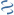MultiGeneBlast hits

Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP007520 : Mycoplasma yeatsii GM274B chromosome Total score: 1.0 Cumulative Blast bit score: 225
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ribosome biogenesis GTP-binding protein
Accession: AJM71880
Location: 471284-471871
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 226
Sequence coverage: 95 %
E-value: 3e-71
NCBI BlastP on this gene
Accession: AJM71880
Location: 471284-471871
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 226
Sequence coverage: 95 %
E-value: 3e-71
NCBI BlastP on this gene
engB
cation-transporting ATPase, E1-E2 family
Accession: AJM71879
Location: 468359-471277
NCBI BlastP on this gene
Accession: AJM71879
Location: 468359-471277
NCBI BlastP on this gene
MYE_01995
MYE_01990
rpsO
MYE_01975
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483504 : Mycoplasma mycoides subsp. capri strain GM12 genome assembly, chromosome: omosome. Total score: 1.0 Cumulative Blast bit score: 224
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MMC68I_00256
transcription elongation factor GreA
Accession: SRX61352
Location: 329891-330364
NCBI BlastP on this gene
Accession: SRX61352
Location: 329891-330364
NCBI BlastP on this gene
MMC68I_00255
SDR family NAD(P)-dependent oxidoreductase
Accession: SRX61351
Location: 329124-329846
NCBI BlastP on this gene
Accession: SRX61351
Location: 329124-329846
NCBI BlastP on this gene
MMC68I_00254
MMC68I_00253
MMC68I_00252
MMC68I_00251
MMC68I_00250
YihA family ribosome biogenesis GTP-binding protein
Accession: SRX61346
Location: 325933-326523
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 9e-71
NCBI BlastP on this gene
Accession: SRX61346
Location: 325933-326523
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 9e-71
NCBI BlastP on this gene
MMC68I_00249
cation-translocating P-type ATPase
Accession: SRX61345
Location: 323018-325924
NCBI BlastP on this gene
Accession: SRX61345
Location: 323018-325924
NCBI BlastP on this gene
MMC68I_00248
MMC68I_00247
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR739237 : Mycoplasma feriruminatoris 14/OD_0492 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
Transcription elongation factor GreA
Accession: VZS00575
Location: 731929-732402
NCBI BlastP on this gene
Accession: VZS00575
Location: 731929-732402
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZS00577
Location: 732450-733172
NCBI BlastP on this gene
Accession: VZS00577
Location: 732450-733172
NCBI BlastP on this gene
ydfG_2
MF5582_00669
MF5582_00670
MF5582_00671
MF5582_00672
putative GTP-binding protein EngB
Accession: VZS00587
Location: 735774-736364
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZS00587
Location: 735774-736364
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR739235 : Mycoplasma feriruminatoris 8756-13 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
Transcription elongation factor GreA
Accession: VZR98175
Location: 732040-732513
NCBI BlastP on this gene
Accession: VZR98175
Location: 732040-732513
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZR98177
Location: 732561-733283
NCBI BlastP on this gene
Accession: VZR98177
Location: 732561-733283
NCBI BlastP on this gene
ydfG_2
MF5295_00660
MF5295_00661
MF5295_00662
MF5295_00663
putative GTP-binding protein EngB
Accession: VZR98188
Location: 735886-736476
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZR98188
Location: 735886-736476
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR739234 : Mycoplasma feriruminatoris G1650 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
Transcription elongation factor GreA
Accession: VZR98111
Location: 704731-705204
NCBI BlastP on this gene
Accession: VZR98111
Location: 704731-705204
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZR98113
Location: 705252-705974
NCBI BlastP on this gene
Accession: VZR98113
Location: 705252-705974
NCBI BlastP on this gene
ydfG_2
MF5293_00644
MF5293_00645
MF5293_00646
MF5293_00647
putative GTP-binding protein EngB
Accession: VZR98123
Location: 708577-709167
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZR98123
Location: 708577-709167
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR739233 : Mycoplasma feriruminatoris G1705 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
Transcription elongation factor GreA
Accession: VZR75612
Location: 704742-705215
NCBI BlastP on this gene
Accession: VZR75612
Location: 704742-705215
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZR75613
Location: 705263-705985
NCBI BlastP on this gene
Accession: VZR75613
Location: 705263-705985
NCBI BlastP on this gene
ydfG_2
MF5294_00646
MF5294_00647
MF5294_00648
MF5294_00649
putative GTP-binding protein EngB
Accession: VZR75618
Location: 708588-709178
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZR75618
Location: 708588-709178
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR738858 : Mycoplasma feriruminatoris G5813/1+2 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
Transcription elongation factor GreA
Accession: VZK65469
Location: 703248-703721
NCBI BlastP on this gene
Accession: VZK65469
Location: 703248-703721
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZK65470
Location: 703769-704491
NCBI BlastP on this gene
Accession: VZK65470
Location: 703769-704491
NCBI BlastP on this gene
ydfG_2
MF5292_00648
MF5292_00649
MF5292_00650
MF5292_00651
putative GTP-binding protein EngB
Accession: VZK65475
Location: 707094-707684
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZK65475
Location: 707094-707684
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483505 : Mycoplasma mycoides subsp. capri strain GM12 genome assembly, chromosome: omosome. Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MMC68D_00257
transcription elongation factor GreA
Accession: SRX62734
Location: 330028-330501
NCBI BlastP on this gene
Accession: SRX62734
Location: 330028-330501
NCBI BlastP on this gene
MMC68D_00256
SDR family NAD(P)-dependent oxidoreductase
Accession: SRX62733
Location: 329261-329983
NCBI BlastP on this gene
Accession: SRX62733
Location: 329261-329983
NCBI BlastP on this gene
MMC68D_00255
MMC68D_00254
MMC68D_00253
MMC68D_00252
MMC68D_00251
YihA family ribosome biogenesis GTP-binding protein
Accession: SRX62728
Location: 326070-326660
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: SRX62728
Location: 326070-326660
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MMC68D_00250
cation-translocating P-type ATPase
Accession: SRX62727
Location: 323155-326061
NCBI BlastP on this gene
Accession: SRX62727
Location: 323155-326061
NCBI BlastP on this gene
MMC68D_00249
MMC68D_00248
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP024964 : Entomoplasma melaleucae strain M1 chromosome Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
GTP-binding protein
Accession: ATZ18040
Location: 465112-465708
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: ATZ18040
Location: 465112-465708
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
ctp
valS
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP014346 : Mycoplasma mycoides subsp. mycoides strain T1/44 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
Transcription elongation factor GreA
Accession: AMK56837
Location: 985643-986116
NCBI BlastP on this gene
Accession: AMK56837
Location: 985643-986116
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenaseYdfG
Accession: AMK56838
Location: 986159-986881
NCBI BlastP on this gene
Accession: AMK56838
Location: 986159-986881
NCBI BlastP on this gene
ydfG_2
MSCT144_09480
MSCT144_09490
MSCT144_09500
MSCT144_09510
putative GTP-binding protein EngB
Accession: AMK56843
Location: 989482-990072
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AMK56843
Location: 989482-990072
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011264 : Mycoplasma mycoides subsp. mycoides strain Ben468 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MmmBen468_0327
transcription elongation factor GreA
Accession: AME14557
Location: 332033-332506
NCBI BlastP on this gene
Accession: AME14557
Location: 332033-332506
NCBI BlastP on this gene
MmmBen468_0326
MmmBen468_0325
MmmBen468_0324
MmmBen468_0323
MmmBen468_0322
MmmBen468_0321
ribosome biogenesis GTP-binding protein YsxC
Accession: AME14551
Location: 328077-328667
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME14551
Location: 328077-328667
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen468_0320
MmmBen468_0319
MmmBen468_0318
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011263 : Mycoplasma mycoides subsp. mycoides strain Ben326 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MmmBen326_0315
transcription elongation factor GreA
Accession: AME13565
Location: 325254-325727
NCBI BlastP on this gene
Accession: AME13565
Location: 325254-325727
NCBI BlastP on this gene
MmmBen326_0314
MmmBen326_0313
MmmBen326_0312
MmmBen326_0311
MmmBen326_0310
ribosome biogenesis GTP-binding protein YsxC
Accession: AME13560
Location: 321292-321882
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME13560
Location: 321292-321882
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen326_0309
MmmBen326_0308
MmmBen326_0307
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011262 : Mycoplasma mycoides subsp. mycoides strain Ben181 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MmmBen181_0327
transcription elongation factor GreA
Accession: AME12532
Location: 329471-329944
NCBI BlastP on this gene
Accession: AME12532
Location: 329471-329944
NCBI BlastP on this gene
MmmBen181_0326
MmmBen181_0325
MmmBen181_0324
MmmBen181_0323
MmmBen181_0322
MmmBen181_0321
ribosome biogenesis GTP-binding protein YsxC
Accession: AME12526
Location: 325515-326105
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME12526
Location: 325515-326105
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen181_0320
MmmBen181_0319
MmmBen181_0318
MmmBen181_0317
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011261 : Mycoplasma mycoides subsp. mycoides strain Ben50 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MmmBen50_0315
transcription elongation factor GreA
Accession: AME11508
Location: 333197-333670
NCBI BlastP on this gene
Accession: AME11508
Location: 333197-333670
NCBI BlastP on this gene
MmmBen50_0314
MmmBen50_0313
MmmBen50_0312
MmmBen50_0311
MmmBen50_0310
MmmBen50_0309
ribosome biogenesis GTP-binding protein YsxC
Accession: AME11502
Location: 329241-329831
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME11502
Location: 329241-329831
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen50_0308
MmmBen50_0307
MmmBen50_0306
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP011260 : Mycoplasma mycoides subsp. mycoides strain Ben1 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
MmmBen_0320
transcription elongation factor GreA
Accession: AME10500
Location: 330148-330621
NCBI BlastP on this gene
Accession: AME10500
Location: 330148-330621
NCBI BlastP on this gene
MmmBen_0319
MmmBen_0318
MmmBen_0317
MmmBen_0316
MmmBen_0315
MmmBen_0314
ribosome biogenesis GTP-binding protein YsxC
Accession: AME10494
Location: 326192-326782
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME10494
Location: 326192-326782
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen_0313
MmmBen_0312
MmmBen_0311
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP010267 : Mycoplasma mycoides subsp. mycoides strain izsam_mm5713 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
transcription elongation factor GreA
Accession: AIZ55150
Location: 347146-347619
NCBI BlastP on this gene
Accession: AIZ55150
Location: 347146-347619
NCBI BlastP on this gene
greA
mycmycITA_00320
mycmycITA_00319
mycmycITA_00318
mycmycITA_00317
mycmycITA_00316
GTPase EngB
Accession: AIZ55144
Location: 343190-343780
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AIZ55144
Location: 343190-343780
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
mycmycITA_00314
mycmycITA_00313
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP004357 : Mycoplasma putrefaciens Mput9231 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
aspS
Phosphotransferase system PTS, lichenan-specific IIa component
Accession: AGJ90703
Location: 334196-334909
NCBI BlastP on this gene
Accession: AGJ90703
Location: 334196-334909
NCBI BlastP on this gene
licA
MPUT9231_2780
MPUT9231_2770
