101 %
E-value: 8e-153
NCBI BlastP on this gene
treA
trehalose operon transcriptional repressor
Accession: AJA46468
Location: 395270-395980
NCBI BlastP on this gene
Accession: AJA46468
Location: 395270-395980
NCBI BlastP on this gene
treR
CPAST_c03670
transcriptional regulator, HxlR family
Accession: AJA46466
Location: 393712-394020
NCBI BlastP on this gene
Accession: AJA46466
Location: 393712-394020
NCBI BlastP on this gene
CPAST_c03660
CPAST_c03650
CPAST_c03640
transcriptional regulator, TetR family
Accession: AJA46463
Location: 391286-391915
NCBI BlastP on this gene
Accession: AJA46463
Location: 391286-391915
NCBI BlastP on this gene
CPAST_c03630
167. : CP022535 Spiroplasma corruscae strain EC-1 chromosome Total score: 1.0 Cumulative Blast bit score: 430
PTS system, trehalose-specific IIBC component
Accession: ASP28428
Location: 765801-767363
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 431
Sequence coverage: 93 %
E-value: 9e-142
NCBI BlastP on this gene
Accession: ASP28428
Location: 765801-767363
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 431
Sequence coverage: 93 %
E-value: 9e-142
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: ASP28427
Location: 764166-765788
NCBI BlastP on this gene
Accession: ASP28427
Location: 764166-765788
NCBI BlastP on this gene
treC
SCORR_v1c06540
SCORR_v1c06530
168. : CP031376 Spiroplasma alleghenense strain PLHS-1 chromosome Total score: 1.0 Cumulative Blast bit score: 430
PTS system, trehalose-specific IIBC component
Accession: AXK51694
Location: 1177891-1179567
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 430
Sequence coverage: 97 %
E-value: 6e-141
NCBI BlastP on this gene
Accession: AXK51694
Location: 1177891-1179567
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 430
Sequence coverage: 97 %
E-value: 6e-141
NCBI BlastP on this gene
treB
SALLE_v1c10230
SALLE_v1c10220
SALLE_v1c10210
SALLE_v1c10200
SALLE_v1c10190
SALLE_v1c10180
169. : CP025257 Mesoplasma syrphidae strain YJS chromosome Total score: 1.0 Cumulative Blast bit score: 426
PTS sugar transporter subunit IIA
Accession: AUF83741
Location: 692942-694519
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 426
Sequence coverage: 95 %
E-value: 6e-140
NCBI BlastP on this gene
Accession: AUF83741
Location: 692942-694519
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 426
Sequence coverage: 95 %
E-value: 6e-140
NCBI BlastP on this gene
CXP39_02940
CXP39_02935
CXP39_02930
phosphoenolpyruvate--protein phosphotransferase
Accession: AUF83738
Location: 689137-690858
NCBI BlastP on this gene
Accession: AUF83738
Location: 689137-690858
NCBI BlastP on this gene
ptsP
PTS glucose transporter subunit IIA
Accession: AUF83737
Location: 688567-689043
NCBI BlastP on this gene
Accession: AUF83737
Location: 688567-689043
NCBI BlastP on this gene
CXP39_02920
CXP39_02915
170. : CP043026 Spiroplasma chinense strain CCH chromosome Total score: 1.0 Cumulative Blast bit score: 422
methionyl-tRNA formyltransferase
Accession: QEH61694
Location: 554037-554987
NCBI BlastP on this gene
Accession: QEH61694
Location: 554037-554987
NCBI BlastP on this gene
fmt
SCHIN_v1c04980
efp
SCHIN_v1c05000
pldB
SCHIN_v1c05020
SCHIN_v1c05030
PTS system, trehalose-specific IIB component
Accession: QEH61701
Location: 559595-561274
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 422
Sequence coverage: 97 %
E-value: 9e-138
NCBI BlastP on this gene
Accession: QEH61701
Location: 559595-561274
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 422
Sequence coverage: 97 %
E-value: 9e-138
NCBI BlastP on this gene
treB
SCHIN_v1c05050
mapA
pgmB
SCHIN_v1c05080
171. : CP024870 Spiroplasma clarkii strain CN-5 chromosome Total score: 1.0 Cumulative Blast bit score: 419
SCLAR_v1c11980
