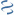MultiGeneBlast hits

Query: S.xylosus malR gene and malA gene.
CP014397 : Staphylococcus aureus strain USA300-SUR10 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM23_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR10683
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQR10683
Location: 1673782-1675188
NCBI BlastP on this gene
AYM23_08435
AYM23_08430
AYM23_08425
LacI family transcriptional regulator
Accession: AQR10680
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR10680
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM23_08420
glucohydrolase
Accession: AQR10679
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR10679
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM23_08415
AraC family transcriptional regulator
Accession: AQR10678
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQR10678
Location: 1669338-1670231
NCBI BlastP on this gene
AYM23_08410
glucose-6-phosphate dehydrogenase
Accession: AQR10677
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQR10677
Location: 1667651-1669135
NCBI BlastP on this gene
AYM23_08405
Query: S.xylosus malR gene and malA gene.
CP014392 : Staphylococcus aureus strain USA300-SUR9 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM22_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR07744
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQR07744
Location: 1673782-1675188
NCBI BlastP on this gene
AYM22_08435
AYM22_08430
AYM22_08425
LacI family transcriptional regulator
Accession: AQR07741
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR07741
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM22_08420
glucohydrolase
Accession: AQR07740
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR07740
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM22_08415
AraC family transcriptional regulator
Accession: AQR07739
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQR07739
Location: 1669338-1670231
NCBI BlastP on this gene
AYM22_08410
glucose-6-phosphate dehydrogenase
Accession: AQR07738
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQR07738
Location: 1667651-1669135
NCBI BlastP on this gene
AYM22_08405
Query: S.xylosus malR gene and malA gene.
CP014387 : Staphylococcus aureus strain USA300-SUR8. Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM21_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR04798
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQR04798
Location: 1673782-1675188
NCBI BlastP on this gene
AYM21_08435
AYM21_08430
AYM21_08425
LacI family transcriptional regulator
Accession: AQR04795
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR04795
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM21_08420
glucohydrolase
Accession: AQR04794
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR04794
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM21_08415
AraC family transcriptional regulator
Accession: AQR04793
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQR04793
Location: 1669338-1670231
NCBI BlastP on this gene
AYM21_08410
glucose-6-phosphate dehydrogenase
Accession: AQR04792
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQR04792
Location: 1667651-1669135
NCBI BlastP on this gene
AYM21_08405
Query: S.xylosus malR gene and malA gene.
CP014384 : Staphylococcus aureus strain USA300-SUR7 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM20_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR01858
Location: 1673706-1675112
NCBI BlastP on this gene
Accession: AQR01858
Location: 1673706-1675112
NCBI BlastP on this gene
AYM20_08435
AYM20_08430
AYM20_08425
LacI family transcriptional regulator
Accession: AQR01855
Location: 1671903-1672922
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR01855
Location: 1671903-1672922
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM20_08420
glucohydrolase
Accession: AQR01854
Location: 1670238-1671887
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR01854
Location: 1670238-1671887
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM20_08415
AraC family transcriptional regulator
Accession: AQR01853
Location: 1669262-1670155
NCBI BlastP on this gene
Accession: AQR01853
Location: 1669262-1670155
NCBI BlastP on this gene
AYM20_08410
glucose-6-phosphate dehydrogenase
Accession: AQR01852
Location: 1667575-1669059
NCBI BlastP on this gene
Accession: AQR01852
Location: 1667575-1669059
NCBI BlastP on this gene
AYM20_08405
Query: S.xylosus malR gene and malA gene.
CP014381 : Staphylococcus aureus strain USA300-SUR6. Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM19_08435
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ98893
Location: 1673702-1675108
NCBI BlastP on this gene
Accession: AQQ98893
Location: 1673702-1675108
NCBI BlastP on this gene
AYM19_08430
AYM19_08425
AYM19_08420
LacI family transcriptional regulator
Accession: AQQ98890
Location: 1671899-1672918
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ98890
Location: 1671899-1672918
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM19_08415
glucohydrolase
Accession: AQQ98889
Location: 1670234-1671883
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ98889
Location: 1670234-1671883
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM19_08410
AraC family transcriptional regulator
Accession: AQQ98888
Location: 1669258-1670151
NCBI BlastP on this gene
Accession: AQQ98888
Location: 1669258-1670151
NCBI BlastP on this gene
AYM19_08405
glucose-6-phosphate dehydrogenase
Accession: AQQ98887
Location: 1667571-1669055
NCBI BlastP on this gene
Accession: AQQ98887
Location: 1667571-1669055
NCBI BlastP on this gene
AYM19_08400
Query: S.xylosus malR gene and malA gene.
CP014376 : Staphylococcus aureus strain USA300-SUR5 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM18_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ95954
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQQ95954
Location: 1673782-1675188
NCBI BlastP on this gene
AYM18_08435
AYM18_08430
AYM18_08425
LacI family transcriptional regulator
Accession: AQQ95951
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ95951
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM18_08420
glucohydrolase
Accession: AQQ95950
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ95950
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM18_08415
AraC family transcriptional regulator
Accession: AQQ95949
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQQ95949
Location: 1669338-1670231
NCBI BlastP on this gene
AYM18_08410
glucose-6-phosphate dehydrogenase
Accession: AQQ95948
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQQ95948
Location: 1667651-1669135
NCBI BlastP on this gene
AYM18_08405
Query: S.xylosus malR gene and malA gene.
CP014371 : Staphylococcus aureus strain USA300-SUR4 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM16_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ93015
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQQ93015
Location: 1673782-1675188
NCBI BlastP on this gene
AYM16_08435
AYM16_08430
AYM16_08425
LacI family transcriptional regulator
Accession: AQQ93012
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ93012
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM16_08420
glucohydrolase
Accession: AQQ93011
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ93011
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM16_08415
AraC family transcriptional regulator
Accession: AQQ93010
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQQ93010
Location: 1669338-1670231
NCBI BlastP on this gene
AYM16_08410
glucose-6-phosphate dehydrogenase
Accession: AQQ93009
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQQ93009
Location: 1667651-1669135
NCBI BlastP on this gene
AYM16_08405
Query: S.xylosus malR gene and malA gene.
CP014368 : Staphylococcus aureus strain USA300-SUR3 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM15_08435
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ90073
Location: 1673690-1675096
NCBI BlastP on this gene
Accession: AQQ90073
Location: 1673690-1675096
NCBI BlastP on this gene
AYM15_08430
AYM15_08425
AYM15_08420
LacI family transcriptional regulator
Accession: AQQ90070
Location: 1671887-1672906
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ90070
Location: 1671887-1672906
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM15_08415
glucohydrolase
Accession: AQQ90069
Location: 1670222-1671871
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ90069
Location: 1670222-1671871
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM15_08410
AraC family transcriptional regulator
Accession: AQQ90068
Location: 1669246-1670139
NCBI BlastP on this gene
Accession: AQQ90068
Location: 1669246-1670139
NCBI BlastP on this gene
AYM15_08405
glucose-6-phosphate dehydrogenase
Accession: AQQ90067
Location: 1667559-1669043
NCBI BlastP on this gene
Accession: AQQ90067
Location: 1667559-1669043
NCBI BlastP on this gene
AYM15_08400
Query: S.xylosus malR gene and malA gene.
CP014365 : Staphylococcus aureus strain USA300-SUR2 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM14_08035
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ87072
Location: 1615774-1617180
NCBI BlastP on this gene
Accession: AQQ87072
Location: 1615774-1617180
NCBI BlastP on this gene
AYM14_08030
AYM14_08025
AYM14_08020
LacI family transcriptional regulator
Accession: AQQ87069
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ87069
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM14_08015
glucohydrolase
Accession: AQQ87068
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ87068
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM14_08010
AraC family transcriptional regulator
Accession: AQQ87067
Location: 1611330-1612223
NCBI BlastP on this gene
Accession: AQQ87067
Location: 1611330-1612223
NCBI BlastP on this gene
AYM14_08005
glucose-6-phosphate dehydrogenase
Accession: AQQ87066
Location: 1609643-1611127
NCBI BlastP on this gene
Accession: AQQ87066
Location: 1609643-1611127
NCBI BlastP on this gene
AYM14_08000
Query: S.xylosus malR gene and malA gene.
CP014362 : Staphylococcus aureus strain USA300-SUR1 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM13_08035
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ84145
Location: 1615774-1617180
NCBI BlastP on this gene
Accession: AQQ84145
Location: 1615774-1617180
NCBI BlastP on this gene
AYM13_08030
AYM13_08025
AYM13_08020
LacI family transcriptional regulator
Accession: AQQ84142
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ84142
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM13_08015
glucohydrolase
Accession: AQQ84141
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ84141
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM13_08010
AraC family transcriptional regulator
Accession: AQQ84140
Location: 1611330-1612223
NCBI BlastP on this gene
Accession: AQQ84140
Location: 1611330-1612223
NCBI BlastP on this gene
AYM13_08005
glucose-6-phosphate dehydrogenase
Accession: AQQ84139
Location: 1609643-1611127
NCBI BlastP on this gene
Accession: AQQ84139
Location: 1609643-1611127
NCBI BlastP on this gene
AYM13_08000
Query: S.xylosus malR gene and malA gene.
CP013959 : Staphylococcus aureus strain V605 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
ASL18_08465
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AND44838
Location: 1684736-1686142
NCBI BlastP on this gene
Accession: AND44838
Location: 1684736-1686142
NCBI BlastP on this gene
ASL18_08460
ASL18_08455
ASL18_08450
LacI family transcriptional regulator
Accession: AND44835
Location: 1682933-1683952
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AND44835
Location: 1682933-1683952
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ASL18_08445
glucohydrolase
Accession: AND44834
Location: 1681268-1682917
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AND44834
Location: 1681268-1682917
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ASL18_08440
AraC family transcriptional regulator
Accession: AND44833
Location: 1680292-1681185
NCBI BlastP on this gene
Accession: AND44833
Location: 1680292-1681185
NCBI BlastP on this gene
ASL18_08435
glucose-6-phosphate dehydrogenase
Accession: AND44832
Location: 1678605-1680089
NCBI BlastP on this gene
Accession: AND44832
Location: 1678605-1680089
NCBI BlastP on this gene
ASL18_08430
Query: S.xylosus malR gene and malA gene.
CP013957 : Staphylococcus aureus strain V521 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
ASL17_08150
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AND36116
Location: 1642456-1643862
NCBI BlastP on this gene
Accession: AND36116
Location: 1642456-1643862
NCBI BlastP on this gene
ASL17_08145
ASL17_08140
ASL17_08135
LacI family transcriptional regulator
Accession: AND36113
Location: 1640653-1641672
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AND36113
Location: 1640653-1641672
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ASL17_08130
glucohydrolase
Accession: AND36112
Location: 1638988-1640637
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AND36112
Location: 1638988-1640637
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ASL17_08125
AraC family transcriptional regulator
Accession: AND36111
Location: 1638012-1638905
NCBI BlastP on this gene
Accession: AND36111
Location: 1638012-1638905
NCBI BlastP on this gene
ASL17_08120
glucose-6-phosphate dehydrogenase
Accession: AND36110
Location: 1636325-1637809
NCBI BlastP on this gene
Accession: AND36110
Location: 1636325-1637809
NCBI BlastP on this gene
ASL17_08115
Query: S.xylosus malR gene and malA gene.
CP013231 : Staphylococcus aureus strain UTSW MRSA 55 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AS858_03285
6-phosphogluconate dehydrogenase
Accession: AMO14155
Location: 656273-657679
NCBI BlastP on this gene
Accession: AMO14155
Location: 656273-657679
NCBI BlastP on this gene
AS858_03280
AS858_03275
AS858_03270
LacI family transcriptional regulator
Accession: AMO14152
Location: 654470-655489
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AMO14152
Location: 654470-655489
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AS858_03265
glucohydrolase
Accession: AMO14151
Location: 652805-654454
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AMO14151
Location: 652805-654454
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AS858_03260
AraC family transcriptional regulator
Accession: AMO14150
Location: 651829-652722
NCBI BlastP on this gene
Accession: AMO14150
Location: 651829-652722
NCBI BlastP on this gene
AS858_03255
glucose-6-phosphate dehydrogenase
Accession: AMO14149
Location: 650142-651626
NCBI BlastP on this gene
Accession: AMO14149
Location: 650142-651626
NCBI BlastP on this gene
AS858_03250
Query: S.xylosus malR gene and malA gene.
CP012120 : Staphylococcus aureus subsp. aureus strain USA300_2014.C02 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
ACO02_2154
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AKR46834
Location: 2616347-2617753
NCBI BlastP on this gene
Accession: AKR46834
Location: 2616347-2617753
NCBI BlastP on this gene
ACO02_2155
glyoxalase/bleomycin resistance/extradiol dioxygenase family protein
Accession: AKR46835
Location: 2617972-2618343
NCBI BlastP on this gene
Accession: AKR46835
Location: 2617972-2618343
NCBI BlastP on this gene
ACO02_2156
ACO02_04385
LacI family transcriptional regulator
Accession: AKR46836
Location: 2618537-2619556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKR46836
Location: 2618537-2619556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ACO02_2157
alpha-glucosidase
Accession: AKR46837
Location: 2619572-2621221
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKR46837
Location: 2619572-2621221
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ACO02_2158
AraC family transcriptional regulator
Accession: AKR46838
Location: 2621304-2622197
NCBI BlastP on this gene
Accession: AKR46838
Location: 2621304-2622197
NCBI BlastP on this gene
ACO02_2159
glucose-6-phosphate dehydrogenase
Accession: AKR46840
Location: 2622400-2623884
NCBI BlastP on this gene
Accession: AKR46840
Location: 2622400-2623884
NCBI BlastP on this gene
ACO02_2161
Query: S.xylosus malR gene and malA gene.