Hypothetical protein, predicted transmembrane protein
Accession: AGJ90700
Location: 332072-332707
NCBI BlastP on this gene
Accession: AGJ90700
Location: 332072-332707
NCBI BlastP on this gene
MPUT9231_2760
Hypothetical protein, predicted transmembrane protein
Accession: AGJ90699
Location: 331498-332082
NCBI BlastP on this gene
Accession: AGJ90699
Location: 331498-332082
NCBI BlastP on this gene
MPUT9231_2750
GTP-binding protein EngB
Accession: AGJ90698
Location: 330906-331493
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AGJ90698
Location: 330906-331493
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
ctp
valS
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP003021 : Mycoplasma putrefaciens KS1 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ribosome biogenesis GTP-binding protein
Accession: AEM68799
Location: 489822-490409
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AEM68799
Location: 489822-490409
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
ysxC
cation-transporting ATPase, E1-E2 family
Accession: AEM68798
Location: 486906-489809
NCBI BlastP on this gene
Accession: AEM68798
Location: 486906-489809
NCBI BlastP on this gene
MPUT_0425
valS
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002107 : Mycoplasma mycoides subsp. mycoides SC str. Gladysdale MU clone SC5 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
putative transcription elongation factor GreA
Accession: ADK70109
Location: 347145-347618
NCBI BlastP on this gene
Accession: ADK70109
Location: 347145-347618
NCBI BlastP on this gene
MMS_A0326
MMS_A0325
MMS_A0324
MMS_A0323
MMS_A0322
MMS_A0321
ribosome biogenesis GTP-binding protein YsxC
Accession: ADK69373
Location: 343189-343779
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: ADK69373
Location: 343189-343779
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
ysxC
MMS_A0319
MMS_A0318
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
BX293980 : Mycoplasma mycoides subsp. mycoides SC str. PG1 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
uvrC
transcription elongation factor
Accession: CAE76937
Location: 336160-336633
NCBI BlastP on this gene
Accession: CAE76937
Location: 336160-336633
NCBI BlastP on this gene
greA
MSC_0295
MSC_0294
MSC_0293
hypothetical transmembrane protein
Accession: CAE76933
Location: 333350-333928
NCBI BlastP on this gene
Accession: CAE76933
Location: 333350-333928
NCBI BlastP on this gene
MSC_0292
hypothetical transmembrane protein
Accession: CAE76932
Location: 332788-333348
NCBI BlastP on this gene
Accession: CAE76932
Location: 332788-333348
NCBI BlastP on this gene
MSC_0291
GTP-binding protein
Accession: CAE76931
Location: 332204-332794
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: CAE76931
Location: 332204-332794
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MSC_0290
ctp
MSC_0288
Conserved hypothetical transmembrane protein
Accession: CAE76928
Location: 327121-329010
NCBI BlastP on this gene
Accession: CAE76928
Location: 327121-329010
NCBI BlastP on this gene
MSC_0287
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP013860 : Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
STURO_v1c05150
STURO_v1c05140
STURO_v1c05130
STURO_v1c05120
STURO_v1c05110
GntR family transcriptional regulator
Accession: ALX70776
Location: 592159-592959
NCBI BlastP on this gene
Accession: ALX70776
Location: 592159-592959
NCBI BlastP on this gene
STURO_v1c05100
STURO_v1c05090
GTP-binding protein YsxC
Accession: ALX70774
Location: 590967-591563
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
Accession: ALX70774
Location: 590967-591563
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
engB
STURO_v1c05070
cell shape determining protein MreB
Accession: ALX70772
Location: 588853-589896
NCBI BlastP on this gene
Accession: ALX70772
Location: 588853-589896
NCBI BlastP on this gene
mreB
STURO_v1c05050
Holliday junction DNA helicase RuvA
Accession: ALX70770
Location: 587822-588370
NCBI BlastP on this gene
Accession: ALX70770
Location: 587822-588370
NCBI BlastP on this gene
ruvA
Holliday junction DNA helicase RuvB
Accession: ALX70769
Location: 586891-587838
NCBI BlastP on this gene
Accession: ALX70769
Location: 586891-587838
NCBI BlastP on this gene
ruvB
STURO_v1c05020
STURO_v1c05010
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP012328 : Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
yqkD
STURON_00516
STURON_00515
STURON_00514
STURON_00513
Putative transcriptional regulator, RpiR family protein
Accession: AKU79758
Location: 592261-593058
NCBI BlastP on this gene
Accession: AKU79758
Location: 592261-593058
NCBI BlastP on this gene
STURON_00512
STURON_00511
GTP-binding protein YsxC
Accession: AKU79756
Location: 591069-591665
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
Accession: AKU79756
Location: 591069-591665
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
engB
STURON_00509
cell shape determining protein MreB
Accession: AKU79754
Location: 588955-589998
NCBI BlastP on this gene
Accession: AKU79754
Location: 588955-589998
NCBI BlastP on this gene
mreB4
STURON_00507
Holliday junction DNA helicase RuvA
Accession: AKU79752
Location: 587924-588472
NCBI BlastP on this gene
Accession: AKU79752
Location: 587924-588472
NCBI BlastP on this gene
ruvA
Holliday junction DNA helicase RuvB
Accession: AKU79751
Location: 586993-587940
NCBI BlastP on this gene
Accession: AKU79751
Location: 586993-587940
NCBI BlastP on this gene
ruvB
STURON_00504
STURON_00503
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP006681 : Spiroplasma culicicola AES-1 Total score: 1.0 Cumulative Blast bit score: 223
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SCULI_v1c06400
SCULI_v1c06390
SCULI_v1c06380
SCULI_v1c06370
SCULI_v1c06360
GTP-binding protein YsxC
Accession: AHI52976
Location: 707436-708020
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 223
Sequence coverage: 96 %
E-value: 5e-70
NCBI BlastP on this gene
Accession: AHI52976
Location: 707436-708020
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 223
Sequence coverage: 96 %
E-value: 5e-70
NCBI BlastP on this gene
engB
SCULI_v1c06340
SCULI_v1c06330
SCULI_v1c06320
SCULI_v1c06310
pldB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP025257 : Mesoplasma syrphidae strain YJS chromosome Total score: 1.0 Cumulative Blast bit score: 218
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CXP39_02220
CXP39_02215
CXP39_02210
CXP39_02205
CXP39_02200
CXP39_02195
CXP39_02190
YihA family ribosome biogenesis GTP-binding protein
Accession: AUF83600
Location: 517713-518300
BlastP hit with AAP55641.1
Percentage identity: 54 %
BlastP bit score: 218
Sequence coverage: 96 %
E-value: 7e-68
NCBI BlastP on this gene
Accession: AUF83600
Location: 517713-518300
BlastP hit with AAP55641.1
Percentage identity: 54 %
BlastP bit score: 218
Sequence coverage: 96 %
E-value: 7e-68
NCBI BlastP on this gene
CXP39_02185
cation-translocating P-type ATPase
Accession: AUF83599
Location: 514788-517691
NCBI BlastP on this gene
Accession: AUF83599
Location: 514788-517691
NCBI BlastP on this gene
CXP39_02180
CXP39_02175
CXP39_02170
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP022535 : Spiroplasma corruscae strain EC-1 chromosome Total score: 1.0 Cumulative Blast bit score: 218
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ABC transporter ATP-binding protein
Accession: ASP28285
Location: 588235-590040
NCBI BlastP on this gene
Accession: ASP28285
Location: 588235-590040
NCBI BlastP on this gene
SCORR_v1c05130
ABC transporter ATP-binding protein
Accession: ASP28284
Location: 586347-588224
NCBI BlastP on this gene
Accession: ASP28284
Location: 586347-588224
NCBI BlastP on this gene
SCORR_v1c05120
SCORR_v1c05110
SCORR_v1c05100
GTP-binding protein YsxC
Accession: ASP28281
Location: 584821-585426
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 218
Sequence coverage: 97 %
E-value: 5e-68
NCBI BlastP on this gene
Accession: ASP28281
Location: 584821-585426
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 218
Sequence coverage: 97 %
E-value: 5e-68
NCBI BlastP on this gene
engB
SCORR_v1c05080
cell shape determining protein MreB
Accession: ASP28279
Location: 582732-583772
NCBI BlastP on this gene
Accession: ASP28279
Location: 582732-583772
NCBI BlastP on this gene
mreB
SCORR_v1c05060
Holliday junction DNA helicase RuvA
Accession: ASP28277
Location: 581697-582242
NCBI BlastP on this gene
Accession: ASP28277
Location: 581697-582242
NCBI BlastP on this gene
ruvA
Holliday junction DNA helicase RuvB
Accession: ASP28276
Location: 580763-581710
NCBI BlastP on this gene
Accession: ASP28276
Location: 580763-581710
NCBI BlastP on this gene
ruvB
SCORR_v1c05030
SCORR_v1c05020
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP024870 : Spiroplasma clarkii strain CN-5 chromosome Total score: 1.0 Cumulative Blast bit score: 215
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
PTS system, beta-glucoside-specific IIABC component
Accession: ATX70920
Location: 683593-686091
NCBI BlastP on this gene
Accession: ATX70920
Location: 683593-686091
NCBI BlastP on this gene
SCLAR_v1c06010
bglA
SCLAR_v1c06030
SCLAR_v1c06040
SCLAR_v1c06050
GTP-binding protein YsxC
Accession: ATX70925
Location: 690554-691150
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 215
Sequence coverage: 96 %
E-value: 6e-67
NCBI BlastP on this gene
Accession: ATX70925
Location: 690554-691150
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 215
Sequence coverage: 96 %
E-value: 6e-67
NCBI BlastP on this gene
engB
SCLAR_v1c06070
SCLAR_v1c06080
SCLAR_v1c06090
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP043026 : Spiroplasma chinense strain CCH chromosome Total score: 1.0 Cumulative Blast bit score: 214
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
GTP-binding protein YsxC
Accession: QEH61914
Location: 815714-816298
BlastP hit with AAP55641.1
Percentage identity: 60 %
BlastP bit score: 214
Sequence coverage: 88 %
E-value: 9e-67
NCBI BlastP on this gene
Accession: QEH61914
Location: 815714-816298
BlastP hit with AAP55641.1
Percentage identity: 60 %
BlastP bit score: 214
Sequence coverage: 88 %
E-value: 9e-67
NCBI BlastP on this gene
engB
SCHIN_v1c07180
SCHIN_v1c07170
SCHIN_v1c07160
SCHIN_v1c07150
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP024967 : Mesoplasma lactucae ATCC 49193 strain 831-C4 chromosome Total score: 1.0 Cumulative Blast bit score: 211
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
GTP-binding protein
Accession: ATZ20177
Location: 431581-432189
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
Accession: ATZ20177
Location: 431581-432189
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
engB
ctp
MLACT_v1c03540
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP023668 : Mesoplasma lactucae ATCC 49193 strain 831-C4 chromosome Total score: 1.0 Cumulative Blast bit score: 211
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
CP520_01150
peptide-methionine (S)-S-oxide reductase
Accession: ATG97365
Location: 274859-275353
NCBI BlastP on this gene
Accession: ATG97365
Location: 274859-275353
NCBI BlastP on this gene
msrA
peptide-methionine (R)-S-oxide reductase
Accession: ATG97366
Location: 275387-275824
NCBI BlastP on this gene
Accession: ATG97366
Location: 275387-275824
NCBI BlastP on this gene
msrB
CP520_01165
CP520_01170
CP520_01175
CP520_01180
YihA family ribosome biogenesis GTP-binding protein
Accession: ATG97371
Location: 278722-279330
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
Accession: ATG97371
Location: 278722-279330
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
CP520_01185
CP520_01190
CP520_01195
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP017015 : Spiroplasma helicoides strain TABS-2 chromosome Total score: 1.0 Cumulative Blast bit score: 211
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SHELI_v1c05220
SHELI_v1c05230
SHELI_v1c05240
SHELI_v1c05250
SHELI_v1c05260
SHELI_v1c05270
SHELI_v1c05280
GTP-binding protein YsxC
Accession: AOG60480
Location: 604570-605169
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 97 %
E-value: 2e-65
NCBI BlastP on this gene
Accession: AOG60480
Location: 604570-605169
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 97 %
E-value: 2e-65
NCBI BlastP on this gene
engB
SHELI_v1c05300
SHELI_v1c05310
SHELI_v1c05320
SHELI_v1c05330
SHELI_v1c05340
SHELI_v1c05350
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP038013 : Spiroplasma gladiatoris strain TG-1 chromosome Total score: 1.0 Cumulative Blast bit score: 197
