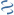MultiGeneBlast hits

Query: Streptococcus suis 05ZYH33, complete genome.
KU665267 : Streptococcus suis strain YS355 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2148
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02767
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02767
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02768
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AOP02768
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02769
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP02769
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02770
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02770
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02771
Location: 4009-5472
NCBI BlastP on this gene
Accession: AOP02771
Location: 4009-5472
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KM972274 : Streptococcus suis strain YS57_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2148
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80272
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80272
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80273
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AKE80273
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80274
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 8e-151
NCBI BlastP on this gene
Accession: AKE80274
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 8e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80275
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AKE80275
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80276
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80276
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE80283
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE80283
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972289 : Streptococcus suis strain YS87_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2147
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80602
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80602
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80603
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80603
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80604
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE80604
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80605
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE80605
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80606
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE80606
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972287 : Streptococcus suis strain YS85_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2147
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80556
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80556
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80557
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80557
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80558
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE80558
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80559
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE80559
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80560
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE80560
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972244 : Streptococcus suis strain YS145_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2147
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79625
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79625
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79626
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79626
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79627
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79627
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79628
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79628
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79629
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79629
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972245 : Streptococcus suis strain YS146_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2146
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79650
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79650
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79651
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79651
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79652
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79652
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79653
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79653
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79654
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79654
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972240 : Streptococcus suis strain YS132_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79527
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79527
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79528
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79528
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79529
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79529
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79530
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79530
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79531
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79531
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972239 : Streptococcus suis strain YS131_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79502
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79502
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79503
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79503
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79504
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79504
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79505
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79505
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79506
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79506
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972237 : Streptococcus suis strain YS125_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79456
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79456
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79459
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79459
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972236 : Streptococcus suis strain YS123_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79430
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79430
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79431
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79431
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79432
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79432
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79433
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79433
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79434
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79434
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972230 : Streptococcus suis strain YS111_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79288
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79288
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79289
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79289
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79290
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79290
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79291
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79291
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79292
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79292
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972229 : Streptococcus suis strain YS110_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79263
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79263
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79264
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79264
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79265
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79265
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79266
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79266
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79267
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79267
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972228 : Streptococcus suis strain YS109_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79238
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79238
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79239
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79239
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79240
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79240
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79241
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79241
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79242
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79242
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972227 : Streptococcus suis strain YS107_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79212
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79212
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79213
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AKE79213
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79214
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79214
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79215
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79215
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79216
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79216
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD-dependent epimerase/dehydratase
Accession: AKE79219
Location: 6885-7739
NCBI BlastP on this gene
Accession: AKE79219
Location: 6885-7739
NCBI BlastP on this gene
cpsH
cpsI
cpsM
cpsN
cpsO
cpsP
Query: Streptococcus suis 05ZYH33, complete genome.
KX870056 : Streptococcus suis strain 1127863 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ79064
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ79064
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79065
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: APZ79065
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79066
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: APZ79066
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79067
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: APZ79067
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
Nucleoside-diphosphate sugar epimerase
Accession: APZ79068
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ79068
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD-dependent epimerase/dehydratase
Accession: APZ79071
Location: 6886-7740
NCBI BlastP on this gene
Accession: APZ79071
Location: 6886-7740
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU665287 : Streptococcus suis strain YS622 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03242
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03242
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03243
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03243
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03244
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP03244
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03245
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 2e-146
NCBI BlastP on this gene
Accession: AOP03245
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 2e-146
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03246
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03246
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972292 : Streptococcus suis strain YS92_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80668
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80668
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80669
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
Accession: AKE80669
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80670
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
Accession: AKE80670
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80671
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
Accession: AKE80671
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80672
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80672
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972291 : Streptococcus suis strain YS91_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80653
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
Accession: AKE80653
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80654
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
Accession: AKE80654
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80655
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
Accession: AKE80655
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80656
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80656
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972271 : Streptococcus suis strain YS46_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE80195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AKE80196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80197
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE80197
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE80204
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE80204
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972242 : Streptococcus suis strain YS140_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79573
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79573
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79574
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79574
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79575
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79575
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79576
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79576
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79577
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79577
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE79584
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE79584
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KC537387 : Streptococcus suis strain YS54 capsular polysaccharide synthesis locus sequence. Total score: 5.5 Cumulative Blast bit score: 2144
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AGS58399
AGS58400
AGS58401
CpsA
Accession: AGS58402
Location: 3330-4769
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGS58402
Location: 3330-4769
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AGS58402
CpsB
Accession: AGS58403
Location: 4786-5475
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AGS58403
Location: 4786-5475
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
AGS58403
CpsC
Accession: AGS58404
Location: 5485-6171
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AGS58404
Location: 5485-6171
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
AGS58404
CpsD
Accession: AGS58405
Location: 6210-6941
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AGS58405
Location: 6210-6941
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
AGS58405
AGS58406
AGS58407
AGS58408
AGS58409
AGS58410
AGS58411
AGS58412
AGS58413
AGS58414
Query: Streptococcus suis 05ZYH33, complete genome.
