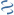MultiGeneBlast hits

Query: Streptococcus suis 05ZYH33, complete genome.
KM972262 : Streptococcus suis strain YS27_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2120
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80009
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80009
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80010
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 384
Sequence coverage: 100 %
E-value: 4e-132
NCBI BlastP on this gene
Accession: AKE80010
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 384
Sequence coverage: 100 %
E-value: 4e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80011
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AKE80011
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80012
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AKE80012
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80013
Location: 3709-5469
NCBI BlastP on this gene
Accession: AKE80013
Location: 3709-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KT163371 : Streptococcus suis strain YS209 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2119
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03744
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 875
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03744
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 875
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03745
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03745
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03746
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP03746
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03747
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
Accession: AOP03747
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03748
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03748
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03754
Location: 10566-11684
NCBI BlastP on this gene
Accession: AOP03754
Location: 10566-11684
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KT163366 : Streptococcus suis strain YS189 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2119
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03598
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 880
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03598
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 880
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03599
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03599
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03600
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03600
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03601
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03601
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03602
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03602
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03608
Location: 10567-11685
NCBI BlastP on this gene
Accession: AOP03608
Location: 10567-11685
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KM972294 : Streptococcus suis strain YS94_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2119
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80700
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 887
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80700
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 887
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80701
Location: 1458-2147
BlastP hit with ABP89531.1
Percentage identity: 85 %
BlastP bit score: 380
Sequence coverage: 100 %
E-value: 6e-131
NCBI BlastP on this gene
Accession: AKE80701
Location: 1458-2147
BlastP hit with ABP89531.1
Percentage identity: 85 %
BlastP bit score: 380
Sequence coverage: 100 %
E-value: 6e-131
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80702
Location: 2157-2843
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AKE80702
Location: 2157-2843
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80703
Location: 2882-3613
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 1e-146
NCBI BlastP on this gene
Accession: AKE80703
Location: 2882-3613
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 1e-146
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80704
Location: 3642-5468
NCBI BlastP on this gene
Accession: AKE80704
Location: 3642-5468
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KJ669337 : Streptococcus suis strain CZ130302 hypothetical protein (ChZ00x1), putative transcripti... Total score: 5.5 Cumulative Blast bit score: 2118
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
ChZ00x1
putative transcriptional regulator of pyridoxine metabolism
Accession: AJA01362
Location: 532-1383
NCBI BlastP on this gene
Accession: AJA01362
Location: 532-1383
NCBI BlastP on this gene
ChZ00x2
putative transcriptional regulator of pyridoxine metabolism
Accession: AJA01363
Location: 1474-2733
NCBI BlastP on this gene
Accession: AJA01363
Location: 1474-2733
NCBI BlastP on this gene
ChZ00x3
ChZ00x4
capsular polysaccharide expression regulator
Accession: AJA01365
Location: 3692-5131
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJA01365
Location: 3692-5131
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
chzA
chain length determinant protein/polysaccharide export protein
Accession: AJA01366
Location: 5148-5837
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AJA01366
Location: 5148-5837
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
chzB
tyrosine-protein kinase
Accession: AJA01367
Location: 5847-6533
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AJA01367
Location: 5847-6533
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
chzC
protein-tyrosine phosphatase
Accession: AJA01368
Location: 6572-7303
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AJA01368
Location: 6572-7303
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
chzD
putative nucleoside-diphosphate sugar epimerase
Accession: AJA01369
Location: 7332-9158
NCBI BlastP on this gene
Accession: AJA01369
Location: 7332-9158
NCBI BlastP on this gene
chzE
chzF
chzG
NAD dependent epimerase/dehydratase family protein
Accession: AJA01372
Location: 10569-11429
NCBI BlastP on this gene
Accession: AJA01372
Location: 10569-11429
NCBI BlastP on this gene
chzH
putative glycosyltransferase of GT1 family
Accession: AJA01373
Location: 11426-12544
NCBI BlastP on this gene
Accession: AJA01373
Location: 11426-12544
NCBI BlastP on this gene
chzI
chzJ
chzK
chzL
chzM
chzN
Query: Streptococcus suis 05ZYH33, complete genome.
CP024974 : Streptococcus suis strain CZ130302 chromosome Total score: 5.5 Cumulative Blast bit score: 2118
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
CVO91_08840
CVO91_08835
CVO91_08830
glutamate-5-semialdehyde dehydrogenase
Accession: ATZ03987
Location: 1734972-1736210
NCBI BlastP on this gene
Accession: ATZ03987
Location: 1734972-1736210
NCBI BlastP on this gene
CVO91_08825
pyrroline-5-carboxylate reductase
Accession: ATZ03986
Location: 1734177-1734962
NCBI BlastP on this gene
Accession: ATZ03986
Location: 1734177-1734962
NCBI BlastP on this gene
proC
CVO91_08815
TetR/AcrR family transcriptional regulator
Accession: ATZ03984
Location: 1732212-1732616
NCBI BlastP on this gene
Accession: ATZ03984
Location: 1732212-1732616
NCBI BlastP on this gene
CVO91_08810
DegV domain-containing protein
Accession: ATZ03983
Location: 1731234-1732085
NCBI BlastP on this gene
Accession: ATZ03983
Location: 1731234-1732085
NCBI BlastP on this gene
CVO91_08805
PLP-dependent aminotransferase family protein
Accession: ATZ03982
Location: 1729884-1731143
NCBI BlastP on this gene
Accession: ATZ03982
Location: 1729884-1731143
NCBI BlastP on this gene
CVO91_08800
CVO91_08795
LytR family transcriptional regulator
Accession: ATZ03980
Location: 1727486-1728925
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ATZ03980
Location: 1727486-1728925
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
CVO91_08790
capsular biosynthesis protein CpsC
Accession: ATZ03979
Location: 1726780-1727469
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: ATZ03979
Location: 1726780-1727469
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
CVO91_08785
tyrosine protein kinase
Accession: ATZ03978
Location: 1726084-1726770
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: ATZ03978
Location: 1726084-1726770
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
CVO91_08780
tyrosine protein phosphatase
Accession: ATZ03977
Location: 1725314-1726045
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: ATZ03977
Location: 1725314-1726045
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
CVO91_08775
polysaccharide biosynthesis protein
Accession: ATZ03976
Location: 1723459-1725285
NCBI BlastP on this gene
Accession: ATZ03976
Location: 1723459-1725285
NCBI BlastP on this gene
CVO91_08770
cell filamentation protein Fic
Accession: ATZ03975
Location: 1722657-1723373
NCBI BlastP on this gene
Accession: ATZ03975
Location: 1722657-1723373
NCBI BlastP on this gene
CVO91_08765
lipid carrier--UDP-N-acetylgalactosaminyltransferase
Accession: ATZ03974
Location: 1722048-1722647
NCBI BlastP on this gene
Accession: ATZ03974
Location: 1722048-1722647
NCBI BlastP on this gene
CVO91_08760
CVO91_08755
CVO91_08750
CVO91_08745
CVO91_08740
glycosyltransferase family 4 protein
Accession: ATZ03970
Location: 1717397-1718476
NCBI BlastP on this gene
Accession: ATZ03970
Location: 1717397-1718476
NCBI BlastP on this gene
CVO91_08735
CVO91_08730
CVO91_08725
Query: Streptococcus suis 05ZYH33, complete genome.