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
SGLAD_v1c05380
SGLAD_v1c05370
SGLAD_v1c05360
SGLAD_v1c05350
SGLAD_v1c05340
GTP-binding protein YsxC
Accession: QBQ07732
Location: 597397-597984
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 197
Sequence coverage: 95 %
E-value: 6e-60
NCBI BlastP on this gene
Accession: QBQ07732
Location: 597397-597984
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 197
Sequence coverage: 95 %
E-value: 6e-60
NCBI BlastP on this gene
engB
SGLAD_v1c05320
SGLAD_v1c05310
SGLAD_v1c05300
SGLAD_v1c05290
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP046276 : Spiroplasma tabanidicola strain TAUS-1 chromosome Total score: 1.0 Cumulative Blast bit score: 195
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
STABA_v1c06480
NAD(P)H-dependent oxidoreductase
Accession: QGS52008
Location: 741079-741660
NCBI BlastP on this gene
Accession: QGS52008
Location: 741079-741660
NCBI BlastP on this gene
STABA_v1c06470
STABA_v1c06460
STABA_v1c06450
STABA_v1c06440
GTP-binding protein
Accession: QGS52004
Location: 738834-739421
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 195
Sequence coverage: 96 %
E-value: 3e-59
NCBI BlastP on this gene
Accession: QGS52004
Location: 738834-739421
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 195
Sequence coverage: 96 %
E-value: 3e-59
NCBI BlastP on this gene
engB
STABA_v1c06420
STABA_v1c06410
STABA_v1c06400
STABA_v1c06390
STABA_v1c06380
P-type Ca2+ transporter type 2C
Accession: QGS51998
Location: 731769-734714
NCBI BlastP on this gene
Accession: QGS51998
Location: 731769-734714
NCBI BlastP on this gene
STABA_v1c06370
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR134274 : Streptococcus salivarius strain NCTC8618 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 192
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: VED87846
Location: 615074-616345
NCBI BlastP on this gene
Accession: VED87846
Location: 615074-616345
NCBI BlastP on this gene
mvaA
hydroxymethylglutaryl-CoA synthase
Accession: VED87848
Location: 616347-617513
NCBI BlastP on this gene
Accession: VED87848
Location: 616347-617513
NCBI BlastP on this gene
mvaS
thyA_2
folA_2
putative uncharacterized conserved protein
Accession: VED87853
Location: 619460-619630
NCBI BlastP on this gene
Accession: VED87853
Location: 619460-619630
NCBI BlastP on this gene
NCTC8618_03036
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: VED87855
Location: 619648-620874
NCBI BlastP on this gene
Accession: VED87855
Location: 619648-620874
NCBI BlastP on this gene
clpX_2
ribosome biogenesis GTP-binding protein YsxC
Accession: VED87857
Location: 620884-621483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
Accession: VED87857
Location: 620884-621483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
engB_2
lysP
homocysteine S-methyltransferase 1
Accession: VED87861
Location: 623002-623952
NCBI BlastP on this gene
Accession: VED87861
Location: 623002-623952
NCBI BlastP on this gene
mmuM
Galactose mutarotase and related enzyme
Accession: VED87862
Location: 624065-624319
NCBI BlastP on this gene
Accession: VED87862
Location: 624065-624319
NCBI BlastP on this gene
NCTC8618_03041
NCTC8618_03042
acyl-phosphate glycerol 3-phosphate acyltransferase
Accession: VED87866
Location: 624889-625530
NCBI BlastP on this gene
Accession: VED87866
Location: 624889-625530
NCBI BlastP on this gene
plsY_2
parE_3
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP014144 : Streptococcus salivarius strain JF Total score: 1.0 Cumulative Blast bit score: 192
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
3-hydroxy-3-methylglutaryl-CoA reductase
Accession: AWB63_02675
Location: 543585-544031
NCBI BlastP on this gene
Accession: AWB63_02675
Location: 543585-544031
NCBI BlastP on this gene
AWB63_02675
hydroxymethylglutaryl-CoA synthase
Accession: AMB82341
Location: 544033-545199
NCBI BlastP on this gene
Accession: AMB82341
Location: 544033-545199
NCBI BlastP on this gene
AWB63_02680
AWB63_02685
AWB63_02690
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: AMB82344
Location: 547334-548560
NCBI BlastP on this gene
Accession: AMB82344
Location: 547334-548560
NCBI BlastP on this gene
AWB63_02695
GTP-binding protein
Accession: AMB82345
Location: 548570-549169
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
Accession: AMB82345
Location: 548570-549169
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
engB
AWB63_02705
homocysteine S-methyltransferase
Accession: AMB82347
Location: 550688-551638
NCBI BlastP on this gene
Accession: AMB82347
Location: 550688-551638
NCBI BlastP on this gene
mmuM
AWB63_02715
AWB63_02720
glycerol-3-phosphate acyltransferase
Accession: AMB82350
Location: 552575-553216
NCBI BlastP on this gene
Accession: AMB82350
Location: 552575-553216
NCBI BlastP on this gene
AWB63_02725
gyrB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP009913 : Streptococcus salivarius strain NCTC 8618 Total score: 1.0 Cumulative Blast bit score: 192
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
3-hydroxy-3-methylglutaryl-CoA reductase
Accession: AIY20743
Location: 615418-616689
NCBI BlastP on this gene
Accession: AIY20743
Location: 615418-616689
NCBI BlastP on this gene
SSAL8618_03035
SSAL8618_03040
SSAL8618_03045
SSAL8618_03050
SSAL8618_03055
SSAL8618_03060
GTP-binding protein
Accession: AIY20749
Location: 621228-621827
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
Accession: AIY20749
Location: 621228-621827
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
engB
SSAL8618_03070
mmuM
SSAL8618_03080
SSAL8618_03085
glycerol-3-phosphate acyltransferase
Accession: AIY20754
Location: 625233-625874
NCBI BlastP on this gene
Accession: AIY20754
Location: 625233-625874
NCBI BlastP on this gene
SSAL8618_03090
gyrB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LS483366 : Streptococcus salivarius strain NCTC7366 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: SQF76373
Location: 1574858-1576129
NCBI BlastP on this gene
Accession: SQF76373
Location: 1574858-1576129
NCBI BlastP on this gene
mvaA
hydroxymethylglutaryl-CoA synthase
Accession: SQF76372
Location: 1573690-1574856
NCBI BlastP on this gene
Accession: SQF76372
Location: 1573690-1574856
NCBI BlastP on this gene
mvaS
thyA
folA
putative uncharacterized conserved protein
Accession: SQF76369
Location: 1571737-1571907
NCBI BlastP on this gene
Accession: SQF76369
Location: 1571737-1571907
NCBI BlastP on this gene
NCTC7366_01457
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: SQF76368
Location: 1570493-1571719
NCBI BlastP on this gene
Accession: SQF76368
Location: 1570493-1571719
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: SQF76367
Location: 1569884-1570483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: SQF76367
Location: 1569884-1570483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
lysP_2
homocysteine S-methyltransferase 1
Accession: SQF76365
Location: 1567416-1568366
NCBI BlastP on this gene
Accession: SQF76365
Location: 1567416-1568366
NCBI BlastP on this gene
mmuM
Galactose mutarotase and related enzyme
Accession: SQF76364
Location: 1567049-1567303
NCBI BlastP on this gene
Accession: SQF76364
Location: 1567049-1567303
NCBI BlastP on this gene
NCTC7366_01452
NCTC7366_01451
acyl-phosphate glycerol 3-phosphate acyltransferase
Accession: SQF76362
Location: 1565838-1566479
NCBI BlastP on this gene
Accession: SQF76362
Location: 1565838-1566479
NCBI BlastP on this gene
plsY
DNA topoisomerase IV subunit B
Accession: SQF76361
Location: 1563745-1565700
NCBI BlastP on this gene
Accession: SQF76361
Location: 1563745-1565700
NCBI BlastP on this gene
parE
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP040804 : Streptococcus salivarius strain LAB813 chromosome Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QEM32286
Location: 788258-789532
NCBI BlastP on this gene
Accession: QEM32286
Location: 788258-789532
NCBI BlastP on this gene
FHI56_05045
hydroxymethylglutaryl-CoA synthase
Accession: QEM32287
Location: 789534-790700
NCBI BlastP on this gene
Accession: QEM32287
Location: 789534-790700
NCBI BlastP on this gene
FHI56_05050
FHI56_05055
FHI56_05060
FHI56_05065
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: QEM32291
Location: 792670-793896
NCBI BlastP on this gene
Accession: QEM32291
Location: 792670-793896
NCBI BlastP on this gene
clpX
YihA family ribosome biogenesis GTP-binding protein
Accession: QEM32292
Location: 793906-794505
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QEM32292
Location: 793906-794505
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
FHI56_05075
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP018189 : Streptococcus salivarius strain ICDC3 chromosome Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QGU83041
Location: 1580717-1581991
NCBI BlastP on this gene
Accession: QGU83041
Location: 1580717-1581991
NCBI BlastP on this gene
BSR20_07285
hydroxymethylglutaryl-CoA synthase
Accession: QGU83040
Location: 1579549-1580715
NCBI BlastP on this gene
Accession: QGU83040
Location: 1579549-1580715
NCBI BlastP on this gene
BSR20_07280
BSR20_07275
BSR20_07270
ATP-dependent protease ATP-binding subunit ClpX
Accession: QGU83037
Location: 1576175-1577401
NCBI BlastP on this gene
Accession: QGU83037
Location: 1576175-1577401
NCBI BlastP on this gene
BSR20_07265
YihA family ribosome biogenesis GTP-binding protein
Accession: QGU83036
Location: 1575566-1576165
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QGU83036
Location: 1575566-1576165
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
BSR20_07255
homocysteine S-methyltransferase
Accession: QGU83034
Location: 1573098-1574048
NCBI BlastP on this gene
Accession: QGU83034
Location: 1573098-1574048
NCBI BlastP on this gene
BSR20_07250
BSR20_07245
BSR20_07240
glycerol-3-phosphate acyltransferase
Accession: QGU83031
Location: 1571520-1572161
NCBI BlastP on this gene
Accession: QGU83031
Location: 1571520-1572161
NCBI BlastP on this gene
BSR20_07235
DNA topoisomerase IV subunit B
Accession: QGU83030
Location: 1569433-1571382
NCBI BlastP on this gene
Accession: QGU83030
Location: 1569433-1571382
NCBI BlastP on this gene
BSR20_07230
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP018187 : Streptococcus salivarius strain ICDC2 Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QGU80161
Location: 596170-597441
NCBI BlastP on this gene
Accession: QGU80161
Location: 596170-597441
NCBI BlastP on this gene
BSR19_03020
hydroxymethylglutaryl-CoA synthase
Accession: QGU80162
Location: 597443-598609
NCBI BlastP on this gene
Accession: QGU80162
Location: 597443-598609
NCBI BlastP on this gene
BSR19_03025
BSR19_03030
BSR19_03035
ATP-dependent protease ATP-binding subunit ClpX
Accession: QGU80165
Location: 600737-601963
NCBI BlastP on this gene
Accession: QGU80165
Location: 600737-601963
NCBI BlastP on this gene
BSR19_03040
YihA family ribosome biogenesis GTP-binding protein
Accession: QGU80166
Location: 601973-602572
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QGU80166
Location: 601973-602572
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
BSR19_03050
homocysteine S-methyltransferase
Accession: QGU80168
Location: 604089-605039
NCBI BlastP on this gene
Accession: QGU80168
Location: 604089-605039
NCBI BlastP on this gene
BSR19_03055
BSR19_03060
BSR19_03065
glycerol-3-phosphate acyltransferase
Accession: QGU80171
Location: 605973-606614
NCBI BlastP on this gene
Accession: QGU80171
Location: 605973-606614
NCBI BlastP on this gene
BSR19_03070
BSR19_03075
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP018186 : Streptococcus salivarius strain ICDC1 chromosome Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QGU79032
Location: 1580742-1582016
NCBI BlastP on this gene
Accession: QGU79032
Location: 1580742-1582016
NCBI BlastP on this gene
BSR14_07285
hydroxymethylglutaryl-CoA synthase
Accession: QGU79031
Location: 1579574-1580740
NCBI BlastP on this gene
Accession: QGU79031
Location: 1579574-1580740
NCBI BlastP on this gene
BSR14_07280
BSR14_07275
BSR14_07270
ATP-dependent protease ATP-binding subunit ClpX
Accession: QGU79028
Location: 1576200-1577426
NCBI BlastP on this gene
Accession: QGU79028
Location: 1576200-1577426
NCBI BlastP on this gene
BSR14_07265
YihA family ribosome biogenesis GTP-binding protein
Accession: QGU79027
Location: 1575591-1576190