KT163368 : Streptococcus suis strain YS191 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03653
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 399
Sequence coverage: 100 %
E-value: 3e-138
NCBI BlastP on this gene
Accession: AOP03653
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 399
Sequence coverage: 100 %
E-value: 3e-138
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03654
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03654
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03655
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 4e-144
NCBI BlastP on this gene
Accession: AOP03655
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 4e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03656
Location: 3640-5466
NCBI BlastP on this gene
Accession: AOP03656
Location: 3640-5466
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KM972258 : Streptococcus suis strain YS173_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79917
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79917
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79920
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79920
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972235 : Streptococcus suis strain YS122_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79405
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79405
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79406
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79406
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79407
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79407
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79408
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79408
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79409
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79409
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972226 : Streptococcus suis strain YS104_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79188
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79188
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79189
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AKE79189
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79190
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79190
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79191
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 1e-145
NCBI BlastP on this gene
Accession: AKE79191
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 1e-145
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79192
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79192
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KU665271 : Streptococcus suis strain YS408 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2142
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02861
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02861
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02862
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02862
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02863
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02863
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02864
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: AOP02864
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02865
Location: 3707-5467
NCBI BlastP on this gene
Accession: AOP02865
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP02871
Location: 10559-11677
NCBI BlastP on this gene
Accession: AOP02871
Location: 10559-11677
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972252 : Streptococcus suis strain YS160_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2142
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79796
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79796
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79797
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79797
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79798
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79798
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79799
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AKE79799
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79800
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79800
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU983475 : Streptococcus suis strain YS555 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03433
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03433
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03434
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03434
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03435
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AOP03435
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03436
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03436
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03437
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03437
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
cpsP
Query: Streptococcus suis 05ZYH33, complete genome.
KU665281 : Streptococcus suis strain YS525 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03101
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03101
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03102
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03102
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03103
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AOP03103
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03104
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03104
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03105
Location: 3707-5467
NCBI BlastP on this gene
Accession: AOP03105
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
cpsP
Query: Streptococcus suis 05ZYH33, complete genome.
KU665272 : Streptococcus suis strain YS443 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02886
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02886
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02887
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02887
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02888
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02888
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02889
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02889
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02890
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02890
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972234 : Streptococcus suis strain YS121_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79376
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79376
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79377
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 3e-136
NCBI BlastP on this gene
Accession: AKE79377
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 3e-136
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79378
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AKE79378
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79379
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79379
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79380
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79380
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE79387
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE79387
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KX870062 : Streptococcus suis strain 1224887 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ79193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ79193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: APZ79194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: APZ79195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: APZ79196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ79197
Location: 4008-5468
NCBI BlastP on this gene
Accession: APZ79197
Location: 4008-5468
NCBI BlastP on this gene
cpsE
cpsF
cpsG
UDP-phosphate galactose phosphotransferase
Accession: APZ79200
Location: 7572-8159
NCBI BlastP on this gene
Accession: APZ79200
Location: 7572-8159
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
Initial sugar transferase(Glycosyltransferase)
Accession: APZ79203
Location: 10886-12148
NCBI BlastP on this gene
Accession: APZ79203
Location: 10886-12148
NCBI BlastP on this gene
cpsK
cpsL
Query: Streptococcus suis 05ZYH33, complete genome.