CP012119 : Staphylococcus aureus subsp. aureus strain USA300_2014.C01 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AC807_0273
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AKR50357
Location: 735340-736746
NCBI BlastP on this gene
Accession: AKR50357
Location: 735340-736746
NCBI BlastP on this gene
AC807_0274
glyoxalase/bleomycin resistance/extradiol dioxygenase family protein
Accession: AKR50358
Location: 734750-735121
NCBI BlastP on this gene
Accession: AKR50358
Location: 734750-735121
NCBI BlastP on this gene
AC807_0275
AC807_03260
LacI family transcriptional regulator
Accession: AKR50359
Location: 733537-734556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKR50359
Location: 733537-734556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AC807_0276
alpha-glucosidase
Accession: AKR50360
Location: 731872-733521
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKR50360
Location: 731872-733521
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AC807_0277
AraC family transcriptional regulator
Accession: AKR50361
Location: 730896-731789
NCBI BlastP on this gene
Accession: AKR50361
Location: 730896-731789
NCBI BlastP on this gene
AC807_0278
glucose-6-phosphate dehydrogenase
Accession: AKR50363
Location: 729209-730693
NCBI BlastP on this gene
Accession: AKR50363
Location: 729209-730693
NCBI BlastP on this gene
AC807_0280
Query: S.xylosus malR gene and malA gene.
CP012018 : Staphylococcus aureus subsp. aureus strain Gv88 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAGV88_01776
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY29166
Location: 1705391-1706797
NCBI BlastP on this gene
Accession: ALY29166
Location: 1705391-1706797
NCBI BlastP on this gene
gnd
SAGV88_01774
HTH-type transcriptional regulator malR
Accession: ALY29164
Location: 1703588-1704607
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY29164
Location: 1703588-1704607
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY29163
Location: 1701923-1703572
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY29163
Location: 1701923-1703572
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAGV88_01772
AraC family transcriptional regulator
Accession: ALY29162
Location: 1700947-1701840
NCBI BlastP on this gene
Accession: ALY29162
Location: 1700947-1701840
NCBI BlastP on this gene
SAGV88_01771
glucose-6-phosphate 1-dehydrogenase
Accession: ALY29161
Location: 1699260-1700744
NCBI BlastP on this gene
Accession: ALY29161
Location: 1699260-1700744
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP012015 : Staphylococcus aureus subsp. aureus strain Gv51 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAGV51_01783
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY25950
Location: 1706128-1707534
NCBI BlastP on this gene
Accession: ALY25950
Location: 1706128-1707534
NCBI BlastP on this gene
gnd
SAGV51_01781
HTH-type transcriptional regulator malR
Accession: ALY25948
Location: 1704325-1705344
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY25948
Location: 1704325-1705344
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY25947
Location: 1702660-1704309
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY25947
Location: 1702660-1704309
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAGV51_01779
AraC family transcriptional regulator
Accession: ALY25946
Location: 1701684-1702577
NCBI BlastP on this gene
Accession: ALY25946
Location: 1701684-1702577
NCBI BlastP on this gene
SAGV51_01778
glucose-6-phosphate 1-dehydrogenase
Accession: ALY25945
Location: 1699997-1701481
NCBI BlastP on this gene
Accession: ALY25945
Location: 1699997-1701481
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP012013 : Staphylococcus aureus subsp. aureus strain Be62 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SABE62_01774
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY23429
Location: 1710624-1712030
NCBI BlastP on this gene
Accession: ALY23429
Location: 1710624-1712030
NCBI BlastP on this gene
gnd
SABE62_01772
HTH-type transcriptional regulator malR
Accession: ALY23427
Location: 1708821-1709840
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY23427
Location: 1708821-1709840
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY23426
Location: 1707156-1708805
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY23426
Location: 1707156-1708805
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SABE62_01770
AraC family transcriptional regulator
Accession: ALY23425
Location: 1706180-1707073
NCBI BlastP on this gene
Accession: ALY23425
Location: 1706180-1707073
NCBI BlastP on this gene
SABE62_01769
glucose-6-phosphate 1-dehydrogenase
Accession: ALY23424
Location: 1704493-1705977
NCBI BlastP on this gene
Accession: ALY23424
Location: 1704493-1705977
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP012012 : Staphylococcus aureus subsp. aureus strain HC1335 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAHC1335_02534
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY21121
Location: 1700261-1701667
NCBI BlastP on this gene
Accession: ALY21121
Location: 1700261-1701667
NCBI BlastP on this gene
gnd
SAHC1335_02532
HTH-type transcriptional regulator malR
Accession: ALY21119
Location: 1698458-1699477
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY21119
Location: 1698458-1699477
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY21118
Location: 1696793-1698442
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY21118
Location: 1696793-1698442
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAHC1335_02530
AraC family transcriptional regulator
Accession: ALY21117
Location: 1695817-1696710
NCBI BlastP on this gene
Accession: ALY21117
Location: 1695817-1696710
NCBI BlastP on this gene
SAHC1335_02529
glucose-6-phosphate 1-dehydrogenase
Accession: ALY21116
Location: 1694130-1695614
NCBI BlastP on this gene
Accession: ALY21116
Location: 1694130-1695614
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP012011 : Staphylococcus aureus subsp. aureus strain HC1340 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAHC1340_02451
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY18296
Location: 1754870-1756276
NCBI BlastP on this gene
Accession: ALY18296
Location: 1754870-1756276
NCBI BlastP on this gene
gnd
SAHC1340_02449
HTH-type transcriptional regulator malR
Accession: ALY18294
Location: 1753067-1754086
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY18294
Location: 1753067-1754086
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY18293
Location: 1751402-1753051
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY18293
Location: 1751402-1753051
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAHC1340_02447
AraC family transcriptional regulator
Accession: ALY18292
Location: 1750426-1751319
NCBI BlastP on this gene
Accession: ALY18292
Location: 1750426-1751319
NCBI BlastP on this gene
SAHC1340_02446
glucose-6-phosphate 1-dehydrogenase
Accession: ALY18291
Location: 1748739-1750223
NCBI BlastP on this gene
Accession: ALY18291
Location: 1748739-1750223
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP011526 : Staphylococcus aureus subsp. aureus DSM 20231 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AA076_07685
6-phosphogluconate dehydrogenase
Accession: AKJ17395
Location: 1548991-1550397
NCBI BlastP on this gene
Accession: AKJ17395
Location: 1548991-1550397
NCBI BlastP on this gene
AA076_07680
AA076_07675
AA076_07670
LacI family transcriptional regulator
Accession: AKJ17392
Location: 1547188-1548207
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKJ17392
Location: 1547188-1548207
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AA076_07665
oligo-1,6-glucosidase
Accession: AKJ17391
Location: 1545523-1547172
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKJ17391
Location: 1545523-1547172
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AA076_07660
AraC family transcriptional regulator
Accession: AKJ17390
Location: 1544547-1545440
NCBI BlastP on this gene
Accession: AKJ17390
Location: 1544547-1545440
NCBI BlastP on this gene
AA076_07655
glucose-6-phosphate dehydrogenase
Accession: AKJ17389
Location: 1542860-1544344
NCBI BlastP on this gene
Accession: AKJ17389
Location: 1542860-1544344
NCBI BlastP on this gene
AA076_07650
Query: S.xylosus malR gene and malA gene.
CP010300 : Staphylococcus aureus strain 27b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
RM28_07960
6-phosphogluconate dehydrogenase
Accession: AJC43093
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC43093
Location: 1613116-1614522
NCBI BlastP on this gene
RM28_07955
RM28_07950
RM28_07945
LacI family transcriptional regulator
Accession: AJC43090
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC43090
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM28_07940
oligo-1,6-glucosidase
Accession: AJC43089
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC43089
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM28_07935
AraC family transcriptional regulator
Accession: AJC43088
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC43088
Location: 1608672-1609565
NCBI BlastP on this gene
RM28_07930
glucose-6-phosphate dehydrogenase
Accession: AJC43087
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC43087
Location: 1606985-1608469
NCBI BlastP on this gene
RM28_07925
Query: S.xylosus malR gene and malA gene.
CP010299 : Staphylococcus aureus strain 25b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
RM27_07960
6-phosphogluconate dehydrogenase
Accession: AJC40247
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC40247
Location: 1613116-1614522
NCBI BlastP on this gene
RM27_07955
RM27_07950
RM27_07945
LacI family transcriptional regulator
Accession: AJC40244
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC40244
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM27_07940
oligo-1,6-glucosidase
Accession: AJC40243
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC40243
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM27_07935
AraC family transcriptional regulator
Accession: AJC40242
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC40242
Location: 1608672-1609565
NCBI BlastP on this gene
RM27_07930
glucose-6-phosphate dehydrogenase
Accession: AJC40241
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC40241
Location: 1606985-1608469
NCBI BlastP on this gene
RM27_07925
Query: S.xylosus malR gene and malA gene.
CP010298 : Staphylococcus aureus strain 26b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
RM31_07970
6-phosphogluconate dehydrogenase
Accession: AJC37407
Location: 1613115-1614521
NCBI BlastP on this gene
Accession: AJC37407
Location: 1613115-1614521
NCBI BlastP on this gene
RM31_07965
RM31_07960
RM31_07955
LacI family transcriptional regulator
Accession: AJC37404
Location: 1611312-1612331
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC37404
Location: 1611312-1612331
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM31_07950
oligo-1,6-glucosidase
Accession: AJC37403
Location: 1609647-1611296
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC37403
Location: 1609647-1611296
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM31_07945
AraC family transcriptional regulator
Accession: AJC37402
Location: 1608671-1609564
NCBI BlastP on this gene
Accession: AJC37402
Location: 1608671-1609564
NCBI BlastP on this gene
RM31_07940
glucose-6-phosphate dehydrogenase
Accession: AJC37401
Location: 1606984-1608468
NCBI BlastP on this gene
Accession: AJC37401
Location: 1606984-1608468
NCBI BlastP on this gene
RM31_07935
Query: S.xylosus malR gene and malA gene.
CP010297 : Staphylococcus aureus strain 33b Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
QT38_07960
6-phosphogluconate dehydrogenase
Accession: AJC34559
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC34559
Location: 1613116-1614522
NCBI BlastP on this gene
QT38_07955
QT38_07950
QT38_07945
LacI family transcriptional regulator
Accession: AJC34556
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC34556
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
QT38_07940
oligo-1,6-glucosidase
Accession: AJC34555
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC34555
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
QT38_07935
AraC family transcriptional regulator
Accession: AJC34554
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC34554
Location: 1608672-1609565
NCBI BlastP on this gene
QT38_07930
glucose-6-phosphate dehydrogenase
Accession: AJC34553
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC34553
Location: 1606985-1608469
NCBI BlastP on this gene
QT38_07925
Query: S.xylosus malR gene and malA gene.
CP010296 : Staphylococcus aureus strain 31b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
RM30_07960
6-phosphogluconate dehydrogenase
Accession: AJC31716
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC31716
Location: 1613116-1614522
NCBI BlastP on this gene
RM30_07955
RM30_07950
RM30_07945
LacI family transcriptional regulator
Accession: AJC31713
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC31713
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM30_07940
oligo-1,6-glucosidase
Accession: AJC31712
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC31712
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM30_07935
AraC family transcriptional regulator
Accession: AJC31711
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC31711
Location: 1608672-1609565
NCBI BlastP on this gene
RM30_07930
glucose-6-phosphate dehydrogenase
Accession: AJC31710
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC31710
Location: 1606985-1608469
NCBI BlastP on this gene
RM30_07925
Query: S.xylosus malR gene and malA gene.
CP010295 : Staphylococcus aureus strain 29b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
RM29_07960
6-phosphogluconate dehydrogenase
Accession: AJC28870
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC28870
Location: 1613116-1614522
NCBI BlastP on this gene
RM29_07955
RM29_07950
RM29_07945
LacI family transcriptional regulator
Accession: AJC28867
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC28867
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM29_07940
oligo-1,6-glucosidase
Accession: AJC28866
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC28866
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM29_07935
AraC family transcriptional regulator
Accession: AJC28865
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC28865
Location: 1608672-1609565
NCBI BlastP on this gene
RM29_07930
glucose-6-phosphate dehydrogenase
Accession: AJC28864
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC28864
Location: 1606985-1608469
NCBI BlastP on this gene
RM29_07925
Query: S.xylosus malR gene and malA gene.
CP009681 : Staphylococcus aureus subsp. aureus strain Gv69 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAGV69_00934
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AIU84723
Location: 891766-893172
NCBI BlastP on this gene
Accession: AIU84723
Location: 891766-893172
NCBI BlastP on this gene
gnd
SAGV69_00936
HTH-type transcriptional regulator malR
Accession: AIU84725
Location: 893956-894975
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AIU84725
Location: 893956-894975
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: AIU84726
Location: 894991-896640
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIU84726
Location: 894991-896640
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAGV69_00938
AraC family transcriptional regulator
Accession: AIU84727
Location: 896723-897616
NCBI BlastP on this gene
Accession: AIU84727
Location: 896723-897616
NCBI BlastP on this gene
SAGV69_00939
glucose-6-phosphate 1-dehydrogenase
Accession: AIU84728
Location: 897819-899303
NCBI BlastP on this gene
Accession: AIU84728
Location: 897819-899303
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP009423 : Staphylococcus aureus subsp. aureus strain USA300_SUR1 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
LG33_08135
6-phosphogluconate dehydrogenase
Accession: APW51020
Location: 1615594-1617000
NCBI BlastP on this gene
Accession: APW51020
Location: 1615594-1617000
NCBI BlastP on this gene
LG33_08130
LG33_08125
LG33_08120
LacI family transcriptional regulator
Accession: APW51017
Location: 1613791-1614810
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: APW51017
Location: 1613791-1614810
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
LG33_08115
oligo-1,6-glucosidase
Accession: APW51016
Location: 1612126-1613775
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APW51016
Location: 1612126-1613775
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
LG33_08110
AraC family transcriptional regulator
Accession: APW51015
Location: 1611150-1612043
NCBI BlastP on this gene
Accession: APW51015
Location: 1611150-1612043
NCBI BlastP on this gene
LG33_08105
glucose-6-phosphate dehydrogenase
Accession: APW51014
Location: 1609463-1610947
NCBI BlastP on this gene
Accession: APW51014
Location: 1609463-1610947
NCBI BlastP on this gene
LG33_08100
Query: S.xylosus malR gene and malA gene.