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QGU79027
Location: 1575591-1576190
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
BSR14_07255
homocysteine S-methyltransferase
Accession: QGU79025
Location: 1573123-1574073
NCBI BlastP on this gene
Accession: QGU79025
Location: 1573123-1574073
NCBI BlastP on this gene
BSR14_07250
BSR14_07245
BSR14_07240
glycerol-3-phosphate acyltransferase
Accession: QGU79022
Location: 1571545-1572186
NCBI BlastP on this gene
Accession: QGU79022
Location: 1571545-1572186
NCBI BlastP on this gene
BSR14_07235
DNA topoisomerase IV subunit B
Accession: QGU79021
Location: 1569458-1571407
NCBI BlastP on this gene
Accession: QGU79021
Location: 1569458-1571407
NCBI BlastP on this gene
BSR14_07230
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP015283 : Streptococcus salivarius strain ATCC 25975 Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
YihA family ribosome biogenesis GTP-binding protein
Accession: ARI60516
Location: 2125010-2125609
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: ARI60516
Location: 2125010-2125609
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
V471_09855
homocysteine S-methyltransferase
Accession: ARI60514
Location: 2122542-2123492
NCBI BlastP on this gene
Accession: ARI60514
Location: 2122542-2123492
NCBI BlastP on this gene
mmuM
V471_09845
V471_09840
glycerol-3-phosphate acyltransferase
Accession: ARI60511
Location: 2120964-2121605
NCBI BlastP on this gene
Accession: ARI60511
Location: 2120964-2121605
NCBI BlastP on this gene
V471_09835
DNA topoisomerase IV subunit B
Accession: ARI60510
Location: 2118877-2120826
NCBI BlastP on this gene
Accession: ARI60510
Location: 2118877-2120826
NCBI BlastP on this gene
gyrB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP015282 : Streptococcus salivarius strain ATCC 27945 Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
3-hydroxy-3-methylglutaryl-CoA reductase
Accession: ARI57667
Location: 972034-973305
NCBI BlastP on this gene
Accession: ARI57667
Location: 972034-973305
NCBI BlastP on this gene
NX99_04050
hydroxymethylglutaryl-CoA synthase
Accession: ARI57666
Location: 970866-972032
NCBI BlastP on this gene
Accession: ARI57666
Location: 970866-972032
NCBI BlastP on this gene
NX99_04045
NX99_04040
NX99_04035
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: ARI57663
Location: 967504-968730
NCBI BlastP on this gene
Accession: ARI57663
Location: 967504-968730
NCBI BlastP on this gene
NX99_04030
YihA family ribosome biogenesis GTP-binding protein
Accession: ARI57662
Location: 966895-967494
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: ARI57662
Location: 966895-967494
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
NX99_04020
homocysteine S-methyltransferase
Accession: ARI57660
Location: 964427-965377
NCBI BlastP on this gene
Accession: ARI57660
Location: 964427-965377
NCBI BlastP on this gene
mmuM
NX99_04010
NX99_04005
glycerol-3-phosphate acyltransferase
Accession: ARI57657
Location: 962845-963486
NCBI BlastP on this gene
Accession: ARI57657
Location: 962845-963486
NCBI BlastP on this gene
NX99_04000
gyrB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP012646 : Streptococcus mitis strain KCOM 1350 (= ChDC B183) Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
RN80_04340
RN80_04345
RN80_04350
RN80_04355
RN80_04360
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: ALD67710
Location: 874218-875450
NCBI BlastP on this gene
Accession: ALD67710
Location: 874218-875450
NCBI BlastP on this gene
RN80_04365
GTP-binding protein
Accession: ALD67711
Location: 875459-876046
BlastP hit with AAP55641.1
Percentage identity: 50 %
BlastP bit score: 190
Sequence coverage: 91 %
E-value: 4e-57
NCBI BlastP on this gene
Accession: ALD67711
Location: 875459-876046
BlastP hit with AAP55641.1
Percentage identity: 50 %
BlastP bit score: 190
Sequence coverage: 91 %
E-value: 4e-57
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002888 : Streptococcus salivarius 57.I Total score: 1.0 Cumulative Blast bit score: 190
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ribosome biogenesis GTP-binding protein YsxC
Accession: AEJ53843
Location: 1550139-1550738
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: AEJ53843
Location: 1550139-1550738
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
rocE
homocysteine S-methyltransferase 1
Accession: AEJ53841
Location: 1547671-1548621
NCBI BlastP on this gene
Accession: AEJ53841
Location: 1547671-1548621
NCBI BlastP on this gene
mmuM
conserved hypothetical protein
Accession: AEJ53840
Location: 1547304-1547558
NCBI BlastP on this gene
Accession: AEJ53840
Location: 1547304-1547558
NCBI BlastP on this gene
Ssal_01575
conserved hypothetical protein
Accession: AEJ53839
Location: 1546852-1547229
NCBI BlastP on this gene
Accession: AEJ53839
Location: 1546852-1547229
NCBI BlastP on this gene
Ssal_01574
acyl-phosphate glycerol 3-phosphate acyltransferase
Accession: AEJ53838
Location: 1546093-1546734
NCBI BlastP on this gene
Accession: AEJ53838
Location: 1546093-1546734
NCBI BlastP on this gene
plsY
DNA topoisomerase IV, B subunit
Accession: AEJ53837
Location: 1544006-1545955
NCBI BlastP on this gene
Accession: AEJ53837
Location: 1544006-1545955
NCBI BlastP on this gene
parE
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
CP002471 : Streptococcus parauberis KCTC 11537 Total score: 1.0 Cumulative Blast bit score: 189
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
3-hydroxy-3-methylglutaryl-coenzyme A reductase
Accession: AEF25052
Location: 643042-644325
NCBI BlastP on this gene
Accession: AEF25052
Location: 643042-644325
NCBI BlastP on this gene
mvaA
3-hydroxy-3-methylglutaryl coenzyme A synthase
Accession: AEF25053
Location: 644322-645488
NCBI BlastP on this gene
Accession: AEF25053
Location: 644322-645488
NCBI BlastP on this gene
mvaS
thyA
dyr
STP_0608
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: AEF25057
Location: 647477-648544
NCBI BlastP on this gene
Accession: AEF25057
Location: 647477-648544
NCBI BlastP on this gene
clpX
GTP-binding protein
Accession: AEF25058
Location: 648553-649152
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 189
Sequence coverage: 91 %
E-value: 9e-57
NCBI BlastP on this gene
Accession: AEF25058
Location: 648553-649152
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 189
Sequence coverage: 91 %
E-value: 9e-57
NCBI BlastP on this gene
STP_0610
STP_0611
ATP-dependent protease ATP-binding subunit ClpL
Accession: AEF25060
Location: 650005-652113
NCBI BlastP on this gene
Accession: AEF25060
Location: 650005-652113
NCBI BlastP on this gene
clpL
rplJ
rplL
STP_0615
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR594040 : Streptococcus australis strain NCTC5338 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
galE
yfdH
NCTC5338_01373
cmk
dhfR
NCTC5338_01370
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VTS72167
Location: 1369417-1370625
NCBI BlastP on this gene
Accession: VTS72167
Location: 1369417-1370625
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VTS72164
Location: 1368821-1369408
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VTS72164
Location: 1368821-1369408
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
NCTC5338_01365
putative sporulation transcription regulator whiA
Accession: VTS72152
Location: 1365607-1366518
NCBI BlastP on this gene
Accession: VTS72152
Location: 1365607-1366518
NCBI BlastP on this gene
whiA
NCTC5338_01363
NCTC5338_01362
NCTC5338_01361
NCTC5338_01360
NCTC5338_01359
Protein of uncharacterised function (DUF2974)
Accession: VTS72134
Location: 1362391-1363635
NCBI BlastP on this gene
Accession: VTS72134
Location: 1362391-1363635
NCBI BlastP on this gene
NCTC5338_01358
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR216061 : Streptococcus pneumoniae strain 2245STDY6178826 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
tpiA
lytC
DPS family peroxide resistance protein
Accession: VFI44001
Location: 1551212-1551730
NCBI BlastP on this gene
Accession: VFI44001
Location: 1551212-1551730
NCBI BlastP on this gene
dps
dhfR
SAMEA3713960_01605
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VFI43976
Location: 1549142-1550374
NCBI BlastP on this gene
Accession: VFI43976
Location: 1549142-1550374
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VFI43968
Location: 1548546-1549133
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VFI43968
Location: 1548546-1549133
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
SAMEA3713960_01600
putative sporulation transcription regulator whiA
Accession: VFI43935
Location: 1545331-1546242
NCBI BlastP on this gene
Accession: VFI43935
Location: 1545331-1546242
NCBI BlastP on this gene
whiA
yumC
DNA integrity scanning protein DisA
Accession: VFI43918
Location: 1543336-1544193
NCBI BlastP on this gene
Accession: VFI43918
Location: 1543336-1544193
NCBI BlastP on this gene
ybbP
SAMEA3713960_01596
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR216040 : Streptococcus pneumoniae strain 2245STDY6030848 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
tpiA
lytC
DPS family peroxide resistance protein
Accession: VFH95156
Location: 1504988-1505506
NCBI BlastP on this gene
Accession: VFH95156
Location: 1504988-1505506
NCBI BlastP on this gene
dps
dhfR
SAMEA3233211_01532
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VFH95153
Location: 1502913-1504121
NCBI BlastP on this gene
Accession: VFH95153
Location: 1502913-1504121
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VFH95152
Location: 1502317-1502904
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VFH95152
Location: 1502317-1502904
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
SAMEA3233211_01527
putative sporulation transcription regulator whiA
Accession: VFH95147
Location: 1499102-1500013
NCBI BlastP on this gene
Accession: VFH95147
Location: 1499102-1500013
NCBI BlastP on this gene
whiA
yumC
DNA integrity scanning protein DisA
Accession: VFH95143
Location: 1497107-1497964
NCBI BlastP on this gene
Accession: VFH95143
Location: 1497107-1497964
NCBI BlastP on this gene
ybbP
SAMEA3233211_01523
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR216032 : Streptococcus pneumoniae strain 2245STDY5775874 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
tpiA
lytC
DPS family peroxide resistance protein
Accession: VFH78713
Location: 1418791-1419309
NCBI BlastP on this gene
Accession: VFH78713
Location: 1418791-1419309
NCBI BlastP on this gene
dps
dhfR
SAMEA2434829_01438
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VFH78690
Location: 1416721-1417953
NCBI BlastP on this gene
Accession: VFH78690
Location: 1416721-1417953
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VFH78682
Location: 1416125-1416712
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VFH78682
Location: 1416125-1416712
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
SAMEA2434829_01433
putative sporulation transcription regulator whiA
Accession: VFH78647
Location: 1412910-1413821
NCBI BlastP on this gene
Accession: VFH78647
Location: 1412910-1413821
NCBI BlastP on this gene
whiA
yumC
DNA integrity scanning protein DisA
Accession: VFH78638
Location: 1410918-1411775
NCBI BlastP on this gene
Accession: VFH78638
Location: 1410918-1411775
NCBI BlastP on this gene
ybbP
SAMEA2434829_01429
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
LR134323 : Streptococcus urinalis strain NCTC13766 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
hydroxymethylglutaryl-CoA reductase, degradative
Accession: VEF31882
Location: 914960-916237
NCBI BlastP on this gene
Accession: VEF31882
Location: 914960-916237
NCBI BlastP on this gene
mvaA
Hydroxymethylglutaryl-CoA synthase
Accession: VEF31883
Location: 916221-917402
NCBI BlastP on this gene
Accession: VEF31883
Location: 916221-917402
NCBI BlastP on this gene
pksG
thyA
dfrA
NCTC13766_00985
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: VEF31887
Location: 919230-920456
NCBI BlastP on this gene
Accession: VEF31887
Location: 919230-920456
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein
Accession: VEF31888
Location: 920468-921067
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 3e-56
NCBI BlastP on this gene
Accession: VEF31888
Location: 920468-921067
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 3e-56
NCBI BlastP on this gene
engB
Query: Spiroplasma citri strain GII-3 trehalose operon, complete sequence.