KU665275 : Streptococcus suis strain YS488 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2117
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02953
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02953
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02954
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 375
Sequence coverage: 100 %
E-value: 1e-128
NCBI BlastP on this gene
Accession: AOP02954
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 375
Sequence coverage: 100 %
E-value: 1e-128
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02955
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP02955
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02956
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AOP02956
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02957
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02957
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP02963
Location: 10566-11684
NCBI BlastP on this gene
Accession: AOP02963
Location: 10566-11684
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KT163375 : Streptococcus suis strain YS241 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2117
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03855
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 882
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03855
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 882
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03856
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
Accession: AOP03856
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03857
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
Accession: AOP03857
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03858
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03858
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03859
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03859
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03865
Location: 10567-11685
NCBI BlastP on this gene
Accession: AOP03865
Location: 10567-11685
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
JF273649 : Streptococcus suis strain 8074 ORF7Z gene Total score: 5.5 Cumulative Blast bit score: 2117
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEH57449
AEH57450
AEH57451
Cps7A
Accession: AEH57452
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57452
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps7A
Cps7B
Accession: AEH57453
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
Accession: AEH57453
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
cps7B
Cps7C
Accession: AEH57454
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AEH57454
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
cps7C
Cps7D
Accession: AEH57455
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEH57455
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cps7D
cps7E
cps7F
cps7G
cps7H
cps7I
cps7J
cps7K
cps7L
cps7M
Query: Streptococcus suis 05ZYH33, complete genome.
CP002641 : Streptococcus suis D9 Total score: 5.5 Cumulative Blast bit score: 2117
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
SSUD9_0732
proB
gamma-glutamyl phosphate reductase
Accession: AER16959
Location: 728256-729494
NCBI BlastP on this gene
Accession: AER16959
Location: 728256-729494
NCBI BlastP on this gene
proA
pyrroline-5-carboxylate reductase
Accession: AER16960
Location: 729504-730289
NCBI BlastP on this gene
Accession: AER16960
Location: 729504-730289
NCBI BlastP on this gene
proC
SSUD9_0736
SSUD9_0737
SSUD9_0738
SSUD9_0739
SSUD9_0740
integral membrane regulatory protein Wzg
Accession: AER16966
Location: 735552-736991
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AER16966
Location: 735552-736991
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
wzg
Cps2B
Accession: AER16967
Location: 737007-737696
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
Accession: AER16967
Location: 737007-737696
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
SSUD9_0742
tyrosine-protein kinase Wze
Accession: AER16968
Location: 737706-738392
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AER16968
Location: 737706-738392
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
wze
protein-tyrosine phosphatase Wzh
Accession: AER16969
Location: 738431-739162
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AER16969
Location: 738431-739162
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
wzh
SSUD9_0745
putative glycosyltransferase Cps7F
Accession: AER16971
Location: 741463-742161
NCBI BlastP on this gene
Accession: AER16971
Location: 741463-742161
NCBI BlastP on this gene
SSUD9_0746
SSUD9_0747
putative glycosyltransferase Cps7H
Accession: AER16973
Location: 743403-744188
NCBI BlastP on this gene
Accession: AER16973
Location: 743403-744188
NCBI BlastP on this gene
SSUD9_0748
SSUD9_0749
SSUD9_0750
polysaccharide polymerase Cps19aI
Accession: AER16976
Location: 746768-748090
NCBI BlastP on this gene
Accession: AER16976
Location: 746768-748090
NCBI BlastP on this gene
SSUD9_0751
SSUD9_0752
Query: Streptococcus suis 05ZYH33, complete genome.
KX870065 : Streptococcus suis strain 1297150 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2115
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ79282
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 879
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ79282
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 879
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79283
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 1e-136
NCBI BlastP on this gene
Accession: APZ79283
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 1e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79284
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 2e-149
NCBI BlastP on this gene
Accession: APZ79284
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 2e-149
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79285
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: APZ79285
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ79286
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ79286
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
Initial sugar transferase(Glycosyl-1-phosphate transferase)
Accession: APZ79288
Location: 6295-6993
NCBI BlastP on this gene
Accession: APZ79288
Location: 6295-6993
NCBI BlastP on this gene
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: APZ79292
Location: 10559-11677
NCBI BlastP on this gene
Accession: APZ79292
Location: 10559-11677
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KT163373 : Streptococcus suis strain YS219 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2115
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03804
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 876
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03804
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 876
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03805
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 6e-136
NCBI BlastP on this gene
Accession: AOP03805
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 6e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03806
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 4e-150
NCBI BlastP on this gene
Accession: AOP03806
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 4e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03807
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AOP03807
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03808
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03808
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03814
Location: 10567-11685
NCBI BlastP on this gene
Accession: AOP03814
Location: 10567-11685
NCBI BlastP on this gene
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU983476 : Streptococcus suis strain YS602 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03456
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03456
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03459
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03459
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03462
Location: 6885-7745
NCBI BlastP on this gene
Accession: AOP03462
Location: 6885-7745
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KT163377 : Streptococcus suis strain YS249 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03917
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03917
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03920
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03920
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03923
Location: 6885-7745
NCBI BlastP on this gene
Accession: AOP03923
Location: 6885-7745
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KT163367 : Streptococcus suis strain YS190 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03627
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03627
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03628
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03628
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03629
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03629
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03630
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03630
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03631
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03631
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03634
Location: 6885-7745
NCBI BlastP on this gene
Accession: AOP03634
Location: 6885-7745
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KM972231 : Streptococcus suis strain YS113_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79313
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79313
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79314
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AKE79314
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79315
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
Accession: AKE79315
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79316
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79316
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79317
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79317
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
MH763820 : Streptococcus suis strain 7619/2012 hypothetical protein gene Total score: 5.5 Cumulative Blast bit score: 2113
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
QBQ85388
integral membrane regulatory protein Wzg
Accession: QBQ85389
Location: 844-2283
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 856
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: QBQ85389
Location: 844-2283
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 856
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
QBQ85389
chain length determinant protein Wzd
Accession: QBQ85390
Location: 2299-2988
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: QBQ85390
Location: 2299-2988
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
QBQ85390
tyrosine-protein kinase Wze
Accession: QBQ85391
Location: 2998-3684
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: QBQ85391
Location: 2998-3684
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
QBQ85391
protein-tyrosine phosphatase Wzh
Accession: QBQ85392
Location: 3723-4454
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: QBQ85392
Location: 3723-4454
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
QBQ85392
Nucleoside-diphosphate sugar epimerase
Accession: QBQ85393
Location: 4484-6310
NCBI BlastP on this gene
Accession: QBQ85393
Location: 4484-6310
NCBI BlastP on this gene
QBQ85393
QBQ85394
pyridoxal phosphate (PLP)-dependent aspartate aminotransferase family protein
Accession: QBQ85395
Location: 7462-8679
NCBI BlastP on this gene
Accession: QBQ85395
Location: 7462-8679
NCBI BlastP on this gene
QBQ85395
QBQ85396
QBQ85397
QBQ85398
QBQ85399
QBQ85400
QBQ85401
QBQ85402
Query: Streptococcus suis 05ZYH33, complete genome.