CP007690 : Staphylococcus aureus strain UA-S391_USA300 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
EX97_07720
6-phosphogluconate dehydrogenase
Accession: AIA28052
Location: 1627063-1628469
NCBI BlastP on this gene
Accession: AIA28052
Location: 1627063-1628469
NCBI BlastP on this gene
EX97_07715
EX97_07710
LacI family transcriptional regulator
Accession: AIA28050
Location: 1625260-1626279
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AIA28050
Location: 1625260-1626279
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EX97_07705
oligo-1,6-glucosidase
Accession: AIA28049
Location: 1623595-1625244
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIA28049
Location: 1623595-1625244
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EX97_07700
AraC family transcriptional regulator
Accession: AIA28048
Location: 1622646-1623512
NCBI BlastP on this gene
Accession: AIA28048
Location: 1622646-1623512
NCBI BlastP on this gene
EX97_07695
glucose-6-phosphate dehydrogenase
Accession: AIA28047
Location: 1620932-1622416
NCBI BlastP on this gene
Accession: AIA28047
Location: 1620932-1622416
NCBI BlastP on this gene
EX97_07690
Query: S.xylosus malR gene and malA gene.
CP007676 : Staphylococcus aureus strain HUV05 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
ER12_007520
6-phosphogluconate dehydrogenase
Accession: ALK36786
Location: 1597399-1598805
NCBI BlastP on this gene
Accession: ALK36786
Location: 1597399-1598805
NCBI BlastP on this gene
ER12_007515
ER12_007510
LacI family transcriptional regulator
Accession: ALK36784
Location: 1595596-1596615
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALK36784
Location: 1595596-1596615
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ER12_007505
oligo-1,6-glucosidase
Accession: ALK36783
Location: 1593931-1595580
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALK36783
Location: 1593931-1595580
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ER12_007500
AraC family transcriptional regulator
Accession: ALK36782
Location: 1592982-1593848
NCBI BlastP on this gene
Accession: ALK36782
Location: 1592982-1593848
NCBI BlastP on this gene
ER12_007495
glucose-6-phosphate dehydrogenase
Accession: ALK36781
Location: 1591268-1592752
NCBI BlastP on this gene
Accession: ALK36781
Location: 1591268-1592752
NCBI BlastP on this gene
ER12_007490
Query: S.xylosus malR gene and malA gene.
CP007674 : Staphylococcus aureus strain CA15 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
6-phosphogluconate dehydrogenase
Accession: AKK61258
Location: 1592653-1594059
NCBI BlastP on this gene
Accession: AKK61258
Location: 1592653-1594059
NCBI BlastP on this gene
EQ90_07525
EQ90_07520
LacI family transcriptional regulator
Accession: AKK61256
Location: 1590850-1591869
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKK61256
Location: 1590850-1591869
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EQ90_07515
oligo-1,6-glucosidase
Accession: AKK61255
Location: 1589185-1590834
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKK61255
Location: 1589185-1590834
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EQ90_07510
AraC family transcriptional regulator
Accession: AKK61254
Location: 1588236-1589102
NCBI BlastP on this gene
Accession: AKK61254
Location: 1588236-1589102
NCBI BlastP on this gene
EQ90_07505
glucose-6-phosphate dehydrogenase
Accession: AKK61253
Location: 1586523-1588007
NCBI BlastP on this gene
Accession: AKK61253
Location: 1586523-1588007
NCBI BlastP on this gene
EQ90_07500
Query: S.xylosus malR gene and malA gene.
CP007672 : Staphylococcus aureus strain CA12 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
EQ80_007530
6-phosphogluconate dehydrogenase
Accession: ALK39386
Location: 1595783-1597189
NCBI BlastP on this gene
Accession: ALK39386
Location: 1595783-1597189
NCBI BlastP on this gene
EQ80_007525
EQ80_007520
LacI family transcriptional regulator
Accession: ALK39384
Location: 1593980-1594999
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALK39384
Location: 1593980-1594999
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EQ80_007515
oligo-1,6-glucosidase
Accession: ALK39383
Location: 1592315-1593964
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALK39383
Location: 1592315-1593964
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EQ80_007510
AraC family transcriptional regulator
Accession: ALK39382
Location: 1591366-1592232
NCBI BlastP on this gene
Accession: ALK39382
Location: 1591366-1592232
NCBI BlastP on this gene
EQ80_007505
glucose-6-phosphate dehydrogenase
Accession: ALK39381
Location: 1589652-1591136
NCBI BlastP on this gene
Accession: ALK39381
Location: 1589652-1591136
NCBI BlastP on this gene
EQ80_007500
Query: S.xylosus malR gene and malA gene.
CP007670 : Staphylococcus aureus strain M121 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
EP54_07615
6-phosphogluconate dehydrogenase
Accession: AKK58628
Location: 1608156-1609562
NCBI BlastP on this gene
Accession: AKK58628
Location: 1608156-1609562
NCBI BlastP on this gene
EP54_07610
EP54_07605
LacI family transcriptional regulator
Accession: AKK58626
Location: 1606353-1607372
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKK58626
Location: 1606353-1607372
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EP54_07600
oligo-1,6-glucosidase
Accession: AKK58625
Location: 1604688-1606337
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKK58625
Location: 1604688-1606337
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EP54_07595
AraC family transcriptional regulator
Accession: AKK58624
Location: 1603739-1604605
NCBI BlastP on this gene
Accession: AKK58624
Location: 1603739-1604605
NCBI BlastP on this gene
EP54_07590
glucose-6-phosphate dehydrogenase
Accession: AKK58623
Location: 1602025-1603509
NCBI BlastP on this gene
Accession: AKK58623
Location: 1602025-1603509
NCBI BlastP on this gene
EP54_07585
Query: S.xylosus malR gene and malA gene.
CP007657 : Staphylococcus aureus strain V2200 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
EP20_007605
6-phosphogluconate dehydrogenase
Accession: ALK41966
Location: 1609496-1610902
NCBI BlastP on this gene
Accession: ALK41966
Location: 1609496-1610902
NCBI BlastP on this gene
EP20_007600
EP20_007595
LacI family transcriptional regulator
Accession: ALK41964
Location: 1607693-1608712
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALK41964
Location: 1607693-1608712
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EP20_007590
oligo-1,6-glucosidase
Accession: ALK41963
Location: 1606028-1607677
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALK41963
Location: 1606028-1607677
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EP20_007585
AraC family transcriptional regulator
Accession: ALK41962
Location: 1605079-1605945
NCBI BlastP on this gene
Accession: ALK41962
Location: 1605079-1605945
NCBI BlastP on this gene
EP20_007580
glucose-6-phosphate dehydrogenase
Accession: ALK41961
Location: 1603365-1604849
NCBI BlastP on this gene
Accession: ALK41961
Location: 1603365-1604849
NCBI BlastP on this gene
EP20_007575
Query: S.xylosus malR gene and malA gene.
CP007539 : Staphylococcus aureus subsp. aureus strain FDAARGOS_5 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
X998_1523
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AHW66583
Location: 792814-794220
NCBI BlastP on this gene
Accession: AHW66583
Location: 792814-794220
NCBI BlastP on this gene
X998_1522
glyoxalase/bleomycin resistance/extradiol dioxygenase family protein
Accession: AHW66796
Location: 794439-794810
NCBI BlastP on this gene
Accession: AHW66796
Location: 794439-794810
NCBI BlastP on this gene
X998_1521
X998_04075
LacI family transcriptional regulator
Accession: AHW65802
Location: 795004-796023
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AHW65802
Location: 795004-796023
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
X998_1520
alpha-glucosidase
Accession: AHW67315
Location: 796039-797688
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AHW67315
Location: 796039-797688
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
X998_1519
AraC family transcriptional regulator
Accession: AHW68225
Location: 797771-798664
NCBI BlastP on this gene
Accession: AHW68225
Location: 797771-798664
NCBI BlastP on this gene
X998_1518
glucose-6-phosphate dehydrogenase
Accession: AHW66631
Location: 798867-800351
NCBI BlastP on this gene
Accession: AHW66631
Location: 798867-800351
NCBI BlastP on this gene
X998_1516
Query: S.xylosus malR gene and malA gene.
CP007499 : Staphylococcus aureus strain 2395 USA500 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
CH51_08235
6-phosphogluconate dehydrogenase
Accession: AIL58055
Location: 1703378-1704784
NCBI BlastP on this gene
Accession: AIL58055
Location: 1703378-1704784
NCBI BlastP on this gene
CH51_08230
CH51_08225
LacI family transcriptional regulator
Accession: AIL58053
Location: 1701575-1702594
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AIL58053
Location: 1701575-1702594
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
CH51_08220
oligo-1,6-glucosidase
Accession: AIL58052
Location: 1699910-1701559
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIL58052
Location: 1699910-1701559
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
CH51_08215
AraC family transcriptional regulator
Accession: AIL58051
Location: 1698961-1699827
NCBI BlastP on this gene
Accession: AIL58051
Location: 1698961-1699827
NCBI BlastP on this gene
CH51_08210
glucose-6-phosphate dehydrogenase
Accession: AIL58050
Location: 1697247-1698731
NCBI BlastP on this gene
Accession: AIL58050
Location: 1697247-1698731
NCBI BlastP on this gene
CH51_08205
Query: S.xylosus malR gene and malA gene.
CP007447 : Staphylococcus aureus strain XN108 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAXN108_1535
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AID39975
Location: 1610381-1611787
NCBI BlastP on this gene
Accession: AID39975
Location: 1610381-1611787
NCBI BlastP on this gene
SAXN108_1534
Glyoxalase/bleomycin resistance
Accession: AID39974
Location: 1609791-1610162
NCBI BlastP on this gene
Accession: AID39974
Location: 1609791-1610162
NCBI BlastP on this gene
SAXN108_1533
Maltose operon transcriptional repressor MalR, LacI family
Accession: AID39973
Location: 1608578-1609597
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AID39973
Location: 1608578-1609597
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAXN108_1532
alpha-D-1,4-glucosidase
Accession: AID39972
Location: 1606913-1608562
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AID39972
Location: 1606913-1608562
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAXN108_1531
Transcription regulator AraC/XylS family
Accession: AID39971
Location: 1605964-1606830
NCBI BlastP on this gene
Accession: AID39971
Location: 1605964-1606830
NCBI BlastP on this gene
SAXN108_1530
SAXN108_1529
Glucose-6-phosphate 1-dehydrogenase
Accession: AID39969
Location: 1604250-1605734
NCBI BlastP on this gene
Accession: AID39969
Location: 1604250-1605734
NCBI BlastP on this gene
SAXN108_1528
Query: S.xylosus malR gene and malA gene.
CP007176 : Staphylococcus aureus USA300-ISMMS1 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AZ30_07710
6-phosphogluconate dehydrogenase
Accession: AHJ07306
Location: 1632832-1634238
NCBI BlastP on this gene
Accession: AHJ07306
Location: 1632832-1634238
NCBI BlastP on this gene
AZ30_07705
AZ30_07700
LacI family transcriptional regulator
Accession: AHJ07304
Location: 1631029-1632048
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AHJ07304
Location: 1631029-1632048
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AZ30_07695
oligo-1,6-glucosidase
Accession: AHJ07303
Location: 1629364-1631013
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AHJ07303
Location: 1629364-1631013
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AZ30_07690
AraC family transcriptional regulator
Accession: AHJ07302
Location: 1628415-1629281
NCBI BlastP on this gene
Accession: AHJ07302
Location: 1628415-1629281
NCBI BlastP on this gene
AZ30_07685
glucose-6-phosphate 1-dehydrogenase
Accession: AHJ07301
Location: 1626701-1628185
NCBI BlastP on this gene
Accession: AHJ07301
Location: 1626701-1628185
NCBI BlastP on this gene
AZ30_07680
Query: S.xylosus malR gene and malA gene.