301. : CP007520 Mycoplasma yeatsii GM274B chromosome Total score: 1.0 Cumulative Blast bit score: 225
AAP55640.1
AAP55641.1
AAP55642.1
DBD-Pfam|LacI,DBD-SUPERFAMILY|0036955
Location: 1786-2775
Location: 1786-2775
treR
gnl|TC-DB|P36672|4.A.1.2.4
Location: 2844-4412
Location: 2844-4412
treP
GH13 29|GH13
Location: 4441-6090
Location: 4441-6090
treA
ribosome biogenesis GTP-binding protein
Accession: AJM71880
Location: 471284-471871
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 226
Sequence coverage: 95 %
E-value: 3e-71
NCBI BlastP on this gene
Accession: AJM71880
Location: 471284-471871
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 226
Sequence coverage: 95 %
E-value: 3e-71
NCBI BlastP on this gene
engB
cation-transporting ATPase, E1-E2 family
Accession: AJM71879
Location: 468359-471277
NCBI BlastP on this gene
Accession: AJM71879
Location: 468359-471277
NCBI BlastP on this gene
MYE_01995
MYE_01990
rpsO
MYE_01975
302. : LS483504 Mycoplasma mycoides subsp. capri strain GM12 genome assembly, chromosome: omosome. Total score: 1.0 Cumulative Blast bit score: 224
MMC68I_00256
transcription elongation factor GreA
Accession: SRX61352
Location: 329891-330364
NCBI BlastP on this gene
Accession: SRX61352
Location: 329891-330364
NCBI BlastP on this gene
MMC68I_00255
SDR family NAD(P)-dependent oxidoreductase
Accession: SRX61351
Location: 329124-329846
NCBI BlastP on this gene
Accession: SRX61351
Location: 329124-329846
NCBI BlastP on this gene
MMC68I_00254
MMC68I_00253
MMC68I_00252
MMC68I_00251
MMC68I_00250
YihA family ribosome biogenesis GTP-binding protein
Accession: SRX61346
Location: 325933-326523
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 9e-71
NCBI BlastP on this gene
Accession: SRX61346
Location: 325933-326523
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 9e-71
NCBI BlastP on this gene
MMC68I_00249
cation-translocating P-type ATPase
Accession: SRX61345
Location: 323018-325924
NCBI BlastP on this gene
Accession: SRX61345
Location: 323018-325924
NCBI BlastP on this gene
MMC68I_00248
MMC68I_00247
303. : LR739237 Mycoplasma feriruminatoris 14/OD_0492 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
uvrC
Transcription elongation factor GreA
Accession: VZS00575
Location: 731929-732402
NCBI BlastP on this gene
Accession: VZS00575
Location: 731929-732402
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZS00577
Location: 732450-733172
NCBI BlastP on this gene
Accession: VZS00577
Location: 732450-733172
NCBI BlastP on this gene
ydfG_2
MF5582_00669
MF5582_00670
MF5582_00671
MF5582_00672
putative GTP-binding protein EngB
Accession: VZS00587
Location: 735774-736364
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZS00587
Location: 735774-736364
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
304. : LR739235 Mycoplasma feriruminatoris 8756-13 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
uvrC
Transcription elongation factor GreA
Accession: VZR98175
Location: 732040-732513
NCBI BlastP on this gene
Accession: VZR98175
Location: 732040-732513
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZR98177
Location: 732561-733283
NCBI BlastP on this gene
Accession: VZR98177
Location: 732561-733283
NCBI BlastP on this gene
ydfG_2
MF5295_00660
MF5295_00661
MF5295_00662
MF5295_00663
putative GTP-binding protein EngB
Accession: VZR98188
Location: 735886-736476
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZR98188
Location: 735886-736476
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
305. : LR739234 Mycoplasma feriruminatoris G1650 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
uvrC
Transcription elongation factor GreA
Accession: VZR98111
Location: 704731-705204
NCBI BlastP on this gene
Accession: VZR98111
Location: 704731-705204
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZR98113
Location: 705252-705974
NCBI BlastP on this gene
Accession: VZR98113
Location: 705252-705974
NCBI BlastP on this gene
ydfG_2
MF5293_00644
MF5293_00645
MF5293_00646
MF5293_00647
putative GTP-binding protein EngB
Accession: VZR98123
Location: 708577-709167
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZR98123
Location: 708577-709167
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
306. : LR739233 Mycoplasma feriruminatoris G1705 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
uvrC
Transcription elongation factor GreA
Accession: VZR75612
Location: 704742-705215
NCBI BlastP on this gene
Accession: VZR75612
Location: 704742-705215
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZR75613
Location: 705263-705985
NCBI BlastP on this gene
Accession: VZR75613
Location: 705263-705985
NCBI BlastP on this gene
ydfG_2
MF5294_00646
MF5294_00647
MF5294_00648
MF5294_00649
putative GTP-binding protein EngB
Accession: VZR75618
Location: 708588-709178
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZR75618
Location: 708588-709178
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
307. : LR738858 Mycoplasma feriruminatoris G5813/1+2 chromosome : 1. Total score: 1.0 Cumulative Blast bit score: 224
uvrC
Transcription elongation factor GreA
Accession: VZK65469
Location: 703248-703721
NCBI BlastP on this gene
Accession: VZK65469
Location: 703248-703721
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenase YdfG
Accession: VZK65470
Location: 703769-704491
NCBI BlastP on this gene
Accession: VZK65470
Location: 703769-704491
NCBI BlastP on this gene
ydfG_2
MF5292_00648
MF5292_00649
MF5292_00650
MF5292_00651
putative GTP-binding protein EngB
Accession: VZK65475
Location: 707094-707684
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
Accession: VZK65475
Location: 707094-707684
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 225
Sequence coverage: 96 %
E-value: 6e-71
NCBI BlastP on this gene
engB
308. : LS483505 Mycoplasma mycoides subsp. capri strain GM12 genome assembly, chromosome: omosome. Total score: 1.0 Cumulative Blast bit score: 223
MMC68D_00257
transcription elongation factor GreA
Accession: SRX62734
Location: 330028-330501
NCBI BlastP on this gene
Accession: SRX62734
Location: 330028-330501
NCBI BlastP on this gene
MMC68D_00256
SDR family NAD(P)-dependent oxidoreductase
Accession: SRX62733
Location: 329261-329983
NCBI BlastP on this gene
Accession: SRX62733
Location: 329261-329983
NCBI BlastP on this gene
MMC68D_00255
MMC68D_00254
MMC68D_00253
MMC68D_00252
MMC68D_00251
YihA family ribosome biogenesis GTP-binding protein
Accession: SRX62728
Location: 326070-326660
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: SRX62728
Location: 326070-326660
BlastP hit with AAP55641.1
Percentage identity: 58 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MMC68D_00250
cation-translocating P-type ATPase
Accession: SRX62727
Location: 323155-326061
NCBI BlastP on this gene
Accession: SRX62727
Location: 323155-326061
NCBI BlastP on this gene
MMC68D_00249
MMC68D_00248
309. : CP024964 Entomoplasma melaleucae strain M1 chromosome Total score: 1.0 Cumulative Blast bit score: 223
GTP-binding protein
Accession: ATZ18040
Location: 465112-465708
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: ATZ18040
Location: 465112-465708
BlastP hit with AAP55641.1
Percentage identity: 56 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
ctp
valS
310. : CP014346 Mycoplasma mycoides subsp. mycoides strain T1/44 Total score: 1.0 Cumulative Blast bit score: 223
uvrC
Transcription elongation factor GreA
Accession: AMK56837
Location: 985643-986116
NCBI BlastP on this gene
Accession: AMK56837
Location: 985643-986116
NCBI BlastP on this gene
greA
NADP-dependent 3-hydroxy acid dehydrogenaseYdfG
Accession: AMK56838
Location: 986159-986881
NCBI BlastP on this gene
Accession: AMK56838
Location: 986159-986881
NCBI BlastP on this gene
ydfG_2
MSCT144_09480
MSCT144_09490
MSCT144_09500
MSCT144_09510
putative GTP-binding protein EngB
Accession: AMK56843
Location: 989482-990072
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AMK56843
Location: 989482-990072
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
311. : CP011264 Mycoplasma mycoides subsp. mycoides strain Ben468 Total score: 1.0 Cumulative Blast bit score: 223
MmmBen468_0327
transcription elongation factor GreA
Accession: AME14557
Location: 332033-332506
NCBI BlastP on this gene
Accession: AME14557
Location: 332033-332506
NCBI BlastP on this gene
MmmBen468_0326
MmmBen468_0325
MmmBen468_0324
MmmBen468_0323
MmmBen468_0322
MmmBen468_0321
ribosome biogenesis GTP-binding protein YsxC
Accession: AME14551
Location: 328077-328667
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME14551
Location: 328077-328667
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen468_0320
MmmBen468_0319
MmmBen468_0318
312. : CP011263 Mycoplasma mycoides subsp. mycoides strain Ben326 Total score: 1.0 Cumulative Blast bit score: 223
MmmBen326_0315
transcription elongation factor GreA
Accession: AME13565
Location: 325254-325727
NCBI BlastP on this gene
Accession: AME13565
Location: 325254-325727
NCBI BlastP on this gene
MmmBen326_0314
MmmBen326_0313
MmmBen326_0312
MmmBen326_0311
MmmBen326_0310
ribosome biogenesis GTP-binding protein YsxC
Accession: AME13560
Location: 321292-321882
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME13560
Location: 321292-321882
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen326_0309
MmmBen326_0308
MmmBen326_0307
313. : CP011262 Mycoplasma mycoides subsp. mycoides strain Ben181 Total score: 1.0 Cumulative Blast bit score: 223
MmmBen181_0327
transcription elongation factor GreA
Accession: AME12532
Location: 329471-329944
NCBI BlastP on this gene
Accession: AME12532
Location: 329471-329944
NCBI BlastP on this gene
MmmBen181_0326
MmmBen181_0325
MmmBen181_0324
MmmBen181_0323
MmmBen181_0322
MmmBen181_0321
ribosome biogenesis GTP-binding protein YsxC
Accession: AME12526
Location: 325515-326105
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME12526
Location: 325515-326105
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen181_0320
MmmBen181_0319
MmmBen181_0318
MmmBen181_0317
314. : CP011261 Mycoplasma mycoides subsp. mycoides strain Ben50 Total score: 1.0 Cumulative Blast bit score: 223
MmmBen50_0315
transcription elongation factor GreA
Accession: AME11508
Location: 333197-333670
NCBI BlastP on this gene
Accession: AME11508
Location: 333197-333670
NCBI BlastP on this gene
MmmBen50_0314
MmmBen50_0313
MmmBen50_0312
MmmBen50_0311
MmmBen50_0310
MmmBen50_0309
ribosome biogenesis GTP-binding protein YsxC
Accession: AME11502
Location: 329241-329831
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME11502
Location: 329241-329831
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen50_0308
MmmBen50_0307
MmmBen50_0306
315. : CP011260 Mycoplasma mycoides subsp. mycoides strain Ben1 Total score: 1.0 Cumulative Blast bit score: 223
MmmBen_0320
transcription elongation factor GreA
Accession: AME10500
Location: 330148-330621
NCBI BlastP on this gene
Accession: AME10500
Location: 330148-330621
NCBI BlastP on this gene
MmmBen_0319
MmmBen_0318
MmmBen_0317
MmmBen_0316
MmmBen_0315
MmmBen_0314
ribosome biogenesis GTP-binding protein YsxC
Accession: AME10494
Location: 326192-326782
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AME10494
Location: 326192-326782
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MmmBen_0313
MmmBen_0312
MmmBen_0311
316. : CP010267 Mycoplasma mycoides subsp. mycoides strain izsam_mm5713 Total score: 1.0 Cumulative Blast bit score: 223
uvrC
transcription elongation factor GreA
Accession: AIZ55150
Location: 347146-347619
NCBI BlastP on this gene
Accession: AIZ55150
Location: 347146-347619
NCBI BlastP on this gene
greA
mycmycITA_00320
mycmycITA_00319
mycmycITA_00318
mycmycITA_00317
mycmycITA_00316
GTPase EngB
Accession: AIZ55144
Location: 343190-343780
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AIZ55144
Location: 343190-343780
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
mycmycITA_00314
mycmycITA_00313
317. : CP004357 Mycoplasma putrefaciens Mput9231 Total score: 1.0 Cumulative Blast bit score: 223
aspS
Phosphotransferase system PTS, lichenan-specific IIa component