KU983474 : Streptococcus suis strain YS535 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AOP03410
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03410
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03411
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AOP03411
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03412
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP03412
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
protein-Tyrosine phosphatase Wzh
Accession: AOP03413
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
Accession: AOP03413
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03414
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03414
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU665280 : Streptococcus suis strain YS511 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03077
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03077
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03078
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03078
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03079
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AOP03079
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03080
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03080
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03081
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03081
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
cpsP
Query: Streptococcus suis 05ZYH33, complete genome.
KM972251 : Streptococcus suis strain YS159_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79771
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79771
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79772
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79772
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79773
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79773
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79774
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
Accession: AKE79774
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79775
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79775
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM972233 : Streptococcus suis strain YS117_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79351
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79351
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79352
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 392
Sequence coverage: 100 %
E-value: 2e-135
NCBI BlastP on this gene
Accession: AKE79352
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 392
Sequence coverage: 100 %
E-value: 2e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79353
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79353
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79354
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79354
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79355
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79355
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KU665274 : Streptococcus suis strain YS487 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02932
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02932
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02933
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02933
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02934
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02934
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02935
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02935
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02936
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02936
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KU665270 : Streptococcus suis strain YS394 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02838
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02838
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02839
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02839
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02840
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02840
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02841
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AOP02841
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02842
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02842
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KU665266 : Streptococcus suis strain YS351 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02743
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02743
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02744
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AOP02744
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02745
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: AOP02745
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02746
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP02746
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02747
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02747
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP02750
Location: 6886-7746
NCBI BlastP on this gene
Accession: AOP02750
Location: 6886-7746
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KM972256 : Streptococcus suis strain YS170_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79883
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79883
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79884
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: AKE79884
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79885
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
Accession: AKE79885
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79886
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79886
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79887
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79887
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972253 : Streptococcus suis strain YS166_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79819
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79819
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79820
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: AKE79820
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79821
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
Accession: AKE79821
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79822
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79822
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79823
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79823
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU983472 : Streptococcus suis strain YS492 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03362
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03362
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03363
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03363
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03364
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03364
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03365
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP03365
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03366
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP03366
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03372
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP03372
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU665279 : Streptococcus suis strain YS501 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03052
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03052
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03053
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03053
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03054
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03054
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03055
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP03055
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03056
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP03056
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03062
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP03062
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU665278 : Streptococcus suis strain YS498 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03027
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03027
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03028
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03028
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03029
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03029
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03030
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP03030
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03031
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP03031
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03037
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP03037
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU665276 : Streptococcus suis strain YS493 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02977
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02977
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02978
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02978
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02979
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02979
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02980
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP02980
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02981
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP02981
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP02987
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP02987
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KT163364 : Streptococcus suis strain YS178 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03552
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03552
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03553
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03553
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03554
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: AOP03554
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03555
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03555
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03556
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03556
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03559
Location: 6889-7743
NCBI BlastP on this gene
Accession: AOP03559
Location: 6889-7743
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KM972257 : Streptococcus suis strain YS171_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79899
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79899
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79900
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AKE79900
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79901
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79901
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79902
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AKE79902
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79903
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79903