LR536835 : Streptococcus pneumoniae strain 2245STDY6020240 genome assembly, chromosome: 1. Total score: 5.5 Cumulative Blast bit score: 2113
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
pbpX
Phospho-N-acetylmuramoyl-pentapeptide- transferase
Accession: VGM74225
Location: 354521-355501
NCBI BlastP on this gene
Accession: VGM74225
Location: 354521-355501
NCBI BlastP on this gene
mraY
SAMEA3175728_00378
Probable ATP-dependent Clp protease ATP-binding subunit
Accession: VGM74227
Location: 356110-358215
NCBI BlastP on this gene
Accession: VGM74227
Location: 356110-358215
NCBI BlastP on this gene
clpC_2
luxS
Uncharacterized protein conserved in bacteria
Accession: VGM74229
Location: 359087-360571
NCBI BlastP on this gene
Accession: VGM74229
Location: 359087-360571
NCBI BlastP on this gene
SAMEA3175728_00381
dexB
Putative transcriptional regulator ywtF
Accession: VGM74231
Location: 363075-364520
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 533
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: VGM74231
Location: 363075-364520
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 533
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ywtF
Tyrosine-protein phosphatase CpsB
Accession: VGM74232
Location: 364522-365253
BlastP hit with ABP89533.1
Percentage identity: 60 %
BlastP bit score: 303
Sequence coverage: 100 %
E-value: 4e-100
NCBI BlastP on this gene
Accession: VGM74232
Location: 364522-365253
BlastP hit with ABP89533.1
Percentage identity: 60 %
BlastP bit score: 303
Sequence coverage: 100 %
E-value: 4e-100
NCBI BlastP on this gene
cpsB
Capsular polysaccharide type 8 biosynthesis protein cap8A
Accession: VGM74233
Location: 365262-365954
NCBI BlastP on this gene
Accession: VGM74233
Location: 365262-365954
NCBI BlastP on this gene
cap8A
Tyrosine-protein kinase CpsD
Accession: VGM74234
Location: 365964-366653
BlastP hit with ABP89532.1
Percentage identity: 62 %
BlastP bit score: 295
Sequence coverage: 96 %
E-value: 3e-97
NCBI BlastP on this gene
Accession: VGM74234
Location: 365964-366653
BlastP hit with ABP89532.1
Percentage identity: 62 %
BlastP bit score: 295
Sequence coverage: 96 %
E-value: 3e-97
NCBI BlastP on this gene
cpsD
Putative colanic biosynthesis UDP-glucose lipid carrier transferase
Accession: VGM74235
Location: 366669-368036
BlastP hit with ABP89534.1
Percentage identity: 50 %
BlastP bit score: 395
Sequence coverage: 99 %
E-value: 3e-130
NCBI BlastP on this gene
Accession: VGM74235
Location: 366669-368036
BlastP hit with ABP89534.1
Percentage identity: 50 %
BlastP bit score: 395
Sequence coverage: 99 %
E-value: 3e-130
NCBI BlastP on this gene
wcaJ_1
Domain of uncharacterised function (DUF1972)
Accession: VGM74236
Location: 368067-369239
BlastP hit with ABP89535.1
Percentage identity: 71 %
BlastP bit score: 587
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: VGM74236
Location: 368067-369239
BlastP hit with ABP89535.1
Percentage identity: 71 %
BlastP bit score: 587
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAMEA3175728_00388
glycogen/starch synthase, ADP-glucose type
Accession: VGM74237
Location: 369236-370519
NCBI BlastP on this gene
Accession: VGM74237
Location: 369236-370519
NCBI BlastP on this gene
SAMEA3175728_00389
Putative acetyltransferase SA2342
Accession: VGM74238
Location: 370497-371186
NCBI BlastP on this gene
Accession: VGM74238
Location: 370497-371186
NCBI BlastP on this gene
SAMEA3175728_00390
ugd_2
D-inositol-3-phosphate glycosyltransferase
Accession: VGM74240
Location: 372473-373597
NCBI BlastP on this gene
Accession: VGM74240
Location: 372473-373597
NCBI BlastP on this gene
mshA_2
SAMEA3175728_00393
SAMEA3175728_00394
SAMEA3175728_00395
rfbX
Query: Streptococcus suis 05ZYH33, complete genome.
KM972249 : Streptococcus suis strain YS150_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2113
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE79733
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 884
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79733
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 884
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79734
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
Accession: AKE79734
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79735
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AKE79735
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 4e-149
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79736
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AKE79736
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79737
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79737
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
Query: Streptococcus suis 05ZYH33, complete genome.