Accession: AGJ90703
Location: 334196-334909
NCBI BlastP on this gene
Accession: AGJ90703
Location: 334196-334909
NCBI BlastP on this gene
licA
MPUT9231_2780
MPUT9231_2770
Hypothetical protein, predicted transmembrane protein
Accession: AGJ90700
Location: 332072-332707
NCBI BlastP on this gene
Accession: AGJ90700
Location: 332072-332707
NCBI BlastP on this gene
MPUT9231_2760
Hypothetical protein, predicted transmembrane protein
Accession: AGJ90699
Location: 331498-332082
NCBI BlastP on this gene
Accession: AGJ90699
Location: 331498-332082
NCBI BlastP on this gene
MPUT9231_2750
GTP-binding protein EngB
Accession: AGJ90698
Location: 330906-331493
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AGJ90698
Location: 330906-331493
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
engB
ctp
valS
318. : CP003021 Mycoplasma putrefaciens KS1 Total score: 1.0 Cumulative Blast bit score: 223
ribosome biogenesis GTP-binding protein
Accession: AEM68799
Location: 489822-490409
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: AEM68799
Location: 489822-490409
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
ysxC
cation-transporting ATPase, E1-E2 family
Accession: AEM68798
Location: 486906-489809
NCBI BlastP on this gene
Accession: AEM68798
Location: 486906-489809
NCBI BlastP on this gene
MPUT_0425
valS
319. : CP002107 Mycoplasma mycoides subsp. mycoides SC str. Gladysdale MU clone SC5 Total score: 1.0 Cumulative Blast bit score: 223
uvrC
putative transcription elongation factor GreA
Accession: ADK70109
Location: 347145-347618
NCBI BlastP on this gene
Accession: ADK70109
Location: 347145-347618
NCBI BlastP on this gene
MMS_A0326
MMS_A0325
MMS_A0324
MMS_A0323
MMS_A0322
MMS_A0321
ribosome biogenesis GTP-binding protein YsxC
Accession: ADK69373
Location: 343189-343779
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: ADK69373
Location: 343189-343779
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
ysxC
MMS_A0319
MMS_A0318
320. : BX293980 Mycoplasma mycoides subsp. mycoides SC str. PG1 Total score: 1.0 Cumulative Blast bit score: 223
uvrC
transcription elongation factor
Accession: CAE76937
Location: 336160-336633
NCBI BlastP on this gene
Accession: CAE76937
Location: 336160-336633
NCBI BlastP on this gene
greA
MSC_0295
MSC_0294
MSC_0293
hypothetical transmembrane protein
Accession: CAE76933
Location: 333350-333928
NCBI BlastP on this gene
Accession: CAE76933
Location: 333350-333928
NCBI BlastP on this gene
MSC_0292
hypothetical transmembrane protein
Accession: CAE76932
Location: 332788-333348
NCBI BlastP on this gene
Accession: CAE76932
Location: 332788-333348
NCBI BlastP on this gene
MSC_0291
GTP-binding protein
Accession: CAE76931
Location: 332204-332794
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
Accession: CAE76931
Location: 332204-332794
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 224
Sequence coverage: 96 %
E-value: 2e-70
NCBI BlastP on this gene
MSC_0290
ctp
MSC_0288
Conserved hypothetical transmembrane protein
Accession: CAE76928
Location: 327121-329010
NCBI BlastP on this gene
Accession: CAE76928
Location: 327121-329010
NCBI BlastP on this gene
MSC_0287
321. : CP013860 Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 223
STURO_v1c05150
STURO_v1c05140
STURO_v1c05130
STURO_v1c05120
STURO_v1c05110
GntR family transcriptional regulator
Accession: ALX70776
Location: 592159-592959
NCBI BlastP on this gene
Accession: ALX70776
Location: 592159-592959
NCBI BlastP on this gene
STURO_v1c05100
STURO_v1c05090
GTP-binding protein YsxC
Accession: ALX70774
Location: 590967-591563
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
Accession: ALX70774
Location: 590967-591563
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
engB
STURO_v1c05070
cell shape determining protein MreB
Accession: ALX70772
Location: 588853-589896
NCBI BlastP on this gene
Accession: ALX70772
Location: 588853-589896
NCBI BlastP on this gene
mreB
STURO_v1c05050
Holliday junction DNA helicase RuvA
Accession: ALX70770
Location: 587822-588370
NCBI BlastP on this gene
Accession: ALX70770
Location: 587822-588370
NCBI BlastP on this gene
ruvA
Holliday junction DNA helicase RuvB
Accession: ALX70769
Location: 586891-587838
NCBI BlastP on this gene
Accession: ALX70769
Location: 586891-587838
NCBI BlastP on this gene
ruvB
STURO_v1c05020
STURO_v1c05010
322. : CP012328 Spiroplasma turonicum strain Tab4c Total score: 1.0 Cumulative Blast bit score: 223
yqkD
STURON_00516
STURON_00515
STURON_00514
STURON_00513
Putative transcriptional regulator, RpiR family protein
Accession: AKU79758
Location: 592261-593058
NCBI BlastP on this gene
Accession: AKU79758
Location: 592261-593058
NCBI BlastP on this gene
STURON_00512
STURON_00511
GTP-binding protein YsxC
Accession: AKU79756
Location: 591069-591665
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
Accession: AKU79756
Location: 591069-591665
BlastP hit with AAP55641.1
Percentage identity: 59 %
BlastP bit score: 223
Sequence coverage: 88 %
E-value: 9e-70
NCBI BlastP on this gene
engB
STURON_00509
cell shape determining protein MreB
Accession: AKU79754
Location: 588955-589998
NCBI BlastP on this gene
Accession: AKU79754
Location: 588955-589998
NCBI BlastP on this gene
mreB4
STURON_00507
Holliday junction DNA helicase RuvA
Accession: AKU79752
Location: 587924-588472
NCBI BlastP on this gene
Accession: AKU79752
Location: 587924-588472
NCBI BlastP on this gene
ruvA
Holliday junction DNA helicase RuvB
Accession: AKU79751
Location: 586993-587940
NCBI BlastP on this gene
Accession: AKU79751
Location: 586993-587940
NCBI BlastP on this gene
ruvB
STURON_00504
STURON_00503
323. : CP006681 Spiroplasma culicicola AES-1 Total score: 1.0 Cumulative Blast bit score: 223
SCULI_v1c06400
SCULI_v1c06390
SCULI_v1c06380
SCULI_v1c06370
SCULI_v1c06360
GTP-binding protein YsxC
Accession: AHI52976
Location: 707436-708020
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 223
Sequence coverage: 96 %
E-value: 5e-70
NCBI BlastP on this gene
Accession: AHI52976
Location: 707436-708020
BlastP hit with AAP55641.1
Percentage identity: 57 %
BlastP bit score: 223
Sequence coverage: 96 %
E-value: 5e-70
NCBI BlastP on this gene
engB
SCULI_v1c06340
SCULI_v1c06330
SCULI_v1c06320
SCULI_v1c06310
pldB
324. : CP025257 Mesoplasma syrphidae strain YJS chromosome Total score: 1.0 Cumulative Blast bit score: 218
CXP39_02220
CXP39_02215
CXP39_02210
CXP39_02205
CXP39_02200
CXP39_02195
CXP39_02190
YihA family ribosome biogenesis GTP-binding protein
Accession: AUF83600
Location: 517713-518300
BlastP hit with AAP55641.1
Percentage identity: 54 %
BlastP bit score: 218
Sequence coverage: 96 %
E-value: 7e-68
NCBI BlastP on this gene
Accession: AUF83600
Location: 517713-518300
BlastP hit with AAP55641.1
Percentage identity: 54 %
BlastP bit score: 218
Sequence coverage: 96 %
E-value: 7e-68
NCBI BlastP on this gene
CXP39_02185
cation-translocating P-type ATPase
Accession: AUF83599
Location: 514788-517691
NCBI BlastP on this gene
Accession: AUF83599
Location: 514788-517691
NCBI BlastP on this gene
CXP39_02180
CXP39_02175
CXP39_02170
325. : CP022535 Spiroplasma corruscae strain EC-1 chromosome Total score: 1.0 Cumulative Blast bit score: 218
ABC transporter ATP-binding protein
Accession: ASP28285
Location: 588235-590040
NCBI BlastP on this gene
Accession: ASP28285
Location: 588235-590040
NCBI BlastP on this gene
SCORR_v1c05130
ABC transporter ATP-binding protein
Accession: ASP28284
Location: 586347-588224
NCBI BlastP on this gene
Accession: ASP28284
Location: 586347-588224
NCBI BlastP on this gene
SCORR_v1c05120
SCORR_v1c05110
SCORR_v1c05100
GTP-binding protein YsxC
Accession: ASP28281
Location: 584821-585426
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 218
Sequence coverage: 97 %
E-value: 5e-68
NCBI BlastP on this gene
Accession: ASP28281
Location: 584821-585426
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 218
Sequence coverage: 97 %
E-value: 5e-68
NCBI BlastP on this gene
engB
SCORR_v1c05080
cell shape determining protein MreB
Accession: ASP28279
Location: 582732-583772
NCBI BlastP on this gene
Accession: ASP28279
Location: 582732-583772
NCBI BlastP on this gene
mreB
SCORR_v1c05060
Holliday junction DNA helicase RuvA
Accession: ASP28277
Location: 581697-582242
NCBI BlastP on this gene
Accession: ASP28277
Location: 581697-582242
NCBI BlastP on this gene
ruvA
Holliday junction DNA helicase RuvB
Accession: ASP28276
Location: 580763-581710
NCBI BlastP on this gene
Accession: ASP28276
Location: 580763-581710
NCBI BlastP on this gene
ruvB
SCORR_v1c05030
SCORR_v1c05020
326. : CP024870 Spiroplasma clarkii strain CN-5 chromosome Total score: 1.0 Cumulative Blast bit score: 215
PTS system, beta-glucoside-specific IIABC component
Accession: ATX70920
Location: 683593-686091
NCBI BlastP on this gene
Accession: ATX70920
Location: 683593-686091
NCBI BlastP on this gene
SCLAR_v1c06010
bglA
SCLAR_v1c06030
SCLAR_v1c06040
SCLAR_v1c06050
GTP-binding protein YsxC
Accession: ATX70925
Location: 690554-691150
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 215
Sequence coverage: 96 %
E-value: 6e-67
NCBI BlastP on this gene
Accession: ATX70925
Location: 690554-691150
BlastP hit with AAP55641.1
Percentage identity: 55 %
BlastP bit score: 215
Sequence coverage: 96 %
E-value: 6e-67
NCBI BlastP on this gene
engB
SCLAR_v1c06070
SCLAR_v1c06080
SCLAR_v1c06090
327. : CP043026 Spiroplasma chinense strain CCH chromosome Total score: 1.0 Cumulative Blast bit score: 214
GTP-binding protein YsxC
Accession: QEH61914
Location: 815714-816298
BlastP hit with AAP55641.1
Percentage identity: 60 %
BlastP bit score: 214
Sequence coverage: 88 %
E-value: 9e-67
NCBI BlastP on this gene
Accession: QEH61914
Location: 815714-816298
BlastP hit with AAP55641.1
Percentage identity: 60 %
BlastP bit score: 214
Sequence coverage: 88 %
E-value: 9e-67
NCBI BlastP on this gene
engB
SCHIN_v1c07180
SCHIN_v1c07170
SCHIN_v1c07160
SCHIN_v1c07150
328. : CP024967 Mesoplasma lactucae ATCC 49193 strain 831-C4 chromosome Total score: 1.0 Cumulative Blast bit score: 211
GTP-binding protein
Accession: ATZ20177
Location: 431581-432189
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
Accession: ATZ20177
Location: 431581-432189
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
engB
ctp
MLACT_v1c03540
329. : CP023668 Mesoplasma lactucae ATCC 49193 strain 831-C4 chromosome Total score: 1.0 Cumulative Blast bit score: 211
CP520_01150
peptide-methionine (S)-S-oxide reductase
Accession: ATG97365
Location: 274859-275353
NCBI BlastP on this gene
Accession: ATG97365
Location: 274859-275353
NCBI BlastP on this gene
msrA
peptide-methionine (R)-S-oxide reductase
Accession: ATG97366
Location: 275387-275824
NCBI BlastP on this gene
Accession: ATG97366
Location: 275387-275824
NCBI BlastP on this gene
msrB
CP520_01165
CP520_01170
CP520_01175
CP520_01180
YihA family ribosome biogenesis GTP-binding protein
Accession: ATG97371
Location: 278722-279330
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
Accession: ATG97371
Location: 278722-279330
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 98 %
E-value: 5e-65
NCBI BlastP on this gene
CP520_01185
CP520_01190
CP520_01195
330. : CP017015 Spiroplasma helicoides strain TABS-2 chromosome Total score: 1.0 Cumulative Blast bit score: 211
SHELI_v1c05220
SHELI_v1c05230
SHELI_v1c05240
SHELI_v1c05250
SHELI_v1c05260
SHELI_v1c05270
SHELI_v1c05280
GTP-binding protein YsxC
Accession: AOG60480
Location: 604570-605169
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 97 %
E-value: 2e-65
NCBI BlastP on this gene
Accession: AOG60480
Location: 604570-605169
BlastP hit with AAP55641.1
Percentage identity: 53 %
BlastP bit score: 211
Sequence coverage: 97 %
E-value: 2e-65
NCBI BlastP on this gene
engB
SHELI_v1c05300
SHELI_v1c05310
SHELI_v1c05320
SHELI_v1c05330
SHELI_v1c05340
SHELI_v1c05350
331. : CP038013 Spiroplasma gladiatoris strain TG-1 chromosome Total score: 1.0 Cumulative Blast bit score: 197
SGLAD_v1c05380
SGLAD_v1c05370
SGLAD_v1c05360
SGLAD_v1c05350
SGLAD_v1c05340
GTP-binding protein YsxC
Accession: QBQ07732