GntR family transcriptional regulator
Accession: ATX71499
Location: 1329155-1329832
NCBI BlastP on this gene
Accession: ATX71499
Location: 1329155-1329832
NCBI BlastP on this gene
SCLAR_v1c11990
SCLAR_v1c12000
SCLAR_v1c12010
PTS system, trehalose-specific IIB component
Accession: ATX71502
Location: 1332100-1333767
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 9e-137
NCBI BlastP on this gene
Accession: ATX71502
Location: 1332100-1333767
BlastP hit with treP
Percentage identity: 44 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 9e-137
NCBI BlastP on this gene
treB
172. : CP013860 Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 412
PTS system, trehalose-specific IIBC component
Accession: ALX70904
Location: 766378-767934
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
Accession: ALX70904
Location: 766378-767934
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: ALX70903
Location: 764747-766363
NCBI BlastP on this gene
Accession: ALX70903
Location: 764747-766363
NCBI BlastP on this gene
treC
STURO_v1c06430
STURO_v1c06420
STURO_v1c06410
ldhA
173. : CP012328 Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 412
PTS system, trehalose-specific IIB component PTS system, trehalose-specific IIC component
Accession: AKU79893
Location: 766480-768036
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
Accession: AKU79893
Location: 766480-768036
BlastP hit with treP
Percentage identity: 46 %
BlastP bit score: 412
Sequence coverage: 95 %
E-value: 1e-134
NCBI BlastP on this gene
STURON_00647
trehalose-6-phosphate hydrolase
Accession: AKU79892
Location: 764849-766465
NCBI BlastP on this gene
Accession: AKU79892
Location: 764849-766465
NCBI BlastP on this gene
treA
STURON_00645
STURON_00644
STURON_00643
ldhA
174. : CP006681 Spiroplasma culicicola AES-1 Total score: 1.0 Cumulative Blast bit score: 408
SCULI_v1c04520
SCULI_v1c04510
SCULI_v1c04500
SCULI_v1c04490
SCULI_v1c04480
SCULI_v1c04470
PTS system trehalose-specific IIBC component
Accession: AHI52787
Location: 492197-493849
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 408
Sequence coverage: 97 %
E-value: 1e-132
NCBI BlastP on this gene
Accession: AHI52787
Location: 492197-493849
BlastP hit with treP
Percentage identity: 45 %
BlastP bit score: 408
Sequence coverage: 97 %
E-value: 1e-132
NCBI BlastP on this gene
treB
SCULI_v1c04450
pgmB
mapA
SCULI_v1c04420
SCULI_v1c04410
175. : CP025420 Streptococcus parauberis strain SPOF3K chromosome Total score: 1.0 Cumulative Blast bit score: 313
SPSF3K_00109
SPSF3K_00110
[Formate-C-acetyltransferase]-activating enzyme
Accession: AUT04853
Location: 96866-97480
NCBI BlastP on this gene
Accession: AUT04853
Location: 96866-97480
NCBI BlastP on this gene
nrdG
SPSF3K_00112
catE
Streptomycin 3''-adenylyltransferase
Accession: AUT04856
Location: 99438-99764
NCBI BlastP on this gene
Accession: AUT04856
Location: 99438-99764
NCBI BlastP on this gene
aadA
HTH-type transcriptional regulator GmuR
Accession: AUT04857
Location: 99788-100522
NCBI BlastP on this gene
Accession: AUT04857
Location: 99788-100522
NCBI BlastP on this gene
SPSF3K_00115
Protein-N(pi)-phosphohistidine--sugar phosphotransferase
Accession: AUT04858
Location: 100723-102732
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: AUT04858
Location: 100723-102732
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 1e-94
NCBI BlastP on this gene
treP
treC
SPSF3K_00118
Protein-N(pi)-phosphohistidine--sugar phosphotransferase
Accession: AUT04861
Location: 106686-106970
NCBI BlastP on this gene
Accession: AUT04861
Location: 106686-106970
NCBI BlastP on this gene
ulaB
Ascorbate-specific permease IIC component UlaA
Accession: AUT04862
Location: 106983-108323
NCBI BlastP on this gene
Accession: AUT04862
Location: 106983-108323