CP006838 : Staphylococcus aureus subsp. aureus Z172 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAZ172_1526
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AGY89625
Location: 1641838-1643244
NCBI BlastP on this gene
Accession: AGY89625
Location: 1641838-1643244
NCBI BlastP on this gene
gnd
Glyoxalase/bleomycin resistance
Accession: AGY89624
Location: 1641248-1641619
NCBI BlastP on this gene
Accession: AGY89624
Location: 1641248-1641619
NCBI BlastP on this gene
SAZ172_1524
Maltose operon transcriptional repressor MalR, LacI family
Accession: AGY89623
Location: 1640035-1641054
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AGY89623
Location: 1640035-1641054
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
Alpha-glucosidase
Accession: AGY89622
Location: 1638370-1640019
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGY89622
Location: 1638370-1640019
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
Transcription regulator AraC/XylS family
Accession: AGY89621
Location: 1637421-1638287
NCBI BlastP on this gene
Accession: AGY89621
Location: 1637421-1638287
NCBI BlastP on this gene
SAZ172_1521
SAZ172_1520
Glucose-6-phosphate 1-dehydrogenase
Accession: AGY89619
Location: 1635707-1637191
NCBI BlastP on this gene
Accession: AGY89619
Location: 1635707-1637191
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP005288 : Staphylococcus aureus strain Bmb9393 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SABB_00435
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AGP28510
Location: 1703240-1704646
NCBI BlastP on this gene
Accession: AGP28510
Location: 1703240-1704646
NCBI BlastP on this gene
gnd
SABB_00433
HTH-type transcriptional regulator malR
Accession: AGP28508
Location: 1701437-1702456
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AGP28508
Location: 1701437-1702456
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: AGP28507
Location: 1699772-1701421
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGP28507
Location: 1699772-1701421
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SABB_00431
AraC family transcriptional regulator
Accession: AGP28506
Location: 1698796-1699689
NCBI BlastP on this gene
Accession: AGP28506
Location: 1698796-1699689
NCBI BlastP on this gene
SABB_00430
glucose-6-phosphate 1-dehydrogenase
Accession: AGP28505
Location: 1697109-1698593
NCBI BlastP on this gene
Accession: AGP28505
Location: 1697109-1698593
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP003033 : Staphylococcus aureus subsp. aureus VC40 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAVC_06805
6-phosphogluconate dehydrogenase
Accession: AEZ37521
Location: 1487307-1488713
NCBI BlastP on this gene
Accession: AEZ37521
Location: 1487307-1488713
NCBI BlastP on this gene
SAVC_06800
SAVC_06795
maltose operon transcriptional repressor
Accession: AEZ37519
Location: 1485504-1486523
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AEZ37519
Location: 1485504-1486523
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAVC_06790
alpha-D-1,4-glucosidase
Accession: AEZ37518
Location: 1483839-1485488
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEZ37518
Location: 1483839-1485488
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAVC_06785
AraC family transcriptional regulator
Accession: AEZ37517
Location: 1482890-1483756
NCBI BlastP on this gene
Accession: AEZ37517
Location: 1482890-1483756
NCBI BlastP on this gene
SAVC_06780
glucose-6-phosphate 1-dehydrogenase
Accession: AEZ37516
Location: 1481176-1482660
NCBI BlastP on this gene
Accession: AEZ37516
Location: 1481176-1482660
NCBI BlastP on this gene
SAVC_06775
Query: S.xylosus malR gene and malA gene.
CP002643 : Staphylococcus aureus subsp. aureus T0131 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
Peptidase M20/M25/M40 family protein
Accession: AEB88602
Location: 1600234-1601367
NCBI BlastP on this gene
Accession: AEB88602
Location: 1600234-1601367
NCBI BlastP on this gene
SAT0131_01605
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AEB88601
Location: 1598760-1600166
NCBI BlastP on this gene
Accession: AEB88601
Location: 1598760-1600166
NCBI BlastP on this gene
SAT0131_01604
SAT0131_01603
Maltose operon repressor
Accession: AEB88599
Location: 1596957-1597976
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AEB88599
Location: 1596957-1597976
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAT0131_01602
Alpha-D-1,4-glucosidase
Accession: AEB88598
Location: 1595292-1596941
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEB88598
Location: 1595292-1596941
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAT0131_01601
Transcriptional regulator, AraC family
Accession: AEB88597
Location: 1594316-1595209
NCBI BlastP on this gene
Accession: AEB88597
Location: 1594316-1595209
NCBI BlastP on this gene
SAT0131_01600
Glucose-6-phosphate 1-dehydrogenase
Accession: AEB88596
Location: 1592629-1594113
NCBI BlastP on this gene
Accession: AEB88596
Location: 1592629-1594113
NCBI BlastP on this gene
SAT0131_01599
Query: S.xylosus malR gene and malA gene.
CP002120 : Staphylococcus aureus subsp. aureus str. JKD6008 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAA6008_01481
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ADL65519
Location: 1598479-1599885
NCBI BlastP on this gene
Accession: ADL65519
Location: 1598479-1599885
NCBI BlastP on this gene
gnd
glyoxalase/bleomycin resistance
Accession: ADL65518
Location: 1597889-1598260
NCBI BlastP on this gene
Accession: ADL65518
Location: 1597889-1598260
NCBI BlastP on this gene
SAA6008_01479
maltose operon transcriptional repressor
Accession: ADL65517
Location: 1596676-1597695
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ADL65517
Location: 1596676-1597695
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha amylase, catalytic region
Accession: ADL65516
Location: 1595011-1596660
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ADL65516
Location: 1595011-1596660
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: ADL65515
Location: 1594062-1594928
NCBI BlastP on this gene
Accession: ADL65515
Location: 1594062-1594928
NCBI BlastP on this gene
SAA6008_01476
putative glucose-6-phosphate 1-dehydrogenase
Accession: ADL65514
Location: 1592348-1593832
NCBI BlastP on this gene
Accession: ADL65514
Location: 1592348-1593832
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP000730 : Staphylococcus aureus subsp. aureus USA300_TCH1516 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
USA300HOU_1513
phosphogluconate dehydrogenase (decarboxylating)
Accession: ABX29519
Location: 1627066-1628472
NCBI BlastP on this gene
Accession: ABX29519
Location: 1627066-1628472
NCBI BlastP on this gene
pgd
USA300HOU_1511
maltose operon transcriptional repressor
Accession: ABX29517
Location: 1625263-1626282
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ABX29517
Location: 1625263-1626282
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ABX29516
Location: 1623598-1625247
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABX29516
Location: 1623598-1625247
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: ABX29515
Location: 1622622-1623515
NCBI BlastP on this gene
Accession: ABX29515
Location: 1622622-1623515
NCBI BlastP on this gene
USA300HOU_1508
glucose-6-phosphate 1-dehydrogenase
Accession: ABX29514
Location: 1620935-1622419
NCBI BlastP on this gene
Accession: ABX29514
Location: 1620935-1622419
NCBI BlastP on this gene
USA300HOU_1507
Query: S.xylosus malR gene and malA gene.
CP000255 : Staphylococcus aureus subsp. aureus USA300_FPR3757 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
peptidase, M20/M25/M40 family
Accession: ABD20395
Location: 1614593-1615726
NCBI BlastP on this gene
Accession: ABD20395
Location: 1614593-1615726
NCBI BlastP on this gene
SAUSA300_1460
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ABD21549
Location: 1613119-1614525
NCBI BlastP on this gene
Accession: ABD21549
Location: 1613119-1614525
NCBI BlastP on this gene
gnd
SAUSA300_1458
maltose operon transcriptional repressor
Accession: ABD21049
Location: 1611316-1612335
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ABD21049
Location: 1611316-1612335
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha glucosidase
Accession: ABD20873
Location: 1609651-1611300
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABD20873
Location: 1609651-1611300
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAUSA300_1456
transcriptional regulator, AraC family
Accession: ABD22885
Location: 1608675-1609568
NCBI BlastP on this gene
Accession: ABD22885
Location: 1608675-1609568
NCBI BlastP on this gene
SAUSA300_1455
glucose-6-phosphate 1-dehydrogenase
Accession: ABD21013
Location: 1606988-1608472
NCBI BlastP on this gene
Accession: ABD21013
Location: 1606988-1608472
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
CP000253 : Staphylococcus aureus subsp. aureus NCTC 8325 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
SAOUHSC_01606
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ABD30684
Location: 1529679-1531085
NCBI BlastP on this gene
Accession: ABD30684
Location: 1529679-1531085
NCBI BlastP on this gene
SAOUHSC_01605
conserved hypothetical protein
Accession: ABD30683
Location: 1529089-1529460
NCBI BlastP on this gene
Accession: ABD30683
Location: 1529089-1529460
NCBI BlastP on this gene
SAOUHSC_01604
SAOUHSC_01603
transcriptional regulator, putative
Accession: ABD30681
Location: 1527876-1528895
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ABD30681
Location: 1527876-1528895
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAOUHSC_01602
alpha-glucosidase, putative
Accession: ABD30680
Location: 1526211-1527860
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABD30680
Location: 1526211-1527860
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAOUHSC_01601
conserved hypothetical protein
Accession: ABD30679
Location: 1525235-1526128
NCBI BlastP on this gene
Accession: ABD30679
Location: 1525235-1526128
NCBI BlastP on this gene
SAOUHSC_01600
glucose-6-phosphate 1-dehydrogenase
Accession: ABD30678
Location: 1523548-1525032
NCBI BlastP on this gene
Accession: ABD30678
Location: 1523548-1525032
NCBI BlastP on this gene
SAOUHSC_01599
Query: S.xylosus malR gene and malA gene.
CP000046 : Staphylococcus aureus subsp. aureus COL Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
peptidase, M20/M25/M40 family
Accession: AAW36747
Location: 1591772-1592905
NCBI BlastP on this gene
Accession: AAW36747
Location: 1591772-1592905
NCBI BlastP on this gene
SACOL1555
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AAW36746
Location: 1590298-1591704
NCBI BlastP on this gene
Accession: AAW36746
Location: 1590298-1591704
NCBI BlastP on this gene
gnd
SACOL1553
maltose operon repressor
Accession: AAW36744
Location: 1588495-1589514
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AAW36744
Location: 1588495-1589514
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: AAW36743
Location: 1586830-1588479
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AAW36743
Location: 1586830-1588479
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
transcriptional regulator, AraC family
Accession: AAW36742
Location: 1585854-1586747
NCBI BlastP on this gene
Accession: AAW36742
Location: 1585854-1586747
NCBI BlastP on this gene
SACOL1550
glucose-6-phosphate 1-dehydrogenase
Accession: AAW36741
Location: 1584167-1585651
NCBI BlastP on this gene
Accession: AAW36741
Location: 1584167-1585651
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
AP020318 : Staphylococcus aureus KUH180038 DNA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
KUH180038_0509
6-phosphogluconate dehydrogenase
Accession: BBN34691
Location: 509735-511141
NCBI BlastP on this gene
Accession: BBN34691
Location: 509735-511141
NCBI BlastP on this gene
gnd
glyoxalase/bleomycin resistance
Accession: BBN34692
Location: 511360-511731
NCBI BlastP on this gene
Accession: BBN34692
Location: 511360-511731
NCBI BlastP on this gene
KUH180038_0511
maltose operon transcriptional repressor
Accession: BBN34693
Location: 511925-512944
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: BBN34693
Location: 511925-512944
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha amylase
Accession: BBN34694
Location: 512960-514609
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBN34694
Location: 512960-514609
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: BBN34695
Location: 514692-515558
NCBI BlastP on this gene
Accession: BBN34695
Location: 514692-515558
NCBI BlastP on this gene
KUH180038_0514
putative glucose-6-phosphate 1-dehydrogenase
Accession: BBN34696
Location: 515788-517272
NCBI BlastP on this gene
Accession: BBN34696
Location: 515788-517272
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
AP020313 : Staphylococcus aureus KUH140046 DNA Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
KUH140046_0509
6-phosphogluconate dehydrogenase
Accession: BBN27038
Location: 511301-512707
NCBI BlastP on this gene
Accession: BBN27038
Location: 511301-512707
NCBI BlastP on this gene
gnd
glyoxalase/bleomycin resistance
Accession: BBN27039
Location: 512926-513297
NCBI BlastP on this gene
Accession: BBN27039
Location: 512926-513297
NCBI BlastP on this gene
KUH140046_0511
maltose operon transcriptional repressor
Accession: BBN27040
Location: 513491-514510
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: BBN27040
Location: 513491-514510
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha amylase
Accession: BBN27041
Location: 514526-516175
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBN27041
Location: 514526-516175
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: BBN27042
Location: 516258-517124
NCBI BlastP on this gene
Accession: BBN27042
Location: 516258-517124
NCBI BlastP on this gene
KUH140046_0514
putative glucose-6-phosphate 1-dehydrogenase
Accession: BBN27043
Location: 517354-518838
NCBI BlastP on this gene
Accession: BBN27043
Location: 517354-518838
NCBI BlastP on this gene
zwf
Query: S.xylosus malR gene and malA gene.