Location: 597397-597984
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 197
Sequence coverage: 95 %
E-value: 6e-60
NCBI BlastP on this gene
Accession: QBQ07732
Location: 597397-597984
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 197
Sequence coverage: 95 %
E-value: 6e-60
NCBI BlastP on this gene
engB
SGLAD_v1c05320
SGLAD_v1c05310
SGLAD_v1c05300
SGLAD_v1c05290
332. : CP046276 Spiroplasma tabanidicola strain TAUS-1 chromosome Total score: 1.0 Cumulative Blast bit score: 195
STABA_v1c06480
NAD(P)H-dependent oxidoreductase
Accession: QGS52008
Location: 741079-741660
NCBI BlastP on this gene
Accession: QGS52008
Location: 741079-741660
NCBI BlastP on this gene
STABA_v1c06470
STABA_v1c06460
STABA_v1c06450
STABA_v1c06440
GTP-binding protein
Accession: QGS52004
Location: 738834-739421
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 195
Sequence coverage: 96 %
E-value: 3e-59
NCBI BlastP on this gene
Accession: QGS52004
Location: 738834-739421
BlastP hit with AAP55641.1
Percentage identity: 51 %
BlastP bit score: 195
Sequence coverage: 96 %
E-value: 3e-59
NCBI BlastP on this gene
engB
STABA_v1c06420
STABA_v1c06410
STABA_v1c06400
STABA_v1c06390
STABA_v1c06380
P-type Ca2+ transporter type 2C
Accession: QGS51998
Location: 731769-734714
NCBI BlastP on this gene
Accession: QGS51998
Location: 731769-734714
NCBI BlastP on this gene
STABA_v1c06370
333. : LR134274 Streptococcus salivarius strain NCTC8618 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 192
hydroxymethylglutaryl-CoA reductase, degradative
Accession: VED87846
Location: 615074-616345
NCBI BlastP on this gene
Accession: VED87846
Location: 615074-616345
NCBI BlastP on this gene
mvaA
hydroxymethylglutaryl-CoA synthase
Accession: VED87848
Location: 616347-617513
NCBI BlastP on this gene
Accession: VED87848
Location: 616347-617513
NCBI BlastP on this gene
mvaS
thyA_2
folA_2
putative uncharacterized conserved protein
Accession: VED87853
Location: 619460-619630
NCBI BlastP on this gene
Accession: VED87853
Location: 619460-619630
NCBI BlastP on this gene
NCTC8618_03036
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: VED87855
Location: 619648-620874
NCBI BlastP on this gene
Accession: VED87855
Location: 619648-620874
NCBI BlastP on this gene
clpX_2
ribosome biogenesis GTP-binding protein YsxC
Accession: VED87857
Location: 620884-621483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
Accession: VED87857
Location: 620884-621483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
engB_2
lysP
homocysteine S-methyltransferase 1
Accession: VED87861
Location: 623002-623952
NCBI BlastP on this gene
Accession: VED87861
Location: 623002-623952
NCBI BlastP on this gene
mmuM
Galactose mutarotase and related enzyme
Accession: VED87862
Location: 624065-624319
NCBI BlastP on this gene
Accession: VED87862
Location: 624065-624319
NCBI BlastP on this gene
NCTC8618_03041
NCTC8618_03042
acyl-phosphate glycerol 3-phosphate acyltransferase
Accession: VED87866
Location: 624889-625530
NCBI BlastP on this gene
Accession: VED87866
Location: 624889-625530
NCBI BlastP on this gene
plsY_2
parE_3
334. : CP014144 Streptococcus salivarius strain JF Total score: 1.0 Cumulative Blast bit score: 192
3-hydroxy-3-methylglutaryl-CoA reductase
Accession: AWB63_02675
Location: 543585-544031
NCBI BlastP on this gene
Accession: AWB63_02675
Location: 543585-544031
NCBI BlastP on this gene
AWB63_02675
hydroxymethylglutaryl-CoA synthase
Accession: AMB82341
Location: 544033-545199
NCBI BlastP on this gene
Accession: AMB82341
Location: 544033-545199
NCBI BlastP on this gene
AWB63_02680
AWB63_02685
AWB63_02690
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: AMB82344
Location: 547334-548560
NCBI BlastP on this gene
Accession: AMB82344
Location: 547334-548560
NCBI BlastP on this gene
AWB63_02695
GTP-binding protein
Accession: AMB82345
Location: 548570-549169
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
Accession: AMB82345
Location: 548570-549169
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
engB
AWB63_02705
homocysteine S-methyltransferase
Accession: AMB82347
Location: 550688-551638
NCBI BlastP on this gene
Accession: AMB82347
Location: 550688-551638
NCBI BlastP on this gene
mmuM
AWB63_02715
AWB63_02720
glycerol-3-phosphate acyltransferase
Accession: AMB82350
Location: 552575-553216
NCBI BlastP on this gene
Accession: AMB82350
Location: 552575-553216
NCBI BlastP on this gene
AWB63_02725
gyrB
335. : CP009913 Streptococcus salivarius strain NCTC 8618 Total score: 1.0 Cumulative Blast bit score: 192
3-hydroxy-3-methylglutaryl-CoA reductase
Accession: AIY20743
Location: 615418-616689
NCBI BlastP on this gene
Accession: AIY20743
Location: 615418-616689
NCBI BlastP on this gene
SSAL8618_03035
SSAL8618_03040
SSAL8618_03045
SSAL8618_03050
SSAL8618_03055
SSAL8618_03060
GTP-binding protein
Accession: AIY20749
Location: 621228-621827
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
Accession: AIY20749
Location: 621228-621827
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 192
Sequence coverage: 95 %
E-value: 5e-58
NCBI BlastP on this gene
engB
SSAL8618_03070
mmuM
SSAL8618_03080
SSAL8618_03085
glycerol-3-phosphate acyltransferase
Accession: AIY20754
Location: 625233-625874
NCBI BlastP on this gene
Accession: AIY20754
Location: 625233-625874
NCBI BlastP on this gene
SSAL8618_03090
gyrB
336. : LS483366 Streptococcus salivarius strain NCTC7366 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 190
hydroxymethylglutaryl-CoA reductase, degradative
Accession: SQF76373
Location: 1574858-1576129
NCBI BlastP on this gene
Accession: SQF76373
Location: 1574858-1576129
NCBI BlastP on this gene
mvaA
hydroxymethylglutaryl-CoA synthase
Accession: SQF76372
Location: 1573690-1574856
NCBI BlastP on this gene
Accession: SQF76372
Location: 1573690-1574856
NCBI BlastP on this gene
mvaS
thyA
folA
putative uncharacterized conserved protein
Accession: SQF76369
Location: 1571737-1571907
NCBI BlastP on this gene
Accession: SQF76369
Location: 1571737-1571907
NCBI BlastP on this gene
NCTC7366_01457
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: SQF76368
Location: 1570493-1571719
NCBI BlastP on this gene
Accession: SQF76368
Location: 1570493-1571719
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: SQF76367
Location: 1569884-1570483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: SQF76367
Location: 1569884-1570483
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
lysP_2
homocysteine S-methyltransferase 1
Accession: SQF76365
Location: 1567416-1568366
NCBI BlastP on this gene
Accession: SQF76365
Location: 1567416-1568366
NCBI BlastP on this gene
mmuM
Galactose mutarotase and related enzyme
Accession: SQF76364
Location: 1567049-1567303
NCBI BlastP on this gene
Accession: SQF76364
Location: 1567049-1567303
NCBI BlastP on this gene
NCTC7366_01452
NCTC7366_01451
acyl-phosphate glycerol 3-phosphate acyltransferase
Accession: SQF76362
Location: 1565838-1566479
NCBI BlastP on this gene
Accession: SQF76362
Location: 1565838-1566479
NCBI BlastP on this gene
plsY
DNA topoisomerase IV subunit B
Accession: SQF76361
Location: 1563745-1565700
NCBI BlastP on this gene
Accession: SQF76361
Location: 1563745-1565700
NCBI BlastP on this gene
parE
337. : CP040804 Streptococcus salivarius strain LAB813 chromosome Total score: 1.0 Cumulative Blast bit score: 190
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QEM32286
Location: 788258-789532
NCBI BlastP on this gene
Accession: QEM32286
Location: 788258-789532
NCBI BlastP on this gene
FHI56_05045
hydroxymethylglutaryl-CoA synthase
Accession: QEM32287
Location: 789534-790700
NCBI BlastP on this gene
Accession: QEM32287
Location: 789534-790700
NCBI BlastP on this gene
FHI56_05050
FHI56_05055
FHI56_05060
FHI56_05065
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: QEM32291
Location: 792670-793896
NCBI BlastP on this gene
Accession: QEM32291
Location: 792670-793896
NCBI BlastP on this gene
clpX
YihA family ribosome biogenesis GTP-binding protein
Accession: QEM32292
Location: 793906-794505
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QEM32292
Location: 793906-794505
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
FHI56_05075
338. : CP018189 Streptococcus salivarius strain ICDC3 chromosome Total score: 1.0 Cumulative Blast bit score: 190
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QGU83041
Location: 1580717-1581991
NCBI BlastP on this gene
Accession: QGU83041
Location: 1580717-1581991
NCBI BlastP on this gene
BSR20_07285
hydroxymethylglutaryl-CoA synthase
Accession: QGU83040
Location: 1579549-1580715
NCBI BlastP on this gene
Accession: QGU83040
Location: 1579549-1580715
NCBI BlastP on this gene
BSR20_07280
BSR20_07275
BSR20_07270
ATP-dependent protease ATP-binding subunit ClpX
Accession: QGU83037
Location: 1576175-1577401
NCBI BlastP on this gene
Accession: QGU83037
Location: 1576175-1577401
NCBI BlastP on this gene
BSR20_07265
YihA family ribosome biogenesis GTP-binding protein
Accession: QGU83036
Location: 1575566-1576165
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QGU83036
Location: 1575566-1576165
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
BSR20_07255
homocysteine S-methyltransferase
Accession: QGU83034
Location: 1573098-1574048
NCBI BlastP on this gene
Accession: QGU83034
Location: 1573098-1574048
NCBI BlastP on this gene
BSR20_07250
BSR20_07245
BSR20_07240
glycerol-3-phosphate acyltransferase
Accession: QGU83031
Location: 1571520-1572161
NCBI BlastP on this gene
Accession: QGU83031
Location: 1571520-1572161
NCBI BlastP on this gene
BSR20_07235
DNA topoisomerase IV subunit B
Accession: QGU83030
Location: 1569433-1571382
NCBI BlastP on this gene
Accession: QGU83030
Location: 1569433-1571382
NCBI BlastP on this gene
BSR20_07230
339. : CP018187 Streptococcus salivarius strain ICDC2 Total score: 1.0 Cumulative Blast bit score: 190
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QGU80161
Location: 596170-597441
NCBI BlastP on this gene
Accession: QGU80161
Location: 596170-597441
NCBI BlastP on this gene
BSR19_03020
hydroxymethylglutaryl-CoA synthase
Accession: QGU80162
Location: 597443-598609
NCBI BlastP on this gene
Accession: QGU80162
Location: 597443-598609
NCBI BlastP on this gene
BSR19_03025
BSR19_03030
BSR19_03035
ATP-dependent protease ATP-binding subunit ClpX
Accession: QGU80165
Location: 600737-601963
NCBI BlastP on this gene
Accession: QGU80165
Location: 600737-601963
NCBI BlastP on this gene
BSR19_03040
YihA family ribosome biogenesis GTP-binding protein
Accession: QGU80166
Location: 601973-602572
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QGU80166
Location: 601973-602572
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
BSR19_03050
homocysteine S-methyltransferase
Accession: QGU80168
Location: 604089-605039
NCBI BlastP on this gene
Accession: QGU80168
Location: 604089-605039
NCBI BlastP on this gene
BSR19_03055
BSR19_03060
BSR19_03065
glycerol-3-phosphate acyltransferase
Accession: QGU80171
Location: 605973-606614
NCBI BlastP on this gene
Accession: QGU80171
Location: 605973-606614
NCBI BlastP on this gene
BSR19_03070
BSR19_03075
340. : CP018186 Streptococcus salivarius strain ICDC1 chromosome Total score: 1.0 Cumulative Blast bit score: 190
hydroxymethylglutaryl-CoA reductase, degradative
Accession: QGU79032
Location: 1580742-1582016
NCBI BlastP on this gene
Accession: QGU79032
Location: 1580742-1582016
NCBI BlastP on this gene
BSR14_07285
hydroxymethylglutaryl-CoA synthase
Accession: QGU79031
Location: 1579574-1580740
NCBI BlastP on this gene
Accession: QGU79031
Location: 1579574-1580740
NCBI BlastP on this gene
BSR14_07280
BSR14_07275
BSR14_07270
ATP-dependent protease ATP-binding subunit ClpX
Accession: QGU79028
Location: 1576200-1577426
NCBI BlastP on this gene
Accession: QGU79028
Location: 1576200-1577426
NCBI BlastP on this gene
BSR14_07265
YihA family ribosome biogenesis GTP-binding protein
Accession: QGU79027
Location: 1575591-1576190
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: QGU79027
Location: 1575591-1576190