NCBI BlastP on this gene
SPSF3K_00120
176. : CP002471 Streptococcus parauberis KCTC 11537 Total score: 1.0 Cumulative Blast bit score: 313
sugar phosphotransferase system (PTS), IIABC component
Accession: AEF26225
Location: 2042784-2044769
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 8e-95
NCBI BlastP on this gene
Accession: AEF26225
Location: 2042784-2044769
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 313
Sequence coverage: 96 %
E-value: 8e-95
NCBI BlastP on this gene
STP_1777
trehalose-6-phosphate hydrolase
Accession: AEF26224
Location: 2041091-2042716
NCBI BlastP on this gene
Accession: AEF26224
Location: 2041091-2042716
NCBI BlastP on this gene
treA
STP_1775
sugar-specific permease, SgaT/UlaA family
Accession: AEF26222
Location: 2037194-2038534
NCBI BlastP on this gene
Accession: AEF26222
Location: 2037194-2038534
NCBI BlastP on this gene
STP_1774
177. : LR595858 Streptococcus sp. NCTC 10228 strain NCTC10228 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 311
PTS system sugar transporter subunit IIABC
Accession: VUD00898
Location: 2040139-2042133
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
Accession: VUD00898
Location: 2040139-2042133
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: VUD00895
Location: 2038442-2040073
NCBI BlastP on this gene
Accession: VUD00895
Location: 2038442-2040073
NCBI BlastP on this gene
treA
pepO
NCTC10228_02064
tsf
178. : LR134341 Streptococcus pseudoporcinus strain NCTC13786 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 311
anaerobic ribonucleoside triphosphate reductase
Accession: VEF93911
Location: 1303615-1305810
NCBI BlastP on this gene
Accession: VEF93911
Location: 1303615-1305810
NCBI BlastP on this gene
nrdD_2
NCTC13786_01278
mviM
GNAT family acetyltransferase
Accession: VEF93914
Location: 1307036-1307539
NCBI BlastP on this gene
Accession: VEF93914
Location: 1307036-1307539
NCBI BlastP on this gene
NCTC13786_01280
anaerobic ribonucleoside-triphosphate reductase activating protein
Accession: VEF93915
Location: 1307546-1308160
NCBI BlastP on this gene
Accession: VEF93915
Location: 1307546-1308160
NCBI BlastP on this gene
pflA_1
hypothetical cytosolic protein
Accession: VEF93916
Location: 1308281-1309012
NCBI BlastP on this gene
Accession: VEF93916
Location: 1308281-1309012
NCBI BlastP on this gene
yaaA
GntR family transcriptional regulator
Accession: VEF93917
Location: 1309145-1309858
NCBI BlastP on this gene
Accession: VEF93917
Location: 1309145-1309858
NCBI BlastP on this gene
treR
PTS system sugar transporter subunit IIABC
Accession: VEF93918
Location: 1310075-1312069
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
Accession: VEF93918
Location: 1310075-1312069
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 311
Sequence coverage: 96 %
E-value: 1e-93
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: VEF93919
Location: 1312135-1313766
NCBI BlastP on this gene
Accession: VEF93919
Location: 1312135-1313766
NCBI BlastP on this gene
treA
pepO
NCTC13786_01287
tsf
179. : LS483383 Streptococcus cristatus strain NCTC12479 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 310
phosphoenolpyruvate carboxykinase (ATP)
Accession: SQG31908
Location: 561604-563250
NCBI BlastP on this gene
Accession: SQG31908
Location: 561604-563250
NCBI BlastP on this gene
NCTC12479_00557
NCTC12479_00558
GntR family transcriptional regulator
Accession: SQG31910
Location: 565036-565749
NCBI BlastP on this gene
Accession: SQG31910
Location: 565036-565749
NCBI BlastP on this gene
treR
trehalose PTS system, IIABC components
Accession: SQG31911
Location: 565936-567903
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 310
Sequence coverage: 95 %
E-value: 2e-93
NCBI BlastP on this gene
Accession: SQG31911
Location: 565936-567903