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KX870053 : Streptococcus suis strain 1093407 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2137
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ78999
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78999
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79000
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 2e-136
NCBI BlastP on this gene
Accession: APZ79000
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 2e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79001
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 96 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 7e-153
NCBI BlastP on this gene
Accession: APZ79001
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 96 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 7e-153
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79002
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 5e-143
NCBI BlastP on this gene
Accession: APZ79002
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 5e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ79003
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ79003
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KU983471 : Streptococcus suis strain YS387 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2137
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03341
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 881
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03341
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 881
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03342
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 407
Sequence coverage: 100 %
E-value: 3e-141
NCBI BlastP on this gene
Accession: AOP03342
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 407
Sequence coverage: 100 %
E-value: 3e-141
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03343
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03343
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03344
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: AOP03344
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03345
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03345
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972286 : Streptococcus suis strain YS84_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2137
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80535
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 891
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80535
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 891
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein
Accession: AKE80536
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80536
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
wze
Accession: AKE80537
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 437
Sequence coverage: 96 %
E-value: 4e-153
NCBI BlastP on this gene
Accession: AKE80537
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 437
Sequence coverage: 96 %
E-value: 4e-153
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80538
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE80538
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
putative chain length determinant protein Wzd
Accession: AKE80539
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80539
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
201. : KU665267 Streptococcus suis strain YS355 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2148
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
Membrane protein involved in the export of
Accession: ABP89543.1
Location: 15369-16775
NCBI BlastP on this gene
Accession: ABP89543.1
Location: 15369-16775
NCBI BlastP on this gene
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02767
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02767
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02768
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AOP02768
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02769
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP02769
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02770
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02770
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02771
Location: 4009-5472
NCBI BlastP on this gene
Accession: AOP02771
Location: 4009-5472
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
202. : KM972274 Streptococcus suis strain YS57_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2148
integral membrane regulatory protein Wzg
Accession: AKE80272
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80272
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80273
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AKE80273
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80274
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 8e-151
NCBI BlastP on this gene
Accession: AKE80274
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 8e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80275
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AKE80275
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80276
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80276
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE80283
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE80283
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
203. : KM972289 Streptococcus suis strain YS87_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2147
integral membrane regulatory protein Wzg
Accession: AKE80602
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80602
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80603
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80603
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80604
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE80604
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80605
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE80605
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80606
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE80606
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
204. : KM972287 Streptococcus suis strain YS85_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2147
integral membrane regulatory protein Wzg
Accession: AKE80556
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80556
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80557
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80557
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80558
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE80558
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80559
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE80559
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80560
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE80560
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
205. : KM972244 Streptococcus suis strain YS145_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2147
integral membrane regulatory protein Wzg
Accession: AKE79625
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79625
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79626
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79626
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79627
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79627
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79628
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79628
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79629
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79629
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
206. : KM972245 Streptococcus suis strain YS146_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2146
integral membrane regulatory protein Wzg
Accession: AKE79650
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79650
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 905
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79651
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79651
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79652
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79652
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79653
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79653
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79654
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79654
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
207. : KM972240 Streptococcus suis strain YS132_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79527
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79527
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79528
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79528
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79529
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79529
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79530
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79530
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79531
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79531
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
208. : KM972239 Streptococcus suis strain YS131_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79502
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79502
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79503
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79503
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79504
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79504
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79505
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79505
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79506
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79506
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
209. : KM972237 Streptococcus suis strain YS125_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79456
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79456
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79459
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79459
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
210. : KM972236 Streptococcus suis strain YS123_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79430
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79430
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79431
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79431
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79432
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79432
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79433
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79433
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79434
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79434
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
211. : KM972230 Streptococcus suis strain YS111_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79288
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79288
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79289
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79289
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79290
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79290
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79291
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79291