KU983473 : Streptococcus suis strain YS494 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2112
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03387
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03387
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03388
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03388
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03389
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03389
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03390
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03390
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03391
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03391
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03394
Location: 6886-7746
NCBI BlastP on this gene
Accession: AOP03394
Location: 6886-7746
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KT163372 : Streptococcus suis strain YS210 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2112
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03779
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03779
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03780
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03780
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03781
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03781
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03782
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03782
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03783
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03783
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03786
Location: 6886-7746
NCBI BlastP on this gene
Accession: AOP03786
Location: 6886-7746
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
JF273655 : Streptococcus suis strain 89-2479 ORF23Z gene Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEH57586
AEH57587
AEH57588
Cps23A
Accession: AEH57589
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 855
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57589
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 855
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps23A
Cps23B
Accession: AEH57590
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 3e-148
NCBI BlastP on this gene
Accession: AEH57590
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 3e-148
NCBI BlastP on this gene
cps23B
Cps23C
Accession: AEH57591
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEH57591
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cps23C
Cps23D
Accession: AEH57592
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEH57592
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cps23D
cps23E
cps23F
cps23G
cps23H
cps23I
cps23J
cps23K
cps23L
cps23M
Query: Streptococcus suis 05ZYH33, complete genome.
JF273648 : Streptococcus suis strain 11538 ORF5Z gene Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEH57426
AEH57427
AEH57428
Cps5A
Accession: AEH57429
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57429
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps5A
Cps5B
Accession: AEH57430
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AEH57430
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cps5B
Cps5C
Accession: AEH57431
Location: 5093-5770
BlastP hit with ABP89532.1
Percentage identity: 90 %
BlastP bit score: 416
Sequence coverage: 97 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AEH57431
Location: 5093-5770
BlastP hit with ABP89532.1
Percentage identity: 90 %
BlastP bit score: 416
Sequence coverage: 97 %
E-value: 7e-145
NCBI BlastP on this gene
cps5C
Cps5D
Accession: AEH57432
Location: 5809-6540
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 9e-144
NCBI BlastP on this gene
Accession: AEH57432
Location: 5809-6540
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 9e-144
NCBI BlastP on this gene
cps5D
cps5E
cps5F
cps5G
cps5H
cps5I
cps5J
cps5K
cps5L
cps5M
cps5N
cps5O
Query: Streptococcus suis 05ZYH33, complete genome.
JF273646 : Streptococcus suis strain 4961 ORF3Z and ORF3Y genes Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEH57389
AEH57390
AEH57391
Cps3A
Accession: AEH57392
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57392
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps3A
Cps3B
Accession: AEH57393
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AEH57393
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cps3B
Cps3C
Accession: AEH57394
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEH57394
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cps3C
Cps3D
Accession: AEH57395
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEH57395
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cps3D
cps3E
cps3F
cps3G
cps3H
cps3I
cps3J
cps3K
cps3L
cps3M
cps3N
Query: Streptococcus suis 05ZYH33, complete genome.
CP041994 : Streptococcus suis strain INT-01 chromosome Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
FPT06_02825
FPT06_02820
FPT06_02815
glutamate-5-semialdehyde dehydrogenase
Accession: QDS23302
Location: 574772-576010
NCBI BlastP on this gene
Accession: QDS23302
Location: 574772-576010
NCBI BlastP on this gene
FPT06_02810
pyrroline-5-carboxylate reductase
Accession: QDS23301
Location: 573977-574762
NCBI BlastP on this gene
Accession: QDS23301
Location: 573977-574762
NCBI BlastP on this gene
proC
TetR/AcrR family transcriptional regulator
Accession: QDS23300
Location: 573550-573954
NCBI BlastP on this gene
Accession: QDS23300
Location: 573550-573954
NCBI BlastP on this gene
FPT06_02800
FPT06_02795
PLP-dependent aminotransferase family protein
Accession: QDS23298
Location: 571214-572473
NCBI BlastP on this gene
Accession: QDS23298
Location: 571214-572473
NCBI BlastP on this gene
FPT06_02790
yaaA
LytR family transcriptional regulator
Accession: QDS23296
Location: 568788-570227
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: QDS23296
Location: 568788-570227
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
FPT06_02780
capsular biosynthesis protein CpsC
Accession: QDS23295
Location: 568083-568772
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: QDS23295
Location: 568083-568772
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
FPT06_02775
tyrosine-protein kinase
Accession: QDS23294
Location: 567387-568073
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: QDS23294
Location: 567387-568073
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
FPT06_02770
tyrosine protein phosphatase
Accession: QDS23293
Location: 566617-567348
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: QDS23293
Location: 566617-567348
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
FPT06_02765
polysaccharide biosynthesis protein
Accession: QDS23292
Location: 564761-566587
NCBI BlastP on this gene
Accession: QDS23292
Location: 564761-566587
NCBI BlastP on this gene
FPT06_02760
FPT06_02755
aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme
Accession: QDS23290
Location: 562682-563932
NCBI BlastP on this gene
Accession: QDS23290
Location: 562682-563932
NCBI BlastP on this gene
FPT06_02750
FPT06_02745
FPT06_02740
FPT06_02735
FPT06_02730
O-antigen ligase family protein
Accession: QDS23285
Location: 557878-559101
NCBI BlastP on this gene
Accession: QDS23285
Location: 557878-559101
NCBI BlastP on this gene
FPT06_02725
FPT06_02720
FPT06_02715
Query: Streptococcus suis 05ZYH33, complete genome.