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 310
Sequence coverage: 95 %
E-value: 2e-93
NCBI BlastP on this gene
treP
dexS
pyruvate formate-lyase-activating enzyme
Accession: SQG31913
Location: 569968-570765
NCBI BlastP on this gene
Accession: SQG31913
Location: 569968-570765
NCBI BlastP on this gene
act
glycosyl transferase family protein
Accession: SQG31914
Location: 571269-572492
NCBI BlastP on this gene
Accession: SQG31914
Location: 571269-572492
NCBI BlastP on this gene
gspA
180. : CP050133 Streptococcus cristatus ATCC 51100 chromosome Total score: 1.0 Cumulative Blast bit score: 308
PTS system trehalose-specific EIIBC component
Accession: QIP49760
Location: 1439035-1441002
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 95 %
E-value: 6e-93
NCBI BlastP on this gene
Accession: QIP49760
Location: 1439035-1441002
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 308
Sequence coverage: 95 %
E-value: 6e-93
NCBI BlastP on this gene
treP
treC
pyruvate formate lyase-activating protein
Accession: QIP49758
Location: 1436172-1436969
NCBI BlastP on this gene
Accession: QIP49758
Location: 1436172-1436969
NCBI BlastP on this gene
pflA
HBA50_07140
HBA50_07135
181. : CP002394 Bacillus cellulosilyticus DSM 2522 Total score: 1.0 Cumulative Blast bit score: 308
SUF system FeS assembly protein, NifU family
Accession: ADU29113
Location: 959073-959522
NCBI BlastP on this gene
Accession: ADU29113
Location: 959073-959522
NCBI BlastP on this gene
Bcell_0834
Bcell_0836
GCN5-related N-acetyltransferase
Accession: ADU29115
Location: 961158-961712
NCBI BlastP on this gene
Accession: ADU29115
Location: 961158-961712
NCBI BlastP on this gene
Bcell_0837
Bcell_0838
Bcell_0839
Bcell_0840
PTS system, trehalose-specific IIBC subunit
Accession: ADU29119
Location: 964229-965665
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 308
Sequence coverage: 86 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: ADU29119
Location: 964229-965665
BlastP hit with treP
Percentage identity: 40 %
BlastP bit score: 308
Sequence coverage: 86 %
E-value: 1e-94
NCBI BlastP on this gene
Bcell_0841
DNA-(apurinic or apyrimidinic site) lyase
Accession: ADU29120
Location: 965819-966613
NCBI BlastP on this gene
Accession: ADU29120
Location: 965819-966613
NCBI BlastP on this gene
Bcell_0842
Bcell_0843
RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: ADU29122
Location: 967481-968005
NCBI BlastP on this gene
Accession: ADU29122
Location: 967481-968005
NCBI BlastP on this gene
Bcell_0844
cell envelope-related transcriptional attenuator
Accession: ADU29123
Location: 967998-968957
NCBI BlastP on this gene
Accession: ADU29123
Location: 967998-968957
NCBI BlastP on this gene
Bcell_0845
small acid-soluble spore protein alpha/beta type
Accession: ADU29124
Location: 969096-969296
NCBI BlastP on this gene
Accession: ADU29124
Location: 969096-969296
NCBI BlastP on this gene
Bcell_0846
PAS/PAC sensor signal transduction histidine kinase
Accession: ADU29125
Location: 969533-971368
NCBI BlastP on this gene
Accession: ADU29125
Location: 969533-971368
NCBI BlastP on this gene
Bcell_0847
182. : LS483408 Streptococcus uberis strain NCTC4674 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 307
PTS system sugar transporter subunit IIABC
Accession: SQG84040
Location: 1923576-1925561
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: SQG84040
Location: 1923576-1925561
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: SQG84039
Location: 1921877-1923508
NCBI BlastP on this gene
Accession: SQG84039
Location: 1921877-1923508
NCBI BlastP on this gene
treA
ulaC_2
PTS system ascorbate-specific transporter subunits IICB
Accession: SQG84037
Location: 1919314-1919598
NCBI BlastP on this gene
Accession: SQG84037