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79292
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79292
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
212. : KM972229 Streptococcus suis strain YS110_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79263
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79263
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79264
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79264
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79265
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79265
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79266
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79266
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79267
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79267
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
213. : KM972228 Streptococcus suis strain YS109_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79238
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79238
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79239
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79239
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79240
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79240
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79241
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79241
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79242
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79242
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
214. : KM972227 Streptococcus suis strain YS107_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2145
integral membrane regulatory protein Wzg
Accession: AKE79212
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79212
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79213
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AKE79213
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79214
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79214
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79215
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79215
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79216
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79216
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD-dependent epimerase/dehydratase
Accession: AKE79219
Location: 6885-7739
NCBI BlastP on this gene
Accession: AKE79219
Location: 6885-7739
NCBI BlastP on this gene
cpsH
cpsI
cpsM
cpsN
cpsO
cpsP
215. : KX870056 Streptococcus suis strain 1127863 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
Integral membrane regulatory protein Wzg
Accession: APZ79064
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ79064
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79065
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: APZ79065
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79066
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: APZ79066
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79067
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: APZ79067
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
Nucleoside-diphosphate sugar epimerase
Accession: APZ79068
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ79068
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD-dependent epimerase/dehydratase
Accession: APZ79071
Location: 6886-7740
NCBI BlastP on this gene
Accession: APZ79071
Location: 6886-7740
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
216. : KU665287 Streptococcus suis strain YS622 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
Integral membrane regulatory protein Wzg
Accession: AOP03242
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03242
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03243
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03243
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03244
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP03244
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03245
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 2e-146
NCBI BlastP on this gene
Accession: AOP03245
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 2e-146
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03246
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03246
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
217. : KM972292 Streptococcus suis strain YS92_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
integral membrane regulatory protein Wzg
Accession: AKE80668
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80668
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80669
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
Accession: AKE80669
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80670
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
Accession: AKE80670
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80671
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
Accession: AKE80671
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80672
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80672
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
218. : KM972291 Streptococcus suis strain YS91_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
integral membrane regulatory protein Wzg
Accession: AKE80652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80653
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
Accession: AKE80653
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80654
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
Accession: AKE80654
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80655
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
Accession: AKE80655
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 438
Sequence coverage: 100 %
E-value: 3e-153
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80656
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80656
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
219. : KM972271 Streptococcus suis strain YS46_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
integral membrane regulatory protein Wzg
Accession: AKE80193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE80195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AKE80196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80197
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE80197
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE80204
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE80204
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
220. : KM972242 Streptococcus suis strain YS140_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2144
integral membrane regulatory protein Wzg
Accession: AKE79573
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79573
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79574
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79574
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79575
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79575
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79576
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79576
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79577
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79577
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE79584
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE79584
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
221. : KC537387 Streptococcus suis strain YS54 capsular polysaccharide synthesis locus sequence. Total score: 5.5 Cumulative Blast bit score: 2144
AGS58399
AGS58400
AGS58401
CpsA
Accession: AGS58402
Location: 3330-4769
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGS58402
Location: 3330-4769
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AGS58402
CpsB
Accession: AGS58403
Location: 4786-5475
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AGS58403
Location: 4786-5475
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
AGS58403
CpsC
Accession: AGS58404
Location: 5485-6171
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AGS58404
Location: 5485-6171
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
AGS58404
CpsD
Accession: AGS58405
Location: 6210-6941
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AGS58405
Location: 6210-6941
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
AGS58405
AGS58406
AGS58407
AGS58408
AGS58409
AGS58410
AGS58411
AGS58412
AGS58413
AGS58414
222. : KT163368 Streptococcus suis strain YS191 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
Integral membrane regulatory protein Wzg
Accession: AOP03652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03652
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03653
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 399
Sequence coverage: 100 %
E-value: 3e-138
NCBI BlastP on this gene
Accession: AOP03653
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 399
Sequence coverage: 100 %
E-value: 3e-138
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03654
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03654
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03655
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 4e-144
NCBI BlastP on this gene
Accession: AOP03655
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 4e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03656
Location: 3640-5466
NCBI BlastP on this gene
Accession: AOP03656
Location: 3640-5466
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
223. : KM972258 Streptococcus suis strain YS173_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
integral membrane regulatory protein Wzg
Accession: AKE79916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79917
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79917
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79920
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79920
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
224. : KM972235 Streptococcus suis strain YS122_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
integral membrane regulatory protein Wzg
Accession: AKE79405
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79405
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79406
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79406
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79407
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79407
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79408
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79408
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79409
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79409
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
225. : KM972226 Streptococcus suis strain YS104_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2143
integral membrane regulatory protein Wzg
Accession: AKE79188
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79188
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79189
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AKE79189
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79190
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79190
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79191
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 1e-145
NCBI BlastP on this gene
Accession: AKE79191
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 1e-145
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79192