251. : CP014397 Staphylococcus aureus strain USA300-SUR10 Total score: 2.5 Cumulative Blast bit score: 1182
STP|LacI,STP|Peripla BP 1,STP|Peripla BP 3
Location: 439-1452
Location: 439-1452
malR
GH13 31|GH13
Location: 1454-3103
Location: 1454-3103
malA
AYM23_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR10683
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQR10683
Location: 1673782-1675188
NCBI BlastP on this gene
AYM23_08435
AYM23_08430
AYM23_08425
LacI family transcriptional regulator
Accession: AQR10680
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR10680
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM23_08420
glucohydrolase
Accession: AQR10679
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR10679
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM23_08415
AraC family transcriptional regulator
Accession: AQR10678
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQR10678
Location: 1669338-1670231
NCBI BlastP on this gene
AYM23_08410
glucose-6-phosphate dehydrogenase
Accession: AQR10677
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQR10677
Location: 1667651-1669135
NCBI BlastP on this gene
AYM23_08405
252. : CP014392 Staphylococcus aureus strain USA300-SUR9 Total score: 2.5 Cumulative Blast bit score: 1182
AYM22_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR07744
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQR07744
Location: 1673782-1675188
NCBI BlastP on this gene
AYM22_08435
AYM22_08430
AYM22_08425
LacI family transcriptional regulator
Accession: AQR07741
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR07741
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM22_08420
glucohydrolase
Accession: AQR07740
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR07740
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM22_08415
AraC family transcriptional regulator
Accession: AQR07739
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQR07739
Location: 1669338-1670231
NCBI BlastP on this gene
AYM22_08410
glucose-6-phosphate dehydrogenase
Accession: AQR07738
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQR07738
Location: 1667651-1669135
NCBI BlastP on this gene
AYM22_08405
253. : CP014387 Staphylococcus aureus strain USA300-SUR8. Total score: 2.5 Cumulative Blast bit score: 1182
AYM21_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR04798
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQR04798
Location: 1673782-1675188
NCBI BlastP on this gene
AYM21_08435
AYM21_08430
AYM21_08425
LacI family transcriptional regulator
Accession: AQR04795
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR04795
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM21_08420
glucohydrolase
Accession: AQR04794
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR04794
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM21_08415
AraC family transcriptional regulator
Accession: AQR04793
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQR04793
Location: 1669338-1670231
NCBI BlastP on this gene
AYM21_08410
glucose-6-phosphate dehydrogenase
Accession: AQR04792
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQR04792
Location: 1667651-1669135
NCBI BlastP on this gene
AYM21_08405
254. : CP014384 Staphylococcus aureus strain USA300-SUR7 Total score: 2.5 Cumulative Blast bit score: 1182
AYM20_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQR01858
Location: 1673706-1675112
NCBI BlastP on this gene
Accession: AQR01858
Location: 1673706-1675112
NCBI BlastP on this gene
AYM20_08435
AYM20_08430
AYM20_08425
LacI family transcriptional regulator
Accession: AQR01855
Location: 1671903-1672922
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQR01855
Location: 1671903-1672922
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM20_08420
glucohydrolase
Accession: AQR01854
Location: 1670238-1671887
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQR01854
Location: 1670238-1671887
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM20_08415
AraC family transcriptional regulator
Accession: AQR01853
Location: 1669262-1670155
NCBI BlastP on this gene
Accession: AQR01853
Location: 1669262-1670155
NCBI BlastP on this gene
AYM20_08410
glucose-6-phosphate dehydrogenase
Accession: AQR01852
Location: 1667575-1669059
NCBI BlastP on this gene
Accession: AQR01852
Location: 1667575-1669059
NCBI BlastP on this gene
AYM20_08405
255. : CP014381 Staphylococcus aureus strain USA300-SUR6. Total score: 2.5 Cumulative Blast bit score: 1182
AYM19_08435
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ98893
Location: 1673702-1675108
NCBI BlastP on this gene
Accession: AQQ98893
Location: 1673702-1675108
NCBI BlastP on this gene
AYM19_08430
AYM19_08425
AYM19_08420
LacI family transcriptional regulator
Accession: AQQ98890
Location: 1671899-1672918
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ98890
Location: 1671899-1672918
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM19_08415
glucohydrolase
Accession: AQQ98889
Location: 1670234-1671883
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ98889
Location: 1670234-1671883
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM19_08410
AraC family transcriptional regulator
Accession: AQQ98888
Location: 1669258-1670151
NCBI BlastP on this gene
Accession: AQQ98888
Location: 1669258-1670151
NCBI BlastP on this gene
AYM19_08405
glucose-6-phosphate dehydrogenase
Accession: AQQ98887
Location: 1667571-1669055
NCBI BlastP on this gene
Accession: AQQ98887
Location: 1667571-1669055
NCBI BlastP on this gene
AYM19_08400
256. : CP014376 Staphylococcus aureus strain USA300-SUR5 Total score: 2.5 Cumulative Blast bit score: 1182
AYM18_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ95954
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQQ95954
Location: 1673782-1675188
NCBI BlastP on this gene
AYM18_08435
AYM18_08430
AYM18_08425
LacI family transcriptional regulator
Accession: AQQ95951
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ95951
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM18_08420
glucohydrolase
Accession: AQQ95950
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ95950
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM18_08415
AraC family transcriptional regulator
Accession: AQQ95949
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQQ95949
Location: 1669338-1670231
NCBI BlastP on this gene
AYM18_08410
glucose-6-phosphate dehydrogenase
Accession: AQQ95948
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQQ95948
Location: 1667651-1669135
NCBI BlastP on this gene
AYM18_08405
257. : CP014371 Staphylococcus aureus strain USA300-SUR4 Total score: 2.5 Cumulative Blast bit score: 1182
AYM16_08440
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ93015
Location: 1673782-1675188
NCBI BlastP on this gene
Accession: AQQ93015
Location: 1673782-1675188
NCBI BlastP on this gene
AYM16_08435
AYM16_08430
AYM16_08425
LacI family transcriptional regulator
Accession: AQQ93012
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ93012
Location: 1671979-1672998
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM16_08420
glucohydrolase
Accession: AQQ93011
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ93011
Location: 1670314-1671963
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM16_08415
AraC family transcriptional regulator
Accession: AQQ93010
Location: 1669338-1670231
NCBI BlastP on this gene
Accession: AQQ93010
Location: 1669338-1670231
NCBI BlastP on this gene
AYM16_08410
glucose-6-phosphate dehydrogenase
Accession: AQQ93009
Location: 1667651-1669135
NCBI BlastP on this gene
Accession: AQQ93009
Location: 1667651-1669135
NCBI BlastP on this gene
AYM16_08405
258. : CP014368 Staphylococcus aureus strain USA300-SUR3 Total score: 2.5 Cumulative Blast bit score: 1182
AYM15_08435
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ90073
Location: 1673690-1675096
NCBI BlastP on this gene
Accession: AQQ90073
Location: 1673690-1675096
NCBI BlastP on this gene
AYM15_08430
AYM15_08425
AYM15_08420
LacI family transcriptional regulator
Accession: AQQ90070
Location: 1671887-1672906
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ90070
Location: 1671887-1672906
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM15_08415
glucohydrolase
Accession: AQQ90069
Location: 1670222-1671871
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ90069
Location: 1670222-1671871
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM15_08410
AraC family transcriptional regulator
Accession: AQQ90068
Location: 1669246-1670139
NCBI BlastP on this gene
Accession: AQQ90068
Location: 1669246-1670139
NCBI BlastP on this gene
AYM15_08405
glucose-6-phosphate dehydrogenase
Accession: AQQ90067
Location: 1667559-1669043
NCBI BlastP on this gene
Accession: AQQ90067
Location: 1667559-1669043
NCBI BlastP on this gene
AYM15_08400
259. : CP014365 Staphylococcus aureus strain USA300-SUR2 Total score: 2.5 Cumulative Blast bit score: 1182
AYM14_08035
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ87072
Location: 1615774-1617180
NCBI BlastP on this gene
Accession: AQQ87072
Location: 1615774-1617180
NCBI BlastP on this gene
AYM14_08030
AYM14_08025
AYM14_08020
LacI family transcriptional regulator
Accession: AQQ87069
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ87069
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM14_08015
glucohydrolase
Accession: AQQ87068
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ87068
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM14_08010
AraC family transcriptional regulator
Accession: AQQ87067
Location: 1611330-1612223
NCBI BlastP on this gene
Accession: AQQ87067
Location: 1611330-1612223
NCBI BlastP on this gene
AYM14_08005
glucose-6-phosphate dehydrogenase
Accession: AQQ87066
Location: 1609643-1611127
NCBI BlastP on this gene
Accession: AQQ87066
Location: 1609643-1611127
NCBI BlastP on this gene
AYM14_08000
260. : CP014362 Staphylococcus aureus strain USA300-SUR1 Total score: 2.5 Cumulative Blast bit score: 1182
AYM13_08035
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AQQ84145
Location: 1615774-1617180
NCBI BlastP on this gene
Accession: AQQ84145
Location: 1615774-1617180
NCBI BlastP on this gene
AYM13_08030
AYM13_08025
AYM13_08020
LacI family transcriptional regulator
Accession: AQQ84142
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AQQ84142
Location: 1613971-1614990
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AYM13_08015
glucohydrolase
Accession: AQQ84141
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AQQ84141
Location: 1612306-1613955
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AYM13_08010
AraC family transcriptional regulator
Accession: AQQ84140
Location: 1611330-1612223
NCBI BlastP on this gene
Accession: AQQ84140
Location: 1611330-1612223
NCBI BlastP on this gene
AYM13_08005
glucose-6-phosphate dehydrogenase
Accession: AQQ84139
Location: 1609643-1611127
NCBI BlastP on this gene
Accession: AQQ84139
Location: 1609643-1611127
NCBI BlastP on this gene
AYM13_08000
261. : CP013959 Staphylococcus aureus strain V605 Total score: 2.5 Cumulative Blast bit score: 1182
ASL18_08465
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AND44838
Location: 1684736-1686142
NCBI BlastP on this gene
Accession: AND44838
Location: 1684736-1686142
NCBI BlastP on this gene
ASL18_08460
ASL18_08455
ASL18_08450
LacI family transcriptional regulator
Accession: AND44835
Location: 1682933-1683952
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AND44835
Location: 1682933-1683952
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ASL18_08445
glucohydrolase
Accession: AND44834
Location: 1681268-1682917
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AND44834
Location: 1681268-1682917
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ASL18_08440
AraC family transcriptional regulator
Accession: AND44833
Location: 1680292-1681185
NCBI BlastP on this gene
Accession: AND44833
Location: 1680292-1681185
NCBI BlastP on this gene
ASL18_08435
glucose-6-phosphate dehydrogenase
Accession: AND44832
Location: 1678605-1680089
NCBI BlastP on this gene
Accession: AND44832
Location: 1678605-1680089
NCBI BlastP on this gene
ASL18_08430
262. : CP013957 Staphylococcus aureus strain V521 Total score: 2.5 Cumulative Blast bit score: 1182
ASL17_08150
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AND36116
Location: 1642456-1643862
NCBI BlastP on this gene
Accession: AND36116
Location: 1642456-1643862
NCBI BlastP on this gene
ASL17_08145
ASL17_08140
ASL17_08135
LacI family transcriptional regulator
Accession: AND36113
Location: 1640653-1641672
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AND36113
Location: 1640653-1641672
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ASL17_08130
glucohydrolase
Accession: AND36112
Location: 1638988-1640637
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AND36112
Location: 1638988-1640637
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ASL17_08125
AraC family transcriptional regulator
Accession: AND36111
Location: 1638012-1638905
NCBI BlastP on this gene
Accession: AND36111
Location: 1638012-1638905
NCBI BlastP on this gene
ASL17_08120
glucose-6-phosphate dehydrogenase
Accession: AND36110
Location: 1636325-1637809
NCBI BlastP on this gene
Accession: AND36110
Location: 1636325-1637809
NCBI BlastP on this gene
ASL17_08115
263. : CP013231 Staphylococcus aureus strain UTSW MRSA 55 Total score: 2.5 Cumulative Blast bit score: 1182
AS858_03285
6-phosphogluconate dehydrogenase
Accession: AMO14155
Location: 656273-657679
NCBI BlastP on this gene
Accession: AMO14155
Location: 656273-657679
NCBI BlastP on this gene
AS858_03280
AS858_03275
AS858_03270
LacI family transcriptional regulator
Accession: AMO14152
Location: 654470-655489
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AMO14152
Location: 654470-655489
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AS858_03265
glucohydrolase
Accession: AMO14151
Location: 652805-654454
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AMO14151
Location: 652805-654454
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AS858_03260
AraC family transcriptional regulator
Accession: AMO14150
Location: 651829-652722
NCBI BlastP on this gene
Accession: AMO14150
Location: 651829-652722
NCBI BlastP on this gene
AS858_03255
glucose-6-phosphate dehydrogenase
Accession: AMO14149
Location: 650142-651626
NCBI BlastP on this gene
Accession: AMO14149
Location: 650142-651626
NCBI BlastP on this gene
AS858_03250
264. : CP012120 Staphylococcus aureus subsp. aureus strain USA300_2014.C02 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
ACO02_2154
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AKR46834