Location: 1919314-1919598
NCBI BlastP on this gene
NCTC4674_02014
SgaT/UlaA family sugar-specific permease
Accession: SQG84036
Location: 1917958-1919301
NCBI BlastP on this gene
Accession: SQG84036
Location: 1917958-1919301
NCBI BlastP on this gene
ulaA_2
183. : CP022435 Streptococcus uberis strain NZ01 chromosome Total score: 1.0 Cumulative Blast bit score: 307
PTS beta-glucoside transporter subunit EIIBCA
Accession: AUC25774
Location: 1774797-1776782
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: AUC25774
Location: 1774797-1776782
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
CGZ53_09000
treC
transcription antiterminator BglG
Accession: AUC25772
Location: 1770823-1772865
NCBI BlastP on this gene
Accession: AUC25772
Location: 1770823-1772865
NCBI BlastP on this gene
CGZ53_08990
PTS lactose transporter subunit IIB
Accession: AUC25771
Location: 1770534-1770818
NCBI BlastP on this gene
Accession: AUC25771
Location: 1770534-1770818
NCBI BlastP on this gene
CGZ53_08985
PTS ascorbate transporter subunit IIC
Accession: AUC25770
Location: 1769178-1770521
NCBI BlastP on this gene
Accession: AUC25770
Location: 1769178-1770521
NCBI BlastP on this gene
CGZ53_08980
184. : AM946015 Streptococcus uberis 0140J complete genome. Total score: 1.0 Cumulative Blast bit score: 307
sugar phosphotransferase system (PTS), IIABC component
Accession: CAR43740
Location: 1750789-1752774
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
Accession: CAR43740
Location: 1750789-1752774
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 307
Sequence coverage: 96 %
E-value: 2e-92
NCBI BlastP on this gene
SUB1763
putative trehalose-6-phosphate hydrolase
Accession: CAR43735
Location: 1749090-1750721
NCBI BlastP on this gene
Accession: CAR43735
Location: 1749090-1750721
NCBI BlastP on this gene
treA
putative PTS multi-domain regulator
Accession: CAR43734
Location: 1746815-1748857
NCBI BlastP on this gene
Accession: CAR43734
Location: 1746815-1748857
NCBI BlastP on this gene
SUB1761
sugar phosphotransferase system (PTS),
Accession: CAR43732
Location: 1746526-1746810
NCBI BlastP on this gene
Accession: CAR43732
Location: 1746526-1746810
NCBI BlastP on this gene
SUB1760
putative sugar-specific permease, SgaT/UlaA family
Accession: CAR43730
Location: 1745170-1746513
NCBI BlastP on this gene
Accession: CAR43730
Location: 1745170-1746513
NCBI BlastP on this gene
SUB1759
185. : LS483397 Streptococcus uberis strain NCTC3858 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 305
PTS system sugar transporter subunit IIABC
Accession: SQG47165
Location: 1886561-1888546
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 96 %
E-value: 1e-91
NCBI BlastP on this gene
Accession: SQG47165
Location: 1886561-1888546
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 96 %
E-value: 1e-91
NCBI BlastP on this gene
treB
trehalose-6-phosphate hydrolase
Accession: SQG47164
Location: 1884862-1886493
NCBI BlastP on this gene
Accession: SQG47164
Location: 1884862-1886493
NCBI BlastP on this gene
treA
ulaC_3
PTS system ascorbate-specific transporter subunits IICB
Accession: SQG47162
Location: 1882298-1882582
NCBI BlastP on this gene
Accession: SQG47162
Location: 1882298-1882582
NCBI BlastP on this gene
NCTC3858_01941
SgaT/UlaA family sugar-specific permease
Accession: SQG47161
Location: 1880942-1882285
NCBI BlastP on this gene
Accession: SQG47161
Location: 1880942-1882285
NCBI BlastP on this gene
ulaA_3
186. : CP028415 Streptococcus mitis strain SK637 chromosome Total score: 1.0 Cumulative Blast bit score: 305
SK637_00328
SK637_00329
ABC transporter substrate binding family protein
Accession: QBZ12892
Location: 320443-321252
NCBI BlastP on this gene
Accession: QBZ12892
Location: 320443-321252
NCBI BlastP on this gene
SK637_00330