CP030124 : Streptococcus suis strain SH1510 chromosome Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
DP111_09535
DP111_09530
DP111_09525
gamma-glutamyl-phosphate reductase
Accession: AXI66227
Location: 1879316-1880554
NCBI BlastP on this gene
Accession: AXI66227
Location: 1879316-1880554
NCBI BlastP on this gene
DP111_09520
pyrroline-5-carboxylate reductase
Accession: AXI66226
Location: 1878521-1879306
NCBI BlastP on this gene
Accession: AXI66226
Location: 1878521-1879306
NCBI BlastP on this gene
proC
TetR/AcrR family transcriptional regulator
Accession: AXI66225
Location: 1878094-1878498
NCBI BlastP on this gene
Accession: AXI66225
Location: 1878094-1878498
NCBI BlastP on this gene
DP111_09510
DegV domain-containing protein
Accession: AXI66224
Location: 1877116-1877967
NCBI BlastP on this gene
Accession: AXI66224
Location: 1877116-1877967
NCBI BlastP on this gene
DP111_09505
PLP-dependent aminotransferase family protein
Accession: AXI66223
Location: 1875758-1877017
NCBI BlastP on this gene
Accession: AXI66223
Location: 1875758-1877017
NCBI BlastP on this gene
DP111_09500
DP111_09495
LytR family transcriptional regulator
Accession: AXI66221
Location: 1873332-1874771
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AXI66221
Location: 1873332-1874771
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
DP111_09490
capsular biosynthesis protein CpsC
Accession: AXI66220
Location: 1872627-1873316
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AXI66220
Location: 1872627-1873316
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
DP111_09485
tyrosine protein kinase
Accession: AXI66219
Location: 1871931-1872617
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AXI66219
Location: 1871931-1872617
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
DP111_09480
tyrosine protein phosphatase
Accession: AXI66218
Location: 1871161-1871892
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AXI66218
Location: 1871161-1871892
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
DP111_09475
polysaccharide biosynthesis protein
Accession: AXI66217
Location: 1869305-1871131
NCBI BlastP on this gene
Accession: AXI66217
Location: 1869305-1871131
NCBI BlastP on this gene
DP111_09470
DP111_09465
DegT/DnrJ/EryC1/StrS family aminotransferase
Accession: AXI66215
Location: 1866932-1868152
NCBI BlastP on this gene
Accession: AXI66215
Location: 1866932-1868152
NCBI BlastP on this gene
DP111_09460
DP111_09455
DP111_09450
GNAT family N-acetyltransferase
Accession: AXI66212
Location: 1864544-1865134
NCBI BlastP on this gene
Accession: AXI66212
Location: 1864544-1865134
NCBI BlastP on this gene
DP111_09445
DP111_09440
glycosyltransferase family 2 protein
Accession: AXI66210
Location: 1862491-1863396
NCBI BlastP on this gene
Accession: AXI66210
Location: 1862491-1863396
NCBI BlastP on this gene
DP111_09435
DP111_09430
Query: Streptococcus suis 05ZYH33, complete genome.
CP017142 : Streptococcus suis strain GZ0565 chromosome Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
BFP66_06400
BFP66_06395
BFP66_06390
glutamate-5-semialdehyde dehydrogenase
Accession: AOM74869
Location: 1292557-1293795
NCBI BlastP on this gene
Accession: AOM74869
Location: 1292557-1293795
NCBI BlastP on this gene
BFP66_06385
pyrroline-5-carboxylate reductase
Accession: AOM74868
Location: 1291762-1292547
NCBI BlastP on this gene
Accession: AOM74868
Location: 1291762-1292547
NCBI BlastP on this gene
BFP66_06380
TetR family transcriptional regulator
Accession: AOM74867
Location: 1291335-1291739
NCBI BlastP on this gene
Accession: AOM74867
Location: 1291335-1291739
NCBI BlastP on this gene
BFP66_06375
fatty acid-binding protein DegV
Accession: AOM74866
Location: 1290357-1291208
NCBI BlastP on this gene
Accession: AOM74866
Location: 1290357-1291208
NCBI BlastP on this gene
BFP66_06370
GntR family transcriptional regulator
Accession: AOM74865
Location: 1288999-1290258
NCBI BlastP on this gene
Accession: AOM74865
Location: 1288999-1290258
NCBI BlastP on this gene
BFP66_06365
BFP66_06360
LytR family transcriptional regulator
Accession: AOM74863
Location: 1286573-1288012
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOM74863
Location: 1286573-1288012
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
BFP66_06355
capsular biosynthesis protein CpsC
Accession: AOM74862
Location: 1285868-1286557
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AOM74862
Location: 1285868-1286557
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
BFP66_06350
tyrosine protein kinase
Accession: AOM74861
Location: 1285172-1285858
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AOM74861
Location: 1285172-1285858
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
BFP66_06345
tyrosine protein phosphatase
Accession: AOM74860
Location: 1284402-1285133
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOM74860
Location: 1284402-1285133
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
BFP66_06340
BFP66_06335
capsular biosynthesis protein
Accession: AOM74858
Location: 1281780-1282382
NCBI BlastP on this gene
Accession: AOM74858
Location: 1281780-1282382
NCBI BlastP on this gene
BFP66_06330
BFP66_06325
BFP66_06320
BFP66_06315
BFP66_06310
BFP66_06305
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: AOM74853
Location: 1276288-1277007
NCBI BlastP on this gene
Accession: AOM74853
Location: 1276288-1277007
NCBI BlastP on this gene
BFP66_06300
BFP66_06295
BFP66_06290
Query: Streptococcus suis 05ZYH33, complete genome.
CP015557 : Streptococcus suis strain DN13 Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
A6M16_06505
A6M16_06500
A6M16_06495
gamma-glutamyl-phosphate reductase
Accession: AND00153
Location: 1315938-1317176
NCBI BlastP on this gene
Accession: AND00153
Location: 1315938-1317176
NCBI BlastP on this gene
A6M16_06490
pyrroline-5-carboxylate reductase
Accession: AND00152
Location: 1315143-1315928
NCBI BlastP on this gene
Accession: AND00152
Location: 1315143-1315928
NCBI BlastP on this gene
A6M16_06485
TetR family transcriptional regulator
Accession: AND00151
Location: 1314716-1315120
NCBI BlastP on this gene
Accession: AND00151
Location: 1314716-1315120
NCBI BlastP on this gene
A6M16_06480
fatty acid-binding protein DegV
Accession: AND00150
Location: 1313738-1314589
NCBI BlastP on this gene
Accession: AND00150
Location: 1313738-1314589
NCBI BlastP on this gene
A6M16_06475
GntR family transcriptional regulator
Accession: AND00149
Location: 1312380-1313639
NCBI BlastP on this gene
Accession: AND00149
Location: 1312380-1313639
NCBI BlastP on this gene
A6M16_06470
A6M16_06465
LytR family transcriptional regulator
Accession: AND00147
Location: 1309954-1311393
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AND00147
Location: 1309954-1311393
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
A6M16_06460
capsular biosynthesis protein CpsC
Accession: AND00146
Location: 1309249-1309938
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AND00146
Location: 1309249-1309938
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
A6M16_06455
tyrosine protein kinase
Accession: AND00145
Location: 1308553-1309239
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AND00145
Location: 1308553-1309239
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
A6M16_06450
tyrosine protein phosphatase
Accession: AND00144
Location: 1307783-1308514
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AND00144
Location: 1307783-1308514
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
A6M16_06445
A6M16_06440
capsular biosynthesis protein
Accession: AND00142
Location: 1305161-1305763
NCBI BlastP on this gene
Accession: AND00142
Location: 1305161-1305763
NCBI BlastP on this gene
A6M16_06435
A6M16_06430
A6M16_06425
A6M16_06420
A6M16_06415
A6M16_06410
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: AND00137
Location: 1299669-1300388
NCBI BlastP on this gene
Accession: AND00137
Location: 1299669-1300388
NCBI BlastP on this gene
A6M16_06405
A6M16_06400
A6M16_06395
Query: Streptococcus suis 05ZYH33, complete genome.