Location: 2616347-2617753
NCBI BlastP on this gene
Accession: AKR46834
Location: 2616347-2617753
NCBI BlastP on this gene
ACO02_2155
glyoxalase/bleomycin resistance/extradiol dioxygenase family protein
Accession: AKR46835
Location: 2617972-2618343
NCBI BlastP on this gene
Accession: AKR46835
Location: 2617972-2618343
NCBI BlastP on this gene
ACO02_2156
ACO02_04385
LacI family transcriptional regulator
Accession: AKR46836
Location: 2618537-2619556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKR46836
Location: 2618537-2619556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ACO02_2157
alpha-glucosidase
Accession: AKR46837
Location: 2619572-2621221
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKR46837
Location: 2619572-2621221
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ACO02_2158
AraC family transcriptional regulator
Accession: AKR46838
Location: 2621304-2622197
NCBI BlastP on this gene
Accession: AKR46838
Location: 2621304-2622197
NCBI BlastP on this gene
ACO02_2159
glucose-6-phosphate dehydrogenase
Accession: AKR46840
Location: 2622400-2623884
NCBI BlastP on this gene
Accession: AKR46840
Location: 2622400-2623884
NCBI BlastP on this gene
ACO02_2161
265. : CP012119 Staphylococcus aureus subsp. aureus strain USA300_2014.C01 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
AC807_0273
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AKR50357
Location: 735340-736746
NCBI BlastP on this gene
Accession: AKR50357
Location: 735340-736746
NCBI BlastP on this gene
AC807_0274
glyoxalase/bleomycin resistance/extradiol dioxygenase family protein
Accession: AKR50358
Location: 734750-735121
NCBI BlastP on this gene
Accession: AKR50358
Location: 734750-735121
NCBI BlastP on this gene
AC807_0275
AC807_03260
LacI family transcriptional regulator
Accession: AKR50359
Location: 733537-734556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKR50359
Location: 733537-734556
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AC807_0276
alpha-glucosidase
Accession: AKR50360
Location: 731872-733521
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKR50360
Location: 731872-733521
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AC807_0277
AraC family transcriptional regulator
Accession: AKR50361
Location: 730896-731789
NCBI BlastP on this gene
Accession: AKR50361
Location: 730896-731789
NCBI BlastP on this gene
AC807_0278
glucose-6-phosphate dehydrogenase
Accession: AKR50363
Location: 729209-730693
NCBI BlastP on this gene
Accession: AKR50363
Location: 729209-730693
NCBI BlastP on this gene
AC807_0280
266. : CP012018 Staphylococcus aureus subsp. aureus strain Gv88 Total score: 2.5 Cumulative Blast bit score: 1182
SAGV88_01776
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY29166
Location: 1705391-1706797
NCBI BlastP on this gene
Accession: ALY29166
Location: 1705391-1706797
NCBI BlastP on this gene
gnd
SAGV88_01774
HTH-type transcriptional regulator malR
Accession: ALY29164
Location: 1703588-1704607
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY29164
Location: 1703588-1704607
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY29163
Location: 1701923-1703572
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY29163
Location: 1701923-1703572
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAGV88_01772
AraC family transcriptional regulator
Accession: ALY29162
Location: 1700947-1701840
NCBI BlastP on this gene
Accession: ALY29162
Location: 1700947-1701840
NCBI BlastP on this gene
SAGV88_01771
glucose-6-phosphate 1-dehydrogenase
Accession: ALY29161
Location: 1699260-1700744
NCBI BlastP on this gene
Accession: ALY29161
Location: 1699260-1700744
NCBI BlastP on this gene
zwf
267. : CP012015 Staphylococcus aureus subsp. aureus strain Gv51 Total score: 2.5 Cumulative Blast bit score: 1182
SAGV51_01783
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY25950
Location: 1706128-1707534
NCBI BlastP on this gene
Accession: ALY25950
Location: 1706128-1707534
NCBI BlastP on this gene
gnd
SAGV51_01781
HTH-type transcriptional regulator malR
Accession: ALY25948
Location: 1704325-1705344
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY25948
Location: 1704325-1705344
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY25947
Location: 1702660-1704309
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY25947
Location: 1702660-1704309
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAGV51_01779
AraC family transcriptional regulator
Accession: ALY25946
Location: 1701684-1702577
NCBI BlastP on this gene
Accession: ALY25946
Location: 1701684-1702577
NCBI BlastP on this gene
SAGV51_01778
glucose-6-phosphate 1-dehydrogenase
Accession: ALY25945
Location: 1699997-1701481
NCBI BlastP on this gene
Accession: ALY25945
Location: 1699997-1701481
NCBI BlastP on this gene
zwf
268. : CP012013 Staphylococcus aureus subsp. aureus strain Be62 Total score: 2.5 Cumulative Blast bit score: 1182
SABE62_01774
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY23429
Location: 1710624-1712030
NCBI BlastP on this gene
Accession: ALY23429
Location: 1710624-1712030
NCBI BlastP on this gene
gnd
SABE62_01772
HTH-type transcriptional regulator malR
Accession: ALY23427
Location: 1708821-1709840
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY23427
Location: 1708821-1709840
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY23426
Location: 1707156-1708805
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY23426
Location: 1707156-1708805
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SABE62_01770
AraC family transcriptional regulator
Accession: ALY23425
Location: 1706180-1707073
NCBI BlastP on this gene
Accession: ALY23425
Location: 1706180-1707073
NCBI BlastP on this gene
SABE62_01769
glucose-6-phosphate 1-dehydrogenase
Accession: ALY23424
Location: 1704493-1705977
NCBI BlastP on this gene
Accession: ALY23424
Location: 1704493-1705977
NCBI BlastP on this gene
zwf
269. : CP012012 Staphylococcus aureus subsp. aureus strain HC1335 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
SAHC1335_02534
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY21121
Location: 1700261-1701667
NCBI BlastP on this gene
Accession: ALY21121
Location: 1700261-1701667
NCBI BlastP on this gene
gnd
SAHC1335_02532
HTH-type transcriptional regulator malR
Accession: ALY21119
Location: 1698458-1699477
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY21119
Location: 1698458-1699477
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY21118
Location: 1696793-1698442
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY21118
Location: 1696793-1698442
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAHC1335_02530
AraC family transcriptional regulator
Accession: ALY21117
Location: 1695817-1696710
NCBI BlastP on this gene
Accession: ALY21117
Location: 1695817-1696710
NCBI BlastP on this gene
SAHC1335_02529
glucose-6-phosphate 1-dehydrogenase
Accession: ALY21116
Location: 1694130-1695614
NCBI BlastP on this gene
Accession: ALY21116
Location: 1694130-1695614
NCBI BlastP on this gene
zwf
270. : CP012011 Staphylococcus aureus subsp. aureus strain HC1340 Total score: 2.5 Cumulative Blast bit score: 1182
SAHC1340_02451
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ALY18296
Location: 1754870-1756276
NCBI BlastP on this gene
Accession: ALY18296
Location: 1754870-1756276
NCBI BlastP on this gene
gnd
SAHC1340_02449
HTH-type transcriptional regulator malR
Accession: ALY18294
Location: 1753067-1754086
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALY18294
Location: 1753067-1754086
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ALY18293
Location: 1751402-1753051
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALY18293
Location: 1751402-1753051
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAHC1340_02447
AraC family transcriptional regulator
Accession: ALY18292
Location: 1750426-1751319
NCBI BlastP on this gene
Accession: ALY18292
Location: 1750426-1751319
NCBI BlastP on this gene
SAHC1340_02446
glucose-6-phosphate 1-dehydrogenase
Accession: ALY18291
Location: 1748739-1750223
NCBI BlastP on this gene
Accession: ALY18291
Location: 1748739-1750223
NCBI BlastP on this gene
zwf
271. : CP011526 Staphylococcus aureus subsp. aureus DSM 20231 Total score: 2.5 Cumulative Blast bit score: 1182
AA076_07685
6-phosphogluconate dehydrogenase
Accession: AKJ17395
Location: 1548991-1550397
NCBI BlastP on this gene
Accession: AKJ17395
Location: 1548991-1550397
NCBI BlastP on this gene
AA076_07680
AA076_07675
AA076_07670
LacI family transcriptional regulator
Accession: AKJ17392
Location: 1547188-1548207
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKJ17392
Location: 1547188-1548207
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AA076_07665
oligo-1,6-glucosidase
Accession: AKJ17391
Location: 1545523-1547172
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKJ17391
Location: 1545523-1547172
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AA076_07660
AraC family transcriptional regulator
Accession: AKJ17390
Location: 1544547-1545440
NCBI BlastP on this gene
Accession: AKJ17390
Location: 1544547-1545440
NCBI BlastP on this gene
AA076_07655
glucose-6-phosphate dehydrogenase
Accession: AKJ17389
Location: 1542860-1544344
NCBI BlastP on this gene
Accession: AKJ17389
Location: 1542860-1544344
NCBI BlastP on this gene
AA076_07650
272. : CP010300 Staphylococcus aureus strain 27b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
RM28_07960
6-phosphogluconate dehydrogenase
Accession: AJC43093
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC43093
Location: 1613116-1614522
NCBI BlastP on this gene
RM28_07955
RM28_07950
RM28_07945
LacI family transcriptional regulator
Accession: AJC43090
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC43090
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM28_07940
oligo-1,6-glucosidase
Accession: AJC43089
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC43089
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM28_07935
AraC family transcriptional regulator
Accession: AJC43088
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC43088
Location: 1608672-1609565
NCBI BlastP on this gene
RM28_07930
glucose-6-phosphate dehydrogenase
Accession: AJC43087
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC43087
Location: 1606985-1608469
NCBI BlastP on this gene
RM28_07925
273. : CP010299 Staphylococcus aureus strain 25b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
RM27_07960
6-phosphogluconate dehydrogenase
Accession: AJC40247
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC40247
Location: 1613116-1614522
NCBI BlastP on this gene
RM27_07955
RM27_07950
RM27_07945
LacI family transcriptional regulator
Accession: AJC40244
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC40244
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM27_07940
oligo-1,6-glucosidase
Accession: AJC40243
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC40243
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM27_07935
AraC family transcriptional regulator
Accession: AJC40242
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC40242
Location: 1608672-1609565
NCBI BlastP on this gene
RM27_07930
glucose-6-phosphate dehydrogenase
Accession: AJC40241
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC40241
Location: 1606985-1608469
NCBI BlastP on this gene
RM27_07925
274. : CP010298 Staphylococcus aureus strain 26b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
RM31_07970
6-phosphogluconate dehydrogenase
Accession: AJC37407
Location: 1613115-1614521
NCBI BlastP on this gene
Accession: AJC37407
Location: 1613115-1614521
NCBI BlastP on this gene
RM31_07965
RM31_07960
RM31_07955
LacI family transcriptional regulator
Accession: AJC37404
Location: 1611312-1612331
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC37404
Location: 1611312-1612331
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM31_07950
oligo-1,6-glucosidase
Accession: AJC37403
Location: 1609647-1611296
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC37403
Location: 1609647-1611296
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM31_07945
AraC family transcriptional regulator
Accession: AJC37402
Location: 1608671-1609564
NCBI BlastP on this gene
Accession: AJC37402
Location: 1608671-1609564
NCBI BlastP on this gene
RM31_07940
glucose-6-phosphate dehydrogenase
Accession: AJC37401
Location: 1606984-1608468
NCBI BlastP on this gene
Accession: AJC37401
Location: 1606984-1608468
NCBI BlastP on this gene
RM31_07935
275. : CP010297 Staphylococcus aureus strain 33b Total score: 2.5 Cumulative Blast bit score: 1182
QT38_07960
6-phosphogluconate dehydrogenase
Accession: AJC34559
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC34559
Location: 1613116-1614522
NCBI BlastP on this gene
QT38_07955
QT38_07950
QT38_07945
LacI family transcriptional regulator
Accession: AJC34556
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC34556
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
QT38_07940
oligo-1,6-glucosidase
Accession: AJC34555
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC34555
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
QT38_07935
AraC family transcriptional regulator
Accession: AJC34554
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC34554
Location: 1608672-1609565
NCBI BlastP on this gene
QT38_07930
glucose-6-phosphate dehydrogenase
Accession: AJC34553
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC34553
Location: 1606985-1608469
NCBI BlastP on this gene
QT38_07925
276. : CP010296 Staphylococcus aureus strain 31b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
RM30_07960
6-phosphogluconate dehydrogenase
Accession: AJC31716
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC31716
Location: 1613116-1614522
NCBI BlastP on this gene
RM30_07955
RM30_07950
RM30_07945
LacI family transcriptional regulator
Accession: AJC31713
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC31713
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM30_07940
oligo-1,6-glucosidase
Accession: AJC31712
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC31712
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM30_07935
AraC family transcriptional regulator
Accession: AJC31711
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC31711
Location: 1608672-1609565
NCBI BlastP on this gene
RM30_07930
glucose-6-phosphate dehydrogenase
Accession: AJC31710
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC31710
Location: 1606985-1608469
NCBI BlastP on this gene
RM30_07925
277. : CP010295 Staphylococcus aureus strain 29b_MRSA Total score: 2.5 Cumulative Blast bit score: 1182