branched-chain amino acid transport system / permease component family protein
Accession: QBZ12893
Location: 321263-322156
NCBI BlastP on this gene
Accession: QBZ12893
Location: 321263-322156
NCBI BlastP on this gene
SK637_00331
SK637_00332
SK637_00333
PTS system trehalose-specific IIBC component
Accession: QBZ12896
Location: 323900-325867
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 93 %
E-value: 1e-91
NCBI BlastP on this gene
Accession: QBZ12896
Location: 323900-325867
BlastP hit with treP
Percentage identity: 38 %
BlastP bit score: 305
Sequence coverage: 93 %
E-value: 1e-91
NCBI BlastP on this gene
SK637_00334
SK637_00335
SK637_00336
SK637_00337
non-canonical purine NTP pyrophosphatase RdgB
Accession: QBZ12900
Location: 328942-329913
NCBI BlastP on this gene
Accession: QBZ12900
Location: 328942-329913
NCBI BlastP on this gene
SK637_00338
SK637_00339
187. : CP018187 Streptococcus salivarius strain ICDC2 Total score: 1.0 Cumulative Blast bit score: 305
PTS beta-glucoside transporter subunit EIIBCA
Accession: QGU81410
Location: 2140805-2142784
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
Accession: QGU81410
Location: 2140805-2142784
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
BSR19_09965
BSR19_09960
BSR19_09955
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: QGU81407
Location: 2134411-2136999
NCBI BlastP on this gene
Accession: QGU81407
Location: 2134411-2136999
NCBI BlastP on this gene
BSR19_09950
188. : CP002888 Streptococcus salivarius 57.I Total score: 1.0 Cumulative Blast bit score: 305
Ssal_00112
fructose-1,6-bisphosphate aldolase, class II
Accession: AEJ52537
Location: 98328-99209
NCBI BlastP on this gene
Accession: AEJ52537
Location: 98328-99209
NCBI BlastP on this gene
fbaA
Ssal_00115
Ssal_00116
Ssal_00117
treR
pts system, trehalose-specific iibc component
Accession: AEJ52542
Location: 101771-103816
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 4e-91
NCBI BlastP on this gene
Accession: AEJ52542
Location: 101771-103816
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 305
Sequence coverage: 95 %
E-value: 4e-91
NCBI BlastP on this gene
treB
treC
pepO
aldehyde-alcohol dehydrogenase 2
Accession: AEJ52545
Location: 107614-110211
NCBI BlastP on this gene
Accession: AEJ52545
Location: 107614-110211
NCBI BlastP on this gene
adhE
189. : CP015282 Streptococcus salivarius strain ATCC 27945 Total score: 1.0 Cumulative Blast bit score: 304
gamma-glutamylcysteine synthetase
Accession: ARI58245
Location: 1616084-1617379
NCBI BlastP on this gene
Accession: ARI58245
Location: 1616084-1617379
NCBI BlastP on this gene
NX99_07305
fructose-bisphosphate aldolase
Accession: ARI58246
Location: 1617803-1618684
NCBI BlastP on this gene
Accession: ARI58246
Location: 1617803-1618684
NCBI BlastP on this gene
NX99_07315
NX99_07320
NX99_07325
NX99_07330
PTS beta-glucoside transporter subunit EIIBCA
Accession: ARI58250
Location: 1621210-1623189
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
Accession: ARI58250
Location: 1621210-1623189
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 95 %
E-value: 2e-91
NCBI BlastP on this gene
NX99_07335
190. : AP018338 Streptococcus oralis subsp. tigurinus DNA Total score: 1.0 Cumulative Blast bit score: 304
PTS system trehalose-specific IIBC component
Accession: BBA09167
Location: 1594737-1596704
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 94 %
E-value: 4e-91
NCBI BlastP on this gene
Accession: BBA09167
Location: 1594737-1596704
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 304
Sequence coverage: 94 %
E-value: 4e-91
NCBI BlastP on this gene
treP
treC
uncharacterised protein family UPF0154
Accession: BBA09165
Location: 1592658-1592903
NCBI BlastP on this gene
Accession: BBA09165
Location: 1592658-1592903