CP002633 : Streptococcus suis ST3 Total score: 5.5 Cumulative Blast bit score: 2109
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Zn-dependent hydrolase, including glyoxylases
Accession: AEB81183
Location: 732324-732962
NCBI BlastP on this gene
Accession: AEB81183
Location: 732324-732962
NCBI BlastP on this gene
SSUST3_0727
SSUST3_0728
SSUST3_0729
gamma-glutamyl phosphate reductase
Accession: AEB81186
Location: 736841-738079
NCBI BlastP on this gene
Accession: AEB81186
Location: 736841-738079
NCBI BlastP on this gene
proA
pyrroline-5-carboxylate reductase
Accession: AEB81187
Location: 738089-738874
NCBI BlastP on this gene
Accession: AEB81187
Location: 738089-738874
NCBI BlastP on this gene
proC
SSUST3_0732
SSUST3_0733
SSUST3_0734
SSUST3_0735
integral membrane regulatory protein Wzg
Accession: AEB81192
Location: 742624-744063
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEB81192
Location: 742624-744063
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
wzg
Cps2B
Accession: AEB81193
Location: 744079-744768
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AEB81193
Location: 744079-744768
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
SSUST3_0737
tyrosine-protein kinase Wze
Accession: AEB81194
Location: 744778-745464
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEB81194
Location: 744778-745464
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
wze
protein-tyrosine phosphatase Wzh
Accession: AEB81195
Location: 745503-746234
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEB81195
Location: 745503-746234
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
wzh
SSUST3_0740
SSUST3_0741
SSUST3_0742
UDP-galactose phosphate transferase
Accession: AEB81199
Location: 750182-750799
NCBI BlastP on this gene
Accession: AEB81199
Location: 750182-750799
NCBI BlastP on this gene
SSUST3_0743
SSUST3_0744
SSUST3_0745
capsular polysaccharide synthesis protein
Accession: AEB81202
Location: 752575-753753
NCBI BlastP on this gene
Accession: AEB81202
Location: 752575-753753
NCBI BlastP on this gene
SSUST3_0746
SSUST3_0747
SSUST3_0748
Query: Streptococcus suis 05ZYH33, complete genome.
KX870051 : Streptococcus suis strain 78468-5 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2107
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ78944
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78944
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ78945
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: APZ78945
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ78946
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: APZ78946
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ78947
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: APZ78947
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ78948
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ78948
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
Nucleoside-diphosphate sugar epimerase
Accession: APZ78955
Location: 10791-11630
NCBI BlastP on this gene
Accession: APZ78955
Location: 10791-11630
NCBI BlastP on this gene
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KX870050 : Streptococcus suis strain 78468-4 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2107
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ78922
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78922
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ78923
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: APZ78923
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ78924
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: APZ78924
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ78925
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: APZ78925
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ78926
Location: 3641-5467
NCBI BlastP on this gene
Accession: APZ78926
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
Nucleoside-diphosphate sugar epimerase
Accession: APZ78933
Location: 10791-11630
NCBI BlastP on this gene
Accession: APZ78933
Location: 10791-11630
NCBI BlastP on this gene
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KX870049 : Streptococcus suis strain 78468-1 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2107
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: APZ78900
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78900
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ78901
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: APZ78901
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ78902
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: APZ78902
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ78903
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: APZ78903
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ78904
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ78904
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
Nucleoside-diphosphate sugar epimerase
Accession: APZ78911
Location: 10791-11630
NCBI BlastP on this gene
Accession: APZ78911
Location: 10791-11630
NCBI BlastP on this gene
cpsL
cpsM
cpsN
cpsO
Query: Streptococcus suis 05ZYH33, complete genome.
KM576773 : Streptococcus suis strain 43640 Cps31A (cps31A), Cps31B (cps31B), Cps31C (cps31C), Cps3... Total score: 5.5 Cumulative Blast bit score: 2107
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Cps31A
Accession: AIZ73034
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 871
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIZ73034
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 871
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps31A
Cps31B
Accession: AIZ73035
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 82 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
Accession: AIZ73035
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 82 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
cps31B
Cps31C
Accession: AIZ73036
Location: 2155-2838
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 423
Sequence coverage: 95 %
E-value: 1e-147
NCBI BlastP on this gene
Accession: AIZ73036
Location: 2155-2838
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 423
Sequence coverage: 95 %
E-value: 1e-147
NCBI BlastP on this gene
cps31C
Cps31D
Accession: AIZ73037
Location: 2879-3610
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 420
Sequence coverage: 100 %
E-value: 5e-146
NCBI BlastP on this gene
Accession: AIZ73037
Location: 2879-3610
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 420
Sequence coverage: 100 %
E-value: 5e-146
NCBI BlastP on this gene
cps31D
cps31E
cps31F
cps31G
cps31H
cps31I
cps31J
cps31K
cps31L
cps31M
Query: Streptococcus suis 05ZYH33, complete genome.
JF273647 : Streptococcus suis strain 6407 ORF4Z gene Total score: 5.5 Cumulative Blast bit score: 2107
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEH57407
AEH57408
AEH57409
Cps4A
Accession: AEH57410
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57410
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps4A
Cps4B
Accession: AEH57411
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
Accession: AEH57411
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
cps4B
Cps4C
Accession: AEH57412
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEH57412
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cps4C
Cps4D
Accession: AEH57413
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
Accession: AEH57413
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
cps4D
cps4E
cps4F
cps4G
cps4H
cps4I
cps4J
cps4K
cps4L
cps4M
Query: Streptococcus suis 05ZYH33, complete genome.