RM29_07960
6-phosphogluconate dehydrogenase
Accession: AJC28870
Location: 1613116-1614522
NCBI BlastP on this gene
Accession: AJC28870
Location: 1613116-1614522
NCBI BlastP on this gene
RM29_07955
RM29_07950
RM29_07945
LacI family transcriptional regulator
Accession: AJC28867
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AJC28867
Location: 1611313-1612332
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
RM29_07940
oligo-1,6-glucosidase
Accession: AJC28866
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJC28866
Location: 1609648-1611297
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RM29_07935
AraC family transcriptional regulator
Accession: AJC28865
Location: 1608672-1609565
NCBI BlastP on this gene
Accession: AJC28865
Location: 1608672-1609565
NCBI BlastP on this gene
RM29_07930
glucose-6-phosphate dehydrogenase
Accession: AJC28864
Location: 1606985-1608469
NCBI BlastP on this gene
Accession: AJC28864
Location: 1606985-1608469
NCBI BlastP on this gene
RM29_07925
278. : CP009681 Staphylococcus aureus subsp. aureus strain Gv69 Total score: 2.5 Cumulative Blast bit score: 1182
SAGV69_00934
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AIU84723
Location: 891766-893172
NCBI BlastP on this gene
Accession: AIU84723
Location: 891766-893172
NCBI BlastP on this gene
gnd
SAGV69_00936
HTH-type transcriptional regulator malR
Accession: AIU84725
Location: 893956-894975
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AIU84725
Location: 893956-894975
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: AIU84726
Location: 894991-896640
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIU84726
Location: 894991-896640
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAGV69_00938
AraC family transcriptional regulator
Accession: AIU84727
Location: 896723-897616
NCBI BlastP on this gene
Accession: AIU84727
Location: 896723-897616
NCBI BlastP on this gene
SAGV69_00939
glucose-6-phosphate 1-dehydrogenase
Accession: AIU84728
Location: 897819-899303
NCBI BlastP on this gene
Accession: AIU84728
Location: 897819-899303
NCBI BlastP on this gene
zwf
279. : CP009423 Staphylococcus aureus subsp. aureus strain USA300_SUR1 Total score: 2.5 Cumulative Blast bit score: 1182
LG33_08135
6-phosphogluconate dehydrogenase
Accession: APW51020
Location: 1615594-1617000
NCBI BlastP on this gene
Accession: APW51020
Location: 1615594-1617000
NCBI BlastP on this gene
LG33_08130
LG33_08125
LG33_08120
LacI family transcriptional regulator
Accession: APW51017
Location: 1613791-1614810
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: APW51017
Location: 1613791-1614810
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
LG33_08115
oligo-1,6-glucosidase
Accession: APW51016
Location: 1612126-1613775
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APW51016
Location: 1612126-1613775
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
LG33_08110
AraC family transcriptional regulator
Accession: APW51015
Location: 1611150-1612043
NCBI BlastP on this gene
Accession: APW51015
Location: 1611150-1612043
NCBI BlastP on this gene
LG33_08105
glucose-6-phosphate dehydrogenase
Accession: APW51014
Location: 1609463-1610947
NCBI BlastP on this gene
Accession: APW51014
Location: 1609463-1610947
NCBI BlastP on this gene
LG33_08100
280. : CP007690 Staphylococcus aureus strain UA-S391_USA300 Total score: 2.5 Cumulative Blast bit score: 1182
EX97_07720
6-phosphogluconate dehydrogenase
Accession: AIA28052
Location: 1627063-1628469
NCBI BlastP on this gene
Accession: AIA28052
Location: 1627063-1628469
NCBI BlastP on this gene
EX97_07715
EX97_07710
LacI family transcriptional regulator
Accession: AIA28050
Location: 1625260-1626279
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AIA28050
Location: 1625260-1626279
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EX97_07705
oligo-1,6-glucosidase
Accession: AIA28049
Location: 1623595-1625244
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIA28049
Location: 1623595-1625244
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EX97_07700
AraC family transcriptional regulator
Accession: AIA28048
Location: 1622646-1623512
NCBI BlastP on this gene
Accession: AIA28048
Location: 1622646-1623512
NCBI BlastP on this gene
EX97_07695
glucose-6-phosphate dehydrogenase
Accession: AIA28047
Location: 1620932-1622416
NCBI BlastP on this gene
Accession: AIA28047
Location: 1620932-1622416
NCBI BlastP on this gene
EX97_07690
281. : CP007676 Staphylococcus aureus strain HUV05 Total score: 2.5 Cumulative Blast bit score: 1182
ER12_007520
6-phosphogluconate dehydrogenase
Accession: ALK36786
Location: 1597399-1598805
NCBI BlastP on this gene
Accession: ALK36786
Location: 1597399-1598805
NCBI BlastP on this gene
ER12_007515
ER12_007510
LacI family transcriptional regulator
Accession: ALK36784
Location: 1595596-1596615
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALK36784
Location: 1595596-1596615
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
ER12_007505
oligo-1,6-glucosidase
Accession: ALK36783
Location: 1593931-1595580
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALK36783
Location: 1593931-1595580
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ER12_007500
AraC family transcriptional regulator
Accession: ALK36782
Location: 1592982-1593848
NCBI BlastP on this gene
Accession: ALK36782
Location: 1592982-1593848
NCBI BlastP on this gene
ER12_007495
glucose-6-phosphate dehydrogenase
Accession: ALK36781
Location: 1591268-1592752
NCBI BlastP on this gene
Accession: ALK36781
Location: 1591268-1592752
NCBI BlastP on this gene
ER12_007490
282. : CP007674 Staphylococcus aureus strain CA15 Total score: 2.5 Cumulative Blast bit score: 1182
6-phosphogluconate dehydrogenase
Accession: AKK61258
Location: 1592653-1594059
NCBI BlastP on this gene
Accession: AKK61258
Location: 1592653-1594059
NCBI BlastP on this gene
EQ90_07525
EQ90_07520
LacI family transcriptional regulator
Accession: AKK61256
Location: 1590850-1591869
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKK61256
Location: 1590850-1591869
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EQ90_07515
oligo-1,6-glucosidase
Accession: AKK61255
Location: 1589185-1590834
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKK61255
Location: 1589185-1590834
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EQ90_07510
AraC family transcriptional regulator
Accession: AKK61254
Location: 1588236-1589102
NCBI BlastP on this gene
Accession: AKK61254
Location: 1588236-1589102
NCBI BlastP on this gene
EQ90_07505
glucose-6-phosphate dehydrogenase
Accession: AKK61253
Location: 1586523-1588007
NCBI BlastP on this gene
Accession: AKK61253
Location: 1586523-1588007
NCBI BlastP on this gene
EQ90_07500
283. : CP007672 Staphylococcus aureus strain CA12 Total score: 2.5 Cumulative Blast bit score: 1182
EQ80_007530
6-phosphogluconate dehydrogenase
Accession: ALK39386
Location: 1595783-1597189
NCBI BlastP on this gene
Accession: ALK39386
Location: 1595783-1597189
NCBI BlastP on this gene
EQ80_007525
EQ80_007520
LacI family transcriptional regulator
Accession: ALK39384
Location: 1593980-1594999
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALK39384
Location: 1593980-1594999
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EQ80_007515
oligo-1,6-glucosidase
Accession: ALK39383
Location: 1592315-1593964
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALK39383
Location: 1592315-1593964
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EQ80_007510
AraC family transcriptional regulator
Accession: ALK39382
Location: 1591366-1592232
NCBI BlastP on this gene
Accession: ALK39382
Location: 1591366-1592232
NCBI BlastP on this gene
EQ80_007505
glucose-6-phosphate dehydrogenase
Accession: ALK39381
Location: 1589652-1591136
NCBI BlastP on this gene
Accession: ALK39381
Location: 1589652-1591136
NCBI BlastP on this gene
EQ80_007500
284. : CP007670 Staphylococcus aureus strain M121 Total score: 2.5 Cumulative Blast bit score: 1182
EP54_07615
6-phosphogluconate dehydrogenase
Accession: AKK58628
Location: 1608156-1609562
NCBI BlastP on this gene
Accession: AKK58628
Location: 1608156-1609562
NCBI BlastP on this gene
EP54_07610
EP54_07605
LacI family transcriptional regulator
Accession: AKK58626
Location: 1606353-1607372
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AKK58626
Location: 1606353-1607372
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EP54_07600
oligo-1,6-glucosidase
Accession: AKK58625
Location: 1604688-1606337
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKK58625
Location: 1604688-1606337
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EP54_07595
AraC family transcriptional regulator
Accession: AKK58624
Location: 1603739-1604605
NCBI BlastP on this gene
Accession: AKK58624
Location: 1603739-1604605
NCBI BlastP on this gene
EP54_07590
glucose-6-phosphate dehydrogenase
Accession: AKK58623
Location: 1602025-1603509
NCBI BlastP on this gene
Accession: AKK58623
Location: 1602025-1603509
NCBI BlastP on this gene
EP54_07585
285. : CP007657 Staphylococcus aureus strain V2200 Total score: 2.5 Cumulative Blast bit score: 1182
EP20_007605
6-phosphogluconate dehydrogenase
Accession: ALK41966
Location: 1609496-1610902
NCBI BlastP on this gene
Accession: ALK41966
Location: 1609496-1610902
NCBI BlastP on this gene
EP20_007600
EP20_007595
LacI family transcriptional regulator
Accession: ALK41964
Location: 1607693-1608712
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ALK41964
Location: 1607693-1608712
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
EP20_007590
oligo-1,6-glucosidase
Accession: ALK41963
Location: 1606028-1607677
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ALK41963
Location: 1606028-1607677
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
EP20_007585
AraC family transcriptional regulator
Accession: ALK41962
Location: 1605079-1605945
NCBI BlastP on this gene
Accession: ALK41962
Location: 1605079-1605945
NCBI BlastP on this gene
EP20_007580
glucose-6-phosphate dehydrogenase
Accession: ALK41961
Location: 1603365-1604849
NCBI BlastP on this gene
Accession: ALK41961
Location: 1603365-1604849
NCBI BlastP on this gene
EP20_007575
286. : CP007539 Staphylococcus aureus subsp. aureus strain FDAARGOS_5 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
X998_1523
phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating)
Accession: AHW66583
Location: 792814-794220
NCBI BlastP on this gene
Accession: AHW66583
Location: 792814-794220
NCBI BlastP on this gene
X998_1522
glyoxalase/bleomycin resistance/extradiol dioxygenase family protein
Accession: AHW66796
Location: 794439-794810
NCBI BlastP on this gene
Accession: AHW66796
Location: 794439-794810
NCBI BlastP on this gene
X998_1521
X998_04075
LacI family transcriptional regulator
Accession: AHW65802
Location: 795004-796023
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AHW65802
Location: 795004-796023
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
X998_1520
alpha-glucosidase
Accession: AHW67315
Location: 796039-797688
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AHW67315
Location: 796039-797688
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
X998_1519
AraC family transcriptional regulator
Accession: AHW68225
Location: 797771-798664
NCBI BlastP on this gene
Accession: AHW68225
Location: 797771-798664
NCBI BlastP on this gene
X998_1518
glucose-6-phosphate dehydrogenase
Accession: AHW66631
Location: 798867-800351
NCBI BlastP on this gene
Accession: AHW66631
Location: 798867-800351
NCBI BlastP on this gene
X998_1516
287. : CP007499 Staphylococcus aureus strain 2395 USA500 Total score: 2.5 Cumulative Blast bit score: 1182
CH51_08235
6-phosphogluconate dehydrogenase
Accession: AIL58055
Location: 1703378-1704784
NCBI BlastP on this gene
Accession: AIL58055
Location: 1703378-1704784
NCBI BlastP on this gene
CH51_08230
CH51_08225
LacI family transcriptional regulator
Accession: AIL58053
Location: 1701575-1702594
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AIL58053
Location: 1701575-1702594
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
CH51_08220
oligo-1,6-glucosidase
Accession: AIL58052
Location: 1699910-1701559
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIL58052
Location: 1699910-1701559
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
CH51_08215
AraC family transcriptional regulator
Accession: AIL58051
Location: 1698961-1699827
NCBI BlastP on this gene
Accession: AIL58051
Location: 1698961-1699827
NCBI BlastP on this gene
CH51_08210
glucose-6-phosphate dehydrogenase
Accession: AIL58050
Location: 1697247-1698731
NCBI BlastP on this gene
Accession: AIL58050
Location: 1697247-1698731
NCBI BlastP on this gene
CH51_08205
288. : CP007447 Staphylococcus aureus strain XN108 Total score: 2.5 Cumulative Blast bit score: 1182
SAXN108_1535
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AID39975
Location: 1610381-1611787
NCBI BlastP on this gene
Accession: AID39975
Location: 1610381-1611787
NCBI BlastP on this gene
SAXN108_1534
Glyoxalase/bleomycin resistance
Accession: AID39974
Location: 1609791-1610162
NCBI BlastP on this gene
Accession: AID39974
Location: 1609791-1610162
NCBI BlastP on this gene
SAXN108_1533
Maltose operon transcriptional repressor MalR, LacI family
Accession: AID39973
Location: 1608578-1609597
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AID39973
Location: 1608578-1609597
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAXN108_1532
alpha-D-1,4-glucosidase
Accession: AID39972
Location: 1606913-1608562
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AID39972
Location: 1606913-1608562
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAXN108_1531
Transcription regulator AraC/XylS family
Accession: AID39971
Location: 1605964-1606830
NCBI BlastP on this gene
Accession: AID39971
Location: 1605964-1606830
NCBI BlastP on this gene
SAXN108_1530
SAXN108_1529
Glucose-6-phosphate 1-dehydrogenase
Accession: AID39969
Location: 1604250-1605734
NCBI BlastP on this gene
Accession: AID39969
Location: 1604250-1605734
NCBI BlastP on this gene
SAXN108_1528
289. : CP007176 Staphylococcus aureus USA300-ISMMS1 Total score: 2.5 Cumulative Blast bit score: 1182
AZ30_07710
6-phosphogluconate dehydrogenase
Accession: AHJ07306
Location: 1632832-1634238
NCBI BlastP on this gene
Accession: AHJ07306
Location: 1632832-1634238
NCBI BlastP on this gene
AZ30_07705
AZ30_07700
LacI family transcriptional regulator
Accession: AHJ07304
Location: 1631029-1632048
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AHJ07304
Location: 1631029-1632048
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
AZ30_07695
oligo-1,6-glucosidase