NCBI BlastP on this gene
STO1_015610
murI
Non-canonical purine NTP pyrophosphatase
Accession: BBA09163
Location: 1590691-1591662
NCBI BlastP on this gene
Accession: BBA09163
Location: 1590691-1591662
NCBI BlastP on this gene
rdgB
STO1_015580
191. : LR134336 Streptococcus oralis strain NCTC11427 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 303
trehalose PTS system, IIABC components
Accession: VEF79619
Location: 1614753-1616720
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 7e-91
NCBI BlastP on this gene
Accession: VEF79619
Location: 1614753-1616720
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 7e-91
NCBI BlastP on this gene
treP
treC
NCTC11427_01597
murI
deoxyribonucleotide triphosphate
Accession: VEF79615
Location: 1610705-1611676
NCBI BlastP on this gene
Accession: VEF79615
Location: 1610705-1611676
NCBI BlastP on this gene
rdgB
NCTC11427_01594
192. : LR134323 Streptococcus urinalis strain NCTC13766 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 303
mviM
NCTC13766_00060
acetyltransferase (GNAT) family protein
Accession: VEF30997
Location: 59832-60329
NCBI BlastP on this gene
Accession: VEF30997
Location: 59832-60329
NCBI BlastP on this gene
NCTC13766_00061
anaerobic ribonucleoside-triphosphate reductase activating protein
Accession: VEF30998
Location: 60347-60955
NCBI BlastP on this gene
Accession: VEF30998
Location: 60347-60955
NCBI BlastP on this gene
pflA
yaaA
catE
trehalose operon transcriptional repressor
Accession: VEF31001
Location: 62803-63516
NCBI BlastP on this gene
Accession: VEF31001
Location: 62803-63516
NCBI BlastP on this gene
treR
PTS system transporter subunit IIABC
Accession: VEF31002
Location: 63735-65717
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: VEF31002
Location: 63735-65717
BlastP hit with treP
Percentage identity: 36 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 6e-91
NCBI BlastP on this gene
treB
treA
NADP or NAD utilizing oxidoreductase
Accession: VEF31004
Location: 67440-68420
NCBI BlastP on this gene
Accession: VEF31004
Location: 67440-68420
NCBI BlastP on this gene
afr
pepO
193. : LR134274 Streptococcus salivarius strain NCTC8618 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 303
PTS system trehalose-specific transporter subunit IIBC
Accession: VED90220
Location: 2101140-2103116
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
Accession: VED90220
Location: 2101140-2103116
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
treB_1
putative trehalose-6-phosphate hydrolase
Accession: VED90219
Location: 2099451-2101076
NCBI BlastP on this gene
Accession: VED90219
Location: 2099451-2101076
NCBI BlastP on this gene
treC
pepO
aldehyde-alcohol dehydrogenase 2
Accession: VED90217
Location: 2094746-2097334
NCBI BlastP on this gene
Accession: VED90217
Location: 2094746-2097334
NCBI BlastP on this gene
adhE_4
194. : CP040804 Streptococcus salivarius strain LAB813 chromosome Total score: 1.0 Cumulative Blast bit score: 303
IS30-like element IS1139 family transposase
Accession: QEM31807
Location: 250403-251428
NCBI BlastP on this gene
Accession: QEM31807
Location: 250403-251428
NCBI BlastP on this gene
FHI56_02420
FHI56_02430
FHI56_02435
FHI56_02440
FHI56_02445
treR
PTS trehalose transporter subunit IIBC
Location: 255186-257165
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 93 %
E-value: 7e-91
Location: 255186-257165
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 93 %
E-value: 7e-91
treB
treC
FHI56_02465
bifunctional acetaldehyde-CoA/alcohol dehydrogenase
Accession: QEM31814
Location: 260963-263560
NCBI BlastP on this gene
Accession: QEM31814
Location: 260963-263560
NCBI BlastP on this gene
adhE
195. : CP023507 Streptococcus oralis strain FDAARGOS_367 chromosome Total score: 1.0 Cumulative Blast bit score: 303