CP008921 : Streptococcus suis 6407 Total score: 5.5 Cumulative Blast bit score: 2107
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
ID09_07160
ID09_07155
ID09_07145
gamma-glutamyl phosphate reductase
Accession: AIG43812
Location: 1433256-1434494
NCBI BlastP on this gene
Accession: AIG43812
Location: 1433256-1434494
NCBI BlastP on this gene
ID09_07140
pyrroline-5-carboxylate reductase
Accession: AIG43811
Location: 1432461-1433246
NCBI BlastP on this gene
Accession: AIG43811
Location: 1432461-1433246
NCBI BlastP on this gene
ID09_07135
TetR family transcriptional regulator
Accession: AIG43810
Location: 1432034-1432438
NCBI BlastP on this gene
Accession: AIG43810
Location: 1432034-1432438
NCBI BlastP on this gene
ID09_07130
ID09_07125
GntR family transcriptional regulator
Accession: AIG43808
Location: 1429698-1430957
NCBI BlastP on this gene
Accession: AIG43808
Location: 1429698-1430957
NCBI BlastP on this gene
ID09_07120
ID09_07115
LytR family transcriptional regulator
Accession: AIG43806
Location: 1427272-1428711
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIG43806
Location: 1427272-1428711
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
ID09_07110
capsular biosynthesis protein CpsC
Accession: AIG43805
Location: 1426567-1427256
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
Accession: AIG43805
Location: 1426567-1427256
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
ID09_07105
tyrosine protein kinase
Accession: AIG43804
Location: 1425871-1426557
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AIG43804
Location: 1425871-1426557
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
ID09_07100
tyrosine protein phosphatase
Accession: AIG43803
Location: 1425101-1425832
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
Accession: AIG43803
Location: 1425101-1425832
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
ID09_07095
ID09_07090
ID09_07085
capsular biosynthesis protein
Accession: AIG43800
Location: 1420872-1422092
NCBI BlastP on this gene
Accession: AIG43800
Location: 1420872-1422092
NCBI BlastP on this gene
ID09_07080
amylovoran biosynthesis protein AmsE
Accession: AIG43799
Location: 1420049-1420879
NCBI BlastP on this gene
Accession: AIG43799
Location: 1420049-1420879
NCBI BlastP on this gene
ID09_07075
ID09_07070
ID09_07065
ID09_07060
ID09_07055
ID09_07050
Query: Streptococcus suis 05ZYH33, complete genome.
KY574603 : Streptococcus suis strain 1388970_seq cps gene cluster Total score: 5.5 Cumulative Blast bit score: 2099
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: ARO73588
Location: 1-1338
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 833
Sequence coverage: 91 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ARO73588
Location: 1-1338
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 833
Sequence coverage: 91 %
E-value: 0.0
NCBI BlastP on this gene
cps9A
chain length determinant protein Wzd
Accession: ARO73589
Location: 1354-2043
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 7e-148
NCBI BlastP on this gene
Accession: ARO73589
Location: 1354-2043
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 7e-148
NCBI BlastP on this gene
cps9B
tyrosine-protein kinase Wze
Accession: ARO73590
Location: 2053-2739
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 426
Sequence coverage: 96 %
E-value: 1e-148
NCBI BlastP on this gene
Accession: ARO73590
Location: 2053-2739
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 426
Sequence coverage: 96 %
E-value: 1e-148
NCBI BlastP on this gene
cps9C
protein-tyrosine phosphatase Wzh
Accession: ARO73591
Location: 2778-3509
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: ARO73591
Location: 2778-3509
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
cps9D
nucleoside-diphosphate sugar epimerase
Accession: ARO73592
Location: 3539-5365
NCBI BlastP on this gene
Accession: ARO73592
Location: 3539-5365
NCBI BlastP on this gene
cps9E
cps9F
cps9G
cps9H
cps9I
cps9J
capsular polysaccharide synthesis protein
Accession: ARO73598
Location: 9925-10884
NCBI BlastP on this gene
Accession: ARO73598
Location: 9925-10884
NCBI BlastP on this gene
cps9K
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: ARO73599
Location: 10903-11622
NCBI BlastP on this gene
Accession: ARO73599
Location: 10903-11622
NCBI BlastP on this gene
cps9L
NAD-dependent epimerase/dehydratase
Accession: ARO73600
Location: 11631-12686
NCBI BlastP on this gene
Accession: ARO73600
Location: 11631-12686
NCBI BlastP on this gene
cps9M
cps9N
Query: Streptococcus suis 05ZYH33, complete genome.
CR931684 : Streptococcus pneumoniae strain 1039/41 (serotype 23b). Total score: 5.5 Cumulative Blast bit score: 2097
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
dexB
integral membrane regulatory protein Wzg
Accession: CAI33834
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 543
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33834
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 543
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wzg
wzh
capsular polysaccharide biosynthesis protein Wzd
Accession: CAI33836
Location: 4774-5466
NCBI BlastP on this gene
Accession: CAI33836
Location: 4774-5466
NCBI BlastP on this gene
wzd
wze
undecaprenylphosphate glucosephosphotransferase WchA (initial sugar transferase)
Accession: CAI33838
Location: 6180-7541
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
Accession: CAI33838
Location: 6180-7541
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
wchA
putative rhamnosyl transferase WchF
Accession: CAI33839
Location: 7528-8751
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33839
Location: 7528-8751
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wchF
oligosaccharide repeat unit polymerase Wzy
Accession: CAI33840
Location: 9052-10239
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
Accession: CAI33840
Location: 9052-10239
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
wzy
putative glycosyl transferase
Accession: CAI33841
Location: 10249-11217
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 178
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
Accession: CAI33841
Location: 10249-11217
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 178
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
wchV
wchW
wzx
putative glycerol phosphotransferase
Accession: CAI33844
Location: 13840-14994
NCBI BlastP on this gene
Accession: CAI33844
Location: 13840-14994
NCBI BlastP on this gene
wchX
putative glycerol-2-phosphate dehydrogenase
Accession: CAI33845
Location: 15030-16058
NCBI BlastP on this gene
Accession: CAI33845
Location: 15030-16058
NCBI BlastP on this gene
gtp1
gtp2
gtp3
glucose-1-phosphate thymidylyltransferase RmlA
Accession: CAI33848
Location: 17786-18655
NCBI BlastP on this gene
Accession: CAI33848
Location: 17786-18655
NCBI BlastP on this gene
rmlA
Query: Streptococcus suis 05ZYH33, complete genome.