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79192
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
226. : KU665271 Streptococcus suis strain YS408 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2142
Integral membrane regulatory protein Wzg
Accession: AOP02861
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02861
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02862
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02862
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02863
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02863
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02864
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: AOP02864
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02865
Location: 3707-5467
NCBI BlastP on this gene
Accession: AOP02865
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP02871
Location: 10559-11677
NCBI BlastP on this gene
Accession: AOP02871
Location: 10559-11677
NCBI BlastP on this gene
cpsK
cpsL
cpsM
227. : KM972252 Streptococcus suis strain YS160_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2142
integral membrane regulatory protein Wzg
Accession: AKE79796
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79796
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79797
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: AKE79797
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79798
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79798
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79799
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AKE79799
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79800
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79800
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
228. : KU983475 Streptococcus suis strain YS555 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
Integral membrane regulatory protein Wzg
Accession: AOP03433
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03433
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03434
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03434
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03435
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AOP03435
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03436
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03436
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03437
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03437
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
cpsP
229. : KU665281 Streptococcus suis strain YS525 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
Integral membrane regulatory protein Wzg
Accession: AOP03101
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03101
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03102
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03102
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03103
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AOP03103
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03104
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03104
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03105
Location: 3707-5467
NCBI BlastP on this gene
Accession: AOP03105
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
cpsP
230. : KU665272 Streptococcus suis strain YS443 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
Integral membrane regulatory protein Wzg
Accession: AOP02886
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02886
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02887
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02887
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02888
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02888
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02889
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02889
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02890
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02890
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
231. : KM972234 Streptococcus suis strain YS121_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2141
integral membrane regulatory protein Wzg
Accession: AKE79376
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79376
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 903
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79377
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 3e-136
NCBI BlastP on this gene
Accession: AKE79377
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 3e-136
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79378
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AKE79378
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79379
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79379
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79380
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79380
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
nodulation acetyltransferase protein
Accession: AKE79387
Location: 11288-12556
NCBI BlastP on this gene
Accession: AKE79387
Location: 11288-12556
NCBI BlastP on this gene
cpsL
cpsM
232. : KX870062 Streptococcus suis strain 1224887 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
Integral membrane regulatory protein Wzg
Accession: APZ79193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ79193
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
Accession: APZ79194
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 398
Sequence coverage: 100 %
E-value: 1e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: APZ79195
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: APZ79196
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ79197
Location: 4008-5468
NCBI BlastP on this gene
Accession: APZ79197
Location: 4008-5468
NCBI BlastP on this gene
cpsE
cpsF
cpsG
UDP-phosphate galactose phosphotransferase
Accession: APZ79200
Location: 7572-8159
NCBI BlastP on this gene
Accession: APZ79200
Location: 7572-8159
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
Initial sugar transferase(Glycosyltransferase)
Accession: APZ79203
Location: 10886-12148
NCBI BlastP on this gene
Accession: APZ79203
Location: 10886-12148
NCBI BlastP on this gene
cpsK
cpsL
233. : KU983474 Streptococcus suis strain YS535 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
integral membrane regulatory protein Wzg
Accession: AOP03410
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03410
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03411
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AOP03411
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03412
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP03412
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
protein-Tyrosine phosphatase Wzh
Accession: AOP03413
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
Accession: AOP03413
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03414
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03414
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
234. : KU665280 Streptococcus suis strain YS511 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
Integral membrane regulatory protein Wzg
Accession: AOP03077
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03077
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03078
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03078
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03079
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AOP03079
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03080
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03080
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03081
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03081
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
cpsP
235. : KM972251 Streptococcus suis strain YS159_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
integral membrane regulatory protein Wzg
Accession: AKE79771
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79771
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 902
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79772
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE79772
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79773
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: AKE79773
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 432
Sequence coverage: 96 %
E-value: 3e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79774
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
Accession: AKE79774
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79775
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79775
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
236. : KM972233 Streptococcus suis strain YS117_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2140
integral membrane regulatory protein Wzg
Accession: AKE79351
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79351
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 899
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79352
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 392
Sequence coverage: 100 %
E-value: 2e-135
NCBI BlastP on this gene
Accession: AKE79352
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 392
Sequence coverage: 100 %
E-value: 2e-135
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79353
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79353
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79354
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE79354
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79355
Location: 3707-5467
NCBI BlastP on this gene
Accession: AKE79355
Location: 3707-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsL
cpsM
cpsN
cpsO
237. : KU665274 Streptococcus suis strain YS487 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
Integral membrane regulatory protein Wzg
Accession: AOP02932
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02932
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 897
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02933
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02933
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02934
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02934
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02935
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02935
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02936
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02936
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsJ
cpsL
cpsM
cpsN
cpsO
238. : KU665270 Streptococcus suis strain YS394 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
Integral membrane regulatory protein Wzg
Accession: AOP02838
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02838
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 900
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02839
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02839
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02840
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02840
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02841
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AOP02841
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02842
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02842
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