CO686_00930
treR
PTS trehalose transporter subunit IIBC
Accession: ATF57782
Location: 180261-182378
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 2e-90
NCBI BlastP on this gene
Accession: ATF57782
Location: 180261-182378
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 2e-90
NCBI BlastP on this gene
CO686_00940
treC
CO686_00950
196. : CP016786 Clostridium isatidis strain DSM 15098 chromosome Total score: 1.0 Cumulative Blast bit score: 303
BEN51_09190
BEN51_09185
energy-coupled thiamine transporter ThiT
Accession: ASW43645
Location: 1943664-1944287
NCBI BlastP on this gene
Accession: ASW43645
Location: 1943664-1944287
NCBI BlastP on this gene
BEN51_09180
BEN51_09175
BEN51_09170
BEN51_09165
PTS maltose transporter subunit IIBC
Accession: ASW43641
Location: 1939146-1940606
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 96 %
E-value: 9e-93
NCBI BlastP on this gene
Accession: ASW43641
Location: 1939146-1940606
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 96 %
E-value: 9e-93
NCBI BlastP on this gene
BEN51_09160
BEN51_09155
BEN51_09145
BEN51_09140
BEN51_09135
BEN51_09130
197. : CP016207 Streptococcus sp. oral taxon 064 strain W10853 sequence. Total score: 1.0 Cumulative Blast bit score: 303
PTS beta-glucoside transporter subunit EIIBCA
Accession: ANR74721
Location: 343050-345017
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: ANR74721
Location: 343050-345017
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 6e-91
NCBI BlastP on this gene
AXF18_01655
AXF18_01650
AXF18_01645
198. : CP014144 Streptococcus salivarius strain JF Total score: 1.0 Cumulative Blast bit score: 303
gamma-glutamylcysteine synthetase
Accession: AMB83681
Location: 2117640-2118935
NCBI BlastP on this gene
Accession: AMB83681
Location: 2117640-2118935
NCBI BlastP on this gene
AWB63_09820
fructose-bisphosphate aldolase
Accession: AMB83682
Location: 2119357-2120238
NCBI BlastP on this gene
Accession: AMB83682
Location: 2119357-2120238
NCBI BlastP on this gene
AWB63_09830
AWB63_09835
AWB63_09840
AWB63_09845
PTS beta-glucoside transporter subunit EIIBCA
Accession: AMB83686
Location: 2122723-2124699
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
Accession: AMB83686
Location: 2122723-2124699
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
AWB63_09850
199. : CP009913 Streptococcus salivarius strain NCTC 8618 Total score: 1.0 Cumulative Blast bit score: 303
PTS system trehalose-specific transporter subunits IIBCA
Accession: AIY21959
Location: 2084011-2085987
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
Accession: AIY21959
Location: 2084011-2085987
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 95 %
E-value: 1e-90
NCBI BlastP on this gene
SSAL8618_09625
SSAL8618_09620
SSAL8618_09615
SSAL8618_09610
200. : CP007628 Streptococcus sp. VT 162 Total score: 1.0 Cumulative Blast bit score: 303
GntR family transcriptional regulator
Accession: AHZ47129
Location: 305639-306352
NCBI BlastP on this gene
Accession: AHZ47129
Location: 305639-306352
NCBI BlastP on this gene
V470_01515
PTS system trehalose-specific transporter subunits IIBCA
Accession: AHZ47130
Location: 306537-308504
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 5e-91
NCBI BlastP on this gene
Accession: AHZ47130
Location: 306537-308504
BlastP hit with treP
Percentage identity: 37 %
BlastP bit score: 303
Sequence coverage: 94 %
E-value: 5e-91
NCBI BlastP on this gene
V470_01520
V470_01525
V470_01530
V470_01535
deoxyribonucleotide triphosphate pyrophosphatase
Accession: AHZ47134
Location: 311583-312554
NCBI BlastP on this gene
Accession: AHZ47134
Location: 311583-312554
NCBI BlastP on this gene
V470_01540
V470_01545


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.