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
BSR14_07255
homocysteine S-methyltransferase
Accession: QGU79025
Location: 1573123-1574073
NCBI BlastP on this gene
Accession: QGU79025
Location: 1573123-1574073
NCBI BlastP on this gene
BSR14_07250
BSR14_07245
BSR14_07240
glycerol-3-phosphate acyltransferase
Accession: QGU79022
Location: 1571545-1572186
NCBI BlastP on this gene
Accession: QGU79022
Location: 1571545-1572186
NCBI BlastP on this gene
BSR14_07235
DNA topoisomerase IV subunit B
Accession: QGU79021
Location: 1569458-1571407
NCBI BlastP on this gene
Accession: QGU79021
Location: 1569458-1571407
NCBI BlastP on this gene
BSR14_07230
341. : CP015283 Streptococcus salivarius strain ATCC 25975 Total score: 1.0 Cumulative Blast bit score: 190
YihA family ribosome biogenesis GTP-binding protein
Accession: ARI60516
Location: 2125010-2125609
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: ARI60516
Location: 2125010-2125609
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
V471_09855
homocysteine S-methyltransferase
Accession: ARI60514
Location: 2122542-2123492
NCBI BlastP on this gene
Accession: ARI60514
Location: 2122542-2123492
NCBI BlastP on this gene
mmuM
V471_09845
V471_09840
glycerol-3-phosphate acyltransferase
Accession: ARI60511
Location: 2120964-2121605
NCBI BlastP on this gene
Accession: ARI60511
Location: 2120964-2121605
NCBI BlastP on this gene
V471_09835
DNA topoisomerase IV subunit B
Accession: ARI60510
Location: 2118877-2120826
NCBI BlastP on this gene
Accession: ARI60510
Location: 2118877-2120826
NCBI BlastP on this gene
gyrB
342. : CP015282 Streptococcus salivarius strain ATCC 27945 Total score: 1.0 Cumulative Blast bit score: 190
3-hydroxy-3-methylglutaryl-CoA reductase
Accession: ARI57667
Location: 972034-973305
NCBI BlastP on this gene
Accession: ARI57667
Location: 972034-973305
NCBI BlastP on this gene
NX99_04050
hydroxymethylglutaryl-CoA synthase
Accession: ARI57666
Location: 970866-972032
NCBI BlastP on this gene
Accession: ARI57666
Location: 970866-972032
NCBI BlastP on this gene
NX99_04045
NX99_04040
NX99_04035
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: ARI57663
Location: 967504-968730
NCBI BlastP on this gene
Accession: ARI57663
Location: 967504-968730
NCBI BlastP on this gene
NX99_04030
YihA family ribosome biogenesis GTP-binding protein
Accession: ARI57662
Location: 966895-967494
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: ARI57662
Location: 966895-967494
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
NX99_04020
homocysteine S-methyltransferase
Accession: ARI57660
Location: 964427-965377
NCBI BlastP on this gene
Accession: ARI57660
Location: 964427-965377
NCBI BlastP on this gene
mmuM
NX99_04010
NX99_04005
glycerol-3-phosphate acyltransferase
Accession: ARI57657
Location: 962845-963486
NCBI BlastP on this gene
Accession: ARI57657
Location: 962845-963486
NCBI BlastP on this gene
NX99_04000
gyrB
343. : CP012646 Streptococcus mitis strain KCOM 1350 (= ChDC B183) Total score: 1.0 Cumulative Blast bit score: 190
RN80_04340
RN80_04345
RN80_04350
RN80_04355
RN80_04360
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: ALD67710
Location: 874218-875450
NCBI BlastP on this gene
Accession: ALD67710
Location: 874218-875450
NCBI BlastP on this gene
RN80_04365
GTP-binding protein
Accession: ALD67711
Location: 875459-876046
BlastP hit with AAP55641.1
Percentage identity: 50 %
BlastP bit score: 190
Sequence coverage: 91 %
E-value: 4e-57
NCBI BlastP on this gene
Accession: ALD67711
Location: 875459-876046
BlastP hit with AAP55641.1
Percentage identity: 50 %
BlastP bit score: 190
Sequence coverage: 91 %
E-value: 4e-57
NCBI BlastP on this gene
engB
344. : CP002888 Streptococcus salivarius 57.I Total score: 1.0 Cumulative Blast bit score: 190
ribosome biogenesis GTP-binding protein YsxC
Accession: AEJ53843
Location: 1550139-1550738
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
Accession: AEJ53843
Location: 1550139-1550738
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 190
Sequence coverage: 95 %
E-value: 5e-57
NCBI BlastP on this gene
engB
rocE
homocysteine S-methyltransferase 1
Accession: AEJ53841
Location: 1547671-1548621
NCBI BlastP on this gene
Accession: AEJ53841
Location: 1547671-1548621
NCBI BlastP on this gene
mmuM
conserved hypothetical protein
Accession: AEJ53840
Location: 1547304-1547558
NCBI BlastP on this gene
Accession: AEJ53840
Location: 1547304-1547558
NCBI BlastP on this gene
Ssal_01575
conserved hypothetical protein
Accession: AEJ53839
Location: 1546852-1547229
NCBI BlastP on this gene
Accession: AEJ53839
Location: 1546852-1547229
NCBI BlastP on this gene
Ssal_01574
acyl-phosphate glycerol 3-phosphate acyltransferase
Accession: AEJ53838
Location: 1546093-1546734
NCBI BlastP on this gene
Accession: AEJ53838
Location: 1546093-1546734
NCBI BlastP on this gene
plsY
DNA topoisomerase IV, B subunit
Accession: AEJ53837
Location: 1544006-1545955
NCBI BlastP on this gene
Accession: AEJ53837
Location: 1544006-1545955
NCBI BlastP on this gene
parE
345. : CP002471 Streptococcus parauberis KCTC 11537 Total score: 1.0 Cumulative Blast bit score: 189
3-hydroxy-3-methylglutaryl-coenzyme A reductase
Accession: AEF25052
Location: 643042-644325
NCBI BlastP on this gene
Accession: AEF25052
Location: 643042-644325
NCBI BlastP on this gene
mvaA
3-hydroxy-3-methylglutaryl coenzyme A synthase
Accession: AEF25053
Location: 644322-645488
NCBI BlastP on this gene
Accession: AEF25053
Location: 644322-645488
NCBI BlastP on this gene
mvaS
thyA
dyr
STP_0608
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: AEF25057
Location: 647477-648544
NCBI BlastP on this gene
Accession: AEF25057
Location: 647477-648544
NCBI BlastP on this gene
clpX
GTP-binding protein
Accession: AEF25058
Location: 648553-649152
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 189
Sequence coverage: 91 %
E-value: 9e-57
NCBI BlastP on this gene
Accession: AEF25058
Location: 648553-649152
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 189
Sequence coverage: 91 %
E-value: 9e-57
NCBI BlastP on this gene
STP_0610
STP_0611
ATP-dependent protease ATP-binding subunit ClpL
Accession: AEF25060
Location: 650005-652113
NCBI BlastP on this gene
Accession: AEF25060
Location: 650005-652113
NCBI BlastP on this gene
clpL
rplJ
rplL
STP_0615
346. : LR594040 Streptococcus australis strain NCTC5338 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
galE
yfdH
NCTC5338_01373
cmk
dhfR
NCTC5338_01370
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VTS72167
Location: 1369417-1370625
NCBI BlastP on this gene
Accession: VTS72167
Location: 1369417-1370625
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VTS72164
Location: 1368821-1369408
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VTS72164
Location: 1368821-1369408
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
NCTC5338_01365
putative sporulation transcription regulator whiA
Accession: VTS72152
Location: 1365607-1366518
NCBI BlastP on this gene
Accession: VTS72152
Location: 1365607-1366518
NCBI BlastP on this gene
whiA
NCTC5338_01363
NCTC5338_01362
NCTC5338_01361
NCTC5338_01360
NCTC5338_01359
Protein of uncharacterised function (DUF2974)
Accession: VTS72134
Location: 1362391-1363635
NCBI BlastP on this gene
Accession: VTS72134
Location: 1362391-1363635
NCBI BlastP on this gene
NCTC5338_01358
347. : LR216061 Streptococcus pneumoniae strain 2245STDY6178826 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
tpiA
lytC
DPS family peroxide resistance protein
Accession: VFI44001
Location: 1551212-1551730
NCBI BlastP on this gene
Accession: VFI44001
Location: 1551212-1551730
NCBI BlastP on this gene
dps
dhfR
SAMEA3713960_01605
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VFI43976
Location: 1549142-1550374
NCBI BlastP on this gene
Accession: VFI43976
Location: 1549142-1550374
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VFI43968
Location: 1548546-1549133
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VFI43968
Location: 1548546-1549133
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
SAMEA3713960_01600
putative sporulation transcription regulator whiA
Accession: VFI43935
Location: 1545331-1546242
NCBI BlastP on this gene
Accession: VFI43935
Location: 1545331-1546242
NCBI BlastP on this gene
whiA
yumC
DNA integrity scanning protein DisA
Accession: VFI43918
Location: 1543336-1544193
NCBI BlastP on this gene
Accession: VFI43918
Location: 1543336-1544193
NCBI BlastP on this gene
ybbP
SAMEA3713960_01596
348. : LR216040 Streptococcus pneumoniae strain 2245STDY6030848 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
tpiA
lytC
DPS family peroxide resistance protein
Accession: VFH95156
Location: 1504988-1505506
NCBI BlastP on this gene
Accession: VFH95156
Location: 1504988-1505506
NCBI BlastP on this gene
dps
dhfR
SAMEA3233211_01532
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VFH95153
Location: 1502913-1504121
NCBI BlastP on this gene
Accession: VFH95153
Location: 1502913-1504121
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VFH95152
Location: 1502317-1502904
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VFH95152
Location: 1502317-1502904
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
SAMEA3233211_01527
putative sporulation transcription regulator whiA
Accession: VFH95147
Location: 1499102-1500013
NCBI BlastP on this gene
Accession: VFH95147
Location: 1499102-1500013
NCBI BlastP on this gene
whiA
yumC
DNA integrity scanning protein DisA
Accession: VFH95143
Location: 1497107-1497964
NCBI BlastP on this gene
Accession: VFH95143
Location: 1497107-1497964
NCBI BlastP on this gene
ybbP
SAMEA3233211_01523
349. : LR216032 Streptococcus pneumoniae strain 2245STDY5775874 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
tpiA
lytC
DPS family peroxide resistance protein
Accession: VFH78713
Location: 1418791-1419309
NCBI BlastP on this gene
Accession: VFH78713
Location: 1418791-1419309
NCBI BlastP on this gene
dps
dhfR
SAMEA2434829_01438
ATP-dependent Clp protease, ATP-binding subunit ClpX
Accession: VFH78690
Location: 1416721-1417953
NCBI BlastP on this gene
Accession: VFH78690
Location: 1416721-1417953
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein YsxC
Accession: VFH78682
Location: 1416125-1416712
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
Accession: VFH78682
Location: 1416125-1416712
BlastP hit with AAP55641.1
Percentage identity: 49 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 2e-56
NCBI BlastP on this gene
engB
tdcF
yvcJ
SAMEA2434829_01433
putative sporulation transcription regulator whiA
Accession: VFH78647
Location: 1412910-1413821
NCBI BlastP on this gene
Accession: VFH78647
Location: 1412910-1413821
NCBI BlastP on this gene
whiA
yumC
DNA integrity scanning protein DisA
Accession: VFH78638
Location: 1410918-1411775
NCBI BlastP on this gene
Accession: VFH78638
Location: 1410918-1411775
NCBI BlastP on this gene
ybbP
SAMEA2434829_01429
350. : LR134323 Streptococcus urinalis strain NCTC13766 genome assembly, chromosome: 1. Total score: 1.0 Cumulative Blast bit score: 188
hydroxymethylglutaryl-CoA reductase, degradative
Accession: VEF31882
Location: 914960-916237
NCBI BlastP on this gene
Accession: VEF31882
Location: 914960-916237
NCBI BlastP on this gene
mvaA
Hydroxymethylglutaryl-CoA synthase
Accession: VEF31883
Location: 916221-917402
NCBI BlastP on this gene
Accession: VEF31883
Location: 916221-917402
NCBI BlastP on this gene
pksG
thyA
dfrA
NCTC13766_00985
ATP-dependent Clp protease ATP-binding subunit ClpX
Accession: VEF31887
Location: 919230-920456
NCBI BlastP on this gene
Accession: VEF31887
Location: 919230-920456
NCBI BlastP on this gene
clpX
ribosome biogenesis GTP-binding protein
Accession: VEF31888
Location: 920468-921067
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 3e-56
NCBI BlastP on this gene
Accession: VEF31888
Location: 920468-921067
BlastP hit with AAP55641.1
Percentage identity: 48 %
BlastP bit score: 188
Sequence coverage: 91 %
E-value: 3e-56
NCBI BlastP on this gene
engB


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.