239. : KU665266 Streptococcus suis strain YS351 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
Integral membrane regulatory protein Wzg
Accession: AOP02743
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02743
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 901
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02744
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AOP02744
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02745
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: AOP02745
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02746
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP02746
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02747
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02747
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP02750
Location: 6886-7746
NCBI BlastP on this gene
Accession: AOP02750
Location: 6886-7746
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
240. : KM972256 Streptococcus suis strain YS170_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
integral membrane regulatory protein Wzg
Accession: AKE79883
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79883
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79884
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: AKE79884
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79885
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
Accession: AKE79885
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79886
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79886
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79887
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79887
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
241. : KM972253 Streptococcus suis strain YS166_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2139
integral membrane regulatory protein Wzg
Accession: AKE79819
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79819
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 93 %
BlastP bit score: 904
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79820
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: AKE79820
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 3e-142
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79821
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
Accession: AKE79821
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79822
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79822
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79823
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79823
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
242. : KU983472 Streptococcus suis strain YS492 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
Integral membrane regulatory protein Wzg
Accession: AOP03362
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03362
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03363
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03363
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03364
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03364
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03365
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP03365
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03366
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP03366
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03372
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP03372
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
243. : KU665279 Streptococcus suis strain YS501 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
Integral membrane regulatory protein Wzg
Accession: AOP03052
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03052
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03053
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03053
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03054
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03054
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03055
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP03055
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03056
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP03056
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03062
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP03062
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
244. : KU665278 Streptococcus suis strain YS498 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
Integral membrane regulatory protein Wzg
Accession: AOP03027
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03027
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03028
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03028
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03029
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03029
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03030
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP03030
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03031
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP03031
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03037
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP03037
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
245. : KU665276 Streptococcus suis strain YS493 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
Integral membrane regulatory protein Wzg
Accession: AOP02977
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02977
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 898
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02978
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP02978
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02979
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02979
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02980
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AOP02980
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02981
Location: 3643-5469
NCBI BlastP on this gene
Accession: AOP02981
Location: 3643-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP02987
Location: 10568-11686
NCBI BlastP on this gene
Accession: AOP02987
Location: 10568-11686
NCBI BlastP on this gene
cpsK
cpsL
cpsM
246. : KT163364 Streptococcus suis strain YS178 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
Integral membrane regulatory protein Wzg
Accession: AOP03552
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03552
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03553
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03553
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03554
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: AOP03554
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 430
Sequence coverage: 96 %
E-value: 2e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03555
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03555
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03556
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03556
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03559
Location: 6889-7743
NCBI BlastP on this gene
Accession: AOP03559
Location: 6889-7743
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
247. : KM972257 Streptococcus suis strain YS171_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2138
integral membrane regulatory protein Wzg
Accession: AKE79899
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79899
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79900
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AKE79900
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79901
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AKE79901
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79902
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AKE79902
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79903
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79903
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
248. : KX870053 Streptococcus suis strain 1093407 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2137
Integral membrane regulatory protein Wzg
Accession: APZ78999
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78999
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 894
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79000
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 2e-136
NCBI BlastP on this gene
Accession: APZ79000
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 2e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79001
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 96 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 7e-153
NCBI BlastP on this gene
Accession: APZ79001
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 96 %
BlastP bit score: 436
Sequence coverage: 96 %
E-value: 7e-153
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79002
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 5e-143
NCBI BlastP on this gene
Accession: APZ79002
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 5e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ79003
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ79003
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
249. : KU983471 Streptococcus suis strain YS387 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2137
Integral membrane regulatory protein Wzg
Accession: AOP03341
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 881
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03341
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 881
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03342
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 407
Sequence coverage: 100 %
E-value: 3e-141
NCBI BlastP on this gene
Accession: AOP03342
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 407
Sequence coverage: 100 %
E-value: 3e-141
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03343
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03343
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03344
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: AOP03344
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03345
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03345
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
250. : KM972286 Streptococcus suis strain YS84_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2137
integral membrane regulatory protein Wzg
Accession: AKE80535
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 891
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80535
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 891
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein
Accession: AKE80536
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AKE80536
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
wze
Accession: AKE80537
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 437
Sequence coverage: 96 %
E-value: 4e-153
NCBI BlastP on this gene
Accession: AKE80537
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 437
Sequence coverage: 96 %
E-value: 4e-153
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80538
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AKE80538
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
putative chain length determinant protein Wzd
Accession: AKE80539
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE80539
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.