Accession: AHJ07303
Location: 1629364-1631013
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AHJ07303
Location: 1629364-1631013
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
AZ30_07690
AraC family transcriptional regulator
Accession: AHJ07302
Location: 1628415-1629281
NCBI BlastP on this gene
Accession: AHJ07302
Location: 1628415-1629281
NCBI BlastP on this gene
AZ30_07685
glucose-6-phosphate 1-dehydrogenase
Accession: AHJ07301
Location: 1626701-1628185
NCBI BlastP on this gene
Accession: AHJ07301
Location: 1626701-1628185
NCBI BlastP on this gene
AZ30_07680
290. : CP006838 Staphylococcus aureus subsp. aureus Z172 Total score: 2.5 Cumulative Blast bit score: 1182
SAZ172_1526
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AGY89625
Location: 1641838-1643244
NCBI BlastP on this gene
Accession: AGY89625
Location: 1641838-1643244
NCBI BlastP on this gene
gnd
Glyoxalase/bleomycin resistance
Accession: AGY89624
Location: 1641248-1641619
NCBI BlastP on this gene
Accession: AGY89624
Location: 1641248-1641619
NCBI BlastP on this gene
SAZ172_1524
Maltose operon transcriptional repressor MalR, LacI family
Accession: AGY89623
Location: 1640035-1641054
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AGY89623
Location: 1640035-1641054
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
Alpha-glucosidase
Accession: AGY89622
Location: 1638370-1640019
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGY89622
Location: 1638370-1640019
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
Transcription regulator AraC/XylS family
Accession: AGY89621
Location: 1637421-1638287
NCBI BlastP on this gene
Accession: AGY89621
Location: 1637421-1638287
NCBI BlastP on this gene
SAZ172_1521
SAZ172_1520
Glucose-6-phosphate 1-dehydrogenase
Accession: AGY89619
Location: 1635707-1637191
NCBI BlastP on this gene
Accession: AGY89619
Location: 1635707-1637191
NCBI BlastP on this gene
zwf
291. : CP005288 Staphylococcus aureus strain Bmb9393 chromosome Total score: 2.5 Cumulative Blast bit score: 1182
SABB_00435
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AGP28510
Location: 1703240-1704646
NCBI BlastP on this gene
Accession: AGP28510
Location: 1703240-1704646
NCBI BlastP on this gene
gnd
SABB_00433
HTH-type transcriptional regulator malR
Accession: AGP28508
Location: 1701437-1702456
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AGP28508
Location: 1701437-1702456
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: AGP28507
Location: 1699772-1701421
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGP28507
Location: 1699772-1701421
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SABB_00431
AraC family transcriptional regulator
Accession: AGP28506
Location: 1698796-1699689
NCBI BlastP on this gene
Accession: AGP28506
Location: 1698796-1699689
NCBI BlastP on this gene
SABB_00430
glucose-6-phosphate 1-dehydrogenase
Accession: AGP28505
Location: 1697109-1698593
NCBI BlastP on this gene
Accession: AGP28505
Location: 1697109-1698593
NCBI BlastP on this gene
zwf
292. : CP003033 Staphylococcus aureus subsp. aureus VC40 Total score: 2.5 Cumulative Blast bit score: 1182
SAVC_06805
6-phosphogluconate dehydrogenase
Accession: AEZ37521
Location: 1487307-1488713
NCBI BlastP on this gene
Accession: AEZ37521
Location: 1487307-1488713
NCBI BlastP on this gene
SAVC_06800
SAVC_06795
maltose operon transcriptional repressor
Accession: AEZ37519
Location: 1485504-1486523
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AEZ37519
Location: 1485504-1486523
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAVC_06790
alpha-D-1,4-glucosidase
Accession: AEZ37518
Location: 1483839-1485488
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEZ37518
Location: 1483839-1485488
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAVC_06785
AraC family transcriptional regulator
Accession: AEZ37517
Location: 1482890-1483756
NCBI BlastP on this gene
Accession: AEZ37517
Location: 1482890-1483756
NCBI BlastP on this gene
SAVC_06780
glucose-6-phosphate 1-dehydrogenase
Accession: AEZ37516
Location: 1481176-1482660
NCBI BlastP on this gene
Accession: AEZ37516
Location: 1481176-1482660
NCBI BlastP on this gene
SAVC_06775
293. : CP002643 Staphylococcus aureus subsp. aureus T0131 Total score: 2.5 Cumulative Blast bit score: 1182
Peptidase M20/M25/M40 family protein
Accession: AEB88602
Location: 1600234-1601367
NCBI BlastP on this gene
Accession: AEB88602
Location: 1600234-1601367
NCBI BlastP on this gene
SAT0131_01605
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AEB88601
Location: 1598760-1600166
NCBI BlastP on this gene
Accession: AEB88601
Location: 1598760-1600166
NCBI BlastP on this gene
SAT0131_01604
SAT0131_01603
Maltose operon repressor
Accession: AEB88599
Location: 1596957-1597976
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AEB88599
Location: 1596957-1597976
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAT0131_01602
Alpha-D-1,4-glucosidase
Accession: AEB88598
Location: 1595292-1596941
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEB88598
Location: 1595292-1596941
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAT0131_01601
Transcriptional regulator, AraC family
Accession: AEB88597
Location: 1594316-1595209
NCBI BlastP on this gene
Accession: AEB88597
Location: 1594316-1595209
NCBI BlastP on this gene
SAT0131_01600
Glucose-6-phosphate 1-dehydrogenase
Accession: AEB88596
Location: 1592629-1594113
NCBI BlastP on this gene
Accession: AEB88596
Location: 1592629-1594113
NCBI BlastP on this gene
SAT0131_01599
294. : CP002120 Staphylococcus aureus subsp. aureus str. JKD6008 Total score: 2.5 Cumulative Blast bit score: 1182
SAA6008_01481
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ADL65519
Location: 1598479-1599885
NCBI BlastP on this gene
Accession: ADL65519
Location: 1598479-1599885
NCBI BlastP on this gene
gnd
glyoxalase/bleomycin resistance
Accession: ADL65518
Location: 1597889-1598260
NCBI BlastP on this gene
Accession: ADL65518
Location: 1597889-1598260
NCBI BlastP on this gene
SAA6008_01479
maltose operon transcriptional repressor
Accession: ADL65517
Location: 1596676-1597695
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ADL65517
Location: 1596676-1597695
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha amylase, catalytic region
Accession: ADL65516
Location: 1595011-1596660
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ADL65516
Location: 1595011-1596660
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: ADL65515
Location: 1594062-1594928
NCBI BlastP on this gene
Accession: ADL65515
Location: 1594062-1594928
NCBI BlastP on this gene
SAA6008_01476
putative glucose-6-phosphate 1-dehydrogenase
Accession: ADL65514
Location: 1592348-1593832
NCBI BlastP on this gene
Accession: ADL65514
Location: 1592348-1593832
NCBI BlastP on this gene
zwf
295. : CP000730 Staphylococcus aureus subsp. aureus USA300_TCH1516 Total score: 2.5 Cumulative Blast bit score: 1182
USA300HOU_1513
phosphogluconate dehydrogenase (decarboxylating)
Accession: ABX29519
Location: 1627066-1628472
NCBI BlastP on this gene
Accession: ABX29519
Location: 1627066-1628472
NCBI BlastP on this gene
pgd
USA300HOU_1511
maltose operon transcriptional repressor
Accession: ABX29517
Location: 1625263-1626282
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ABX29517
Location: 1625263-1626282
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: ABX29516
Location: 1623598-1625247
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABX29516
Location: 1623598-1625247
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: ABX29515
Location: 1622622-1623515
NCBI BlastP on this gene
Accession: ABX29515
Location: 1622622-1623515
NCBI BlastP on this gene
USA300HOU_1508
glucose-6-phosphate 1-dehydrogenase
Accession: ABX29514
Location: 1620935-1622419
NCBI BlastP on this gene
Accession: ABX29514
Location: 1620935-1622419
NCBI BlastP on this gene
USA300HOU_1507
296. : CP000255 Staphylococcus aureus subsp. aureus USA300_FPR3757 Total score: 2.5 Cumulative Blast bit score: 1182
peptidase, M20/M25/M40 family
Accession: ABD20395
Location: 1614593-1615726
NCBI BlastP on this gene
Accession: ABD20395
Location: 1614593-1615726
NCBI BlastP on this gene
SAUSA300_1460
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ABD21549
Location: 1613119-1614525
NCBI BlastP on this gene
Accession: ABD21549
Location: 1613119-1614525
NCBI BlastP on this gene
gnd
SAUSA300_1458
maltose operon transcriptional repressor
Accession: ABD21049
Location: 1611316-1612335
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ABD21049
Location: 1611316-1612335
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha glucosidase
Accession: ABD20873
Location: 1609651-1611300
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABD20873
Location: 1609651-1611300
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAUSA300_1456
transcriptional regulator, AraC family
Accession: ABD22885
Location: 1608675-1609568
NCBI BlastP on this gene
Accession: ABD22885
Location: 1608675-1609568
NCBI BlastP on this gene
SAUSA300_1455
glucose-6-phosphate 1-dehydrogenase
Accession: ABD21013
Location: 1606988-1608472
NCBI BlastP on this gene
Accession: ABD21013
Location: 1606988-1608472
NCBI BlastP on this gene
zwf
297. : CP000253 Staphylococcus aureus subsp. aureus NCTC 8325 Total score: 2.5 Cumulative Blast bit score: 1182
SAOUHSC_01606
6-phosphogluconate dehydrogenase, decarboxylating
Accession: ABD30684
Location: 1529679-1531085
NCBI BlastP on this gene
Accession: ABD30684
Location: 1529679-1531085
NCBI BlastP on this gene
SAOUHSC_01605
conserved hypothetical protein
Accession: ABD30683
Location: 1529089-1529460
NCBI BlastP on this gene
Accession: ABD30683
Location: 1529089-1529460
NCBI BlastP on this gene
SAOUHSC_01604
SAOUHSC_01603
transcriptional regulator, putative
Accession: ABD30681
Location: 1527876-1528895
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: ABD30681
Location: 1527876-1528895
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
SAOUHSC_01602
alpha-glucosidase, putative
Accession: ABD30680
Location: 1526211-1527860
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ABD30680
Location: 1526211-1527860
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAOUHSC_01601
conserved hypothetical protein
Accession: ABD30679
Location: 1525235-1526128
NCBI BlastP on this gene
Accession: ABD30679
Location: 1525235-1526128
NCBI BlastP on this gene
SAOUHSC_01600
glucose-6-phosphate 1-dehydrogenase
Accession: ABD30678
Location: 1523548-1525032
NCBI BlastP on this gene
Accession: ABD30678
Location: 1523548-1525032
NCBI BlastP on this gene
SAOUHSC_01599
298. : CP000046 Staphylococcus aureus subsp. aureus COL Total score: 2.5 Cumulative Blast bit score: 1182
peptidase, M20/M25/M40 family
Accession: AAW36747
Location: 1591772-1592905
NCBI BlastP on this gene
Accession: AAW36747
Location: 1591772-1592905
NCBI BlastP on this gene
SACOL1555
6-phosphogluconate dehydrogenase, decarboxylating
Accession: AAW36746
Location: 1590298-1591704
NCBI BlastP on this gene
Accession: AAW36746
Location: 1590298-1591704
NCBI BlastP on this gene
gnd
SACOL1553
maltose operon repressor
Accession: AAW36744
Location: 1588495-1589514
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: AAW36744
Location: 1588495-1589514
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha-glucosidase
Accession: AAW36743
Location: 1586830-1588479
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AAW36743
Location: 1586830-1588479
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
transcriptional regulator, AraC family
Accession: AAW36742
Location: 1585854-1586747
NCBI BlastP on this gene
Accession: AAW36742
Location: 1585854-1586747
NCBI BlastP on this gene
SACOL1550
glucose-6-phosphate 1-dehydrogenase
Accession: AAW36741
Location: 1584167-1585651
NCBI BlastP on this gene
Accession: AAW36741
Location: 1584167-1585651
NCBI BlastP on this gene
zwf
299. : AP020318 Staphylococcus aureus KUH180038 DNA Total score: 2.5 Cumulative Blast bit score: 1182
KUH180038_0509
6-phosphogluconate dehydrogenase
Accession: BBN34691
Location: 509735-511141
NCBI BlastP on this gene
Accession: BBN34691
Location: 509735-511141
NCBI BlastP on this gene
gnd
glyoxalase/bleomycin resistance
Accession: BBN34692
Location: 511360-511731
NCBI BlastP on this gene
Accession: BBN34692
Location: 511360-511731
NCBI BlastP on this gene
KUH180038_0511
maltose operon transcriptional repressor
Accession: BBN34693
Location: 511925-512944
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: BBN34693
Location: 511925-512944
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha amylase
Accession: BBN34694
Location: 512960-514609
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBN34694
Location: 512960-514609
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: BBN34695
Location: 514692-515558
NCBI BlastP on this gene
Accession: BBN34695
Location: 514692-515558
NCBI BlastP on this gene
KUH180038_0514
putative glucose-6-phosphate 1-dehydrogenase
Accession: BBN34696
Location: 515788-517272
NCBI BlastP on this gene
Accession: BBN34696
Location: 515788-517272
NCBI BlastP on this gene
zwf
300. : AP020313 Staphylococcus aureus KUH140046 DNA Total score: 2.5 Cumulative Blast bit score: 1182
KUH140046_0509
6-phosphogluconate dehydrogenase
Accession: BBN27038
Location: 511301-512707
NCBI BlastP on this gene
Accession: BBN27038
Location: 511301-512707
NCBI BlastP on this gene
gnd
glyoxalase/bleomycin resistance
Accession: BBN27039
Location: 512926-513297
NCBI BlastP on this gene
Accession: BBN27039
Location: 512926-513297
NCBI BlastP on this gene
KUH140046_0511
maltose operon transcriptional repressor
Accession: BBN27040
Location: 513491-514510
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
Accession: BBN27040
Location: 513491-514510
BlastP hit with malR
Percentage identity: 45 %
BlastP bit score: 273
Sequence coverage: 93 %
E-value: 2e-85
NCBI BlastP on this gene
malR
alpha amylase
Accession: BBN27041
Location: 514526-516175
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBN27041
Location: 514526-516175
BlastP hit with malA
Percentage identity: 78 %
BlastP bit score: 909
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
malA
AraC family transcriptional regulator
Accession: BBN27042
Location: 516258-517124
NCBI BlastP on this gene
Accession: BBN27042
Location: 516258-517124
NCBI BlastP on this gene
KUH140046_0514
putative glucose-6-phosphate 1-dehydrogenase
Accession: BBN27043
Location: 517354-518838
NCBI BlastP on this gene
Accession: BBN27043
Location: 517354-518838
NCBI BlastP on this gene
zwf


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.