CR931674 : Streptococcus pneumoniae strain Gethens (serotype 18f). Total score: 5.5 Cumulative Blast bit score: 2094
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
dexB
integral membrane regulatory protein Wzg
Accession: CAI33599
Location: 1560-3005
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 536
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33599
Location: 1560-3005
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 536
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wzg
protein-tyrosine phosphatase Wzh
Accession: CAI33600
Location: 3007-3738
BlastP hit with ABP89533.1
Percentage identity: 59 %
BlastP bit score: 297
Sequence coverage: 100 %
E-value: 1e-97
NCBI BlastP on this gene
Accession: CAI33600
Location: 3007-3738
BlastP hit with ABP89533.1
Percentage identity: 59 %
BlastP bit score: 297
Sequence coverage: 100 %
E-value: 1e-97
NCBI BlastP on this gene
wzh
capsular polysaccharide biosynthesis protein Wzd
Accession: CAI33601
Location: 3747-4439
NCBI BlastP on this gene
Accession: CAI33601
Location: 3747-4439
NCBI BlastP on this gene
wzd
tyrosine-protein kinase Wze
Accession: CAI33602
Location: 4449-5132
BlastP hit with ABP89532.1
Percentage identity: 61 %
BlastP bit score: 294
Sequence coverage: 96 %
E-value: 6e-97
NCBI BlastP on this gene
Accession: CAI33602
Location: 4449-5132
BlastP hit with ABP89532.1
Percentage identity: 61 %
BlastP bit score: 294
Sequence coverage: 96 %
E-value: 6e-97
NCBI BlastP on this gene
wze
undecaprenylphosphate glucosephosphotransferase WchA (initial sugar transferase)
Accession: CAI33603
Location: 5148-6515
BlastP hit with ABP89534.1
Percentage identity: 48 %
BlastP bit score: 385
Sequence coverage: 98 %
E-value: 2e-126
NCBI BlastP on this gene
Accession: CAI33603
Location: 5148-6515
BlastP hit with ABP89534.1
Percentage identity: 48 %
BlastP bit score: 385
Sequence coverage: 98 %
E-value: 2e-126
NCBI BlastP on this gene
wchA
putative rhamnosyl transferase WchF
Accession: CAI33604
Location: 6546-7718
BlastP hit with ABP89535.1
Percentage identity: 69 %
BlastP bit score: 582
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33604
Location: 6546-7718
BlastP hit with ABP89535.1
Percentage identity: 69 %
BlastP bit score: 582
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wchF
wciU
wcxM
wciV
wciW
wzx
oligosaccharide repeat unit polymerase Wzy
Accession: CAI33610
Location: 13129-14403
NCBI BlastP on this gene
Accession: CAI33610
Location: 13129-14403
NCBI BlastP on this gene
wzy
wciX
putative glycerol phosphotransferase
Accession: CAI33612
Location: 15455-16615
NCBI BlastP on this gene
Accession: CAI33612
Location: 15455-16615
NCBI BlastP on this gene
wciY
Query: Streptococcus suis 05ZYH33, complete genome.
LT594598 : Streptococcus pneumoniae capsular locus (cps) operon, strain 23B1. Total score: 5.5 Cumulative Blast bit score: 2091
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
tnp
tnp
tnp
integral membrane regulatory protein Wzg
Accession: SBT85355
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 538
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: SBT85355
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 538
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wzg
wzh
capsular polysaccharide biosynthesis protein Wzd
Accession: SBT85357
Location: 4774-5466
NCBI BlastP on this gene
Accession: SBT85357
Location: 4774-5466
NCBI BlastP on this gene
wzd
wze
galactosyl trasnferase WchA
Accession: SBT85359
Location: 6171-7532
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
Accession: SBT85359
Location: 6171-7532
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
wchA
putative rhamnosyl transferase WchF
Accession: SBT85360
Location: 7519-8742
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: SBT85360
Location: 7519-8742
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wchF
oligosaccharide repeat unit polymerase Wzy
Accession: SBT85361
Location: 9043-10230
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
Accession: SBT85361
Location: 9043-10230
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
wzy
glycosyl transferase wchV
Accession: SBT85362
Location: 10240-11208
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 177
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
Accession: SBT85362
Location: 10240-11208
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 177
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
wchV
wchW
capsule biosynthesis repeating unit flippase Wzx
Accession: SBT85364
Location: 12446-13834
NCBI BlastP on this gene
Accession: SBT85364
Location: 12446-13834
NCBI BlastP on this gene
wzx
wchX
glycerol-2-phosphate dehydrogenase Gtp1
Accession: SBT85366
Location: 15021-16049
NCBI BlastP on this gene
Accession: SBT85366
Location: 15021-16049
NCBI BlastP on this gene
gtp1
gtp2
gtp3
glucose-1-phosphate thymidylyltransferase RmlA
Accession: SBT85369
Location: 17758-18627
NCBI BlastP on this gene
Accession: SBT85369
Location: 17758-18627
NCBI BlastP on this gene
rmlA
Query: Streptococcus suis 05ZYH33, complete genome.
JF791167 : Streptococcus suis strain 88-1861 ORFY gene Total score: 5.5 Cumulative Blast bit score: 2085
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEP02726
CpsA
Accession: AEP02727
Location: 3092-4531
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 861
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02727
Location: 3092-4531
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 861
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02727
CpsB
Accession: AEP02728
Location: 4548-5237
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 408
Sequence coverage: 100 %
E-value: 1e-141
NCBI BlastP on this gene
Accession: AEP02728
Location: 4548-5237
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 408
Sequence coverage: 100 %
E-value: 1e-141
NCBI BlastP on this gene
AEP02728
CpsC
Accession: AEP02729
Location: 5247-5933
BlastP hit with ABP89532.1
Percentage identity: 86 %
BlastP bit score: 397
Sequence coverage: 96 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AEP02729
Location: 5247-5933
BlastP hit with ABP89532.1
Percentage identity: 86 %
BlastP bit score: 397
Sequence coverage: 96 %
E-value: 2e-137
NCBI BlastP on this gene
AEP02729
CpsD
Accession: AEP02730
Location: 5971-6702
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 2e-145
NCBI BlastP on this gene
Accession: AEP02730
Location: 5971-6702
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 2e-145
NCBI BlastP on this gene
AEP02730
Query: Streptococcus suis 05ZYH33, complete genome.
CP017092 : Streptococcus suis strain ISU2812 chromosome Total score: 5.5 Cumulative Blast bit score: 2085
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
glutamate-5-semialdehyde dehydrogenase
Accession: ASW52408
Location: 2078301-2079539
NCBI BlastP on this gene
Accession: ASW52408
Location: 2078301-2079539
NCBI BlastP on this gene
A7J09_09945
pyrroline-5-carboxylate reductase
Accession: ASW52407
Location: 2077506-2078291
NCBI BlastP on this gene
Accession: ASW52407
Location: 2077506-2078291
NCBI BlastP on this gene
A7J09_09940
A7J09_09935
A7J09_09930
A7J09_09925
TetR family transcriptional regulator
Accession: ASW52403
Location: 2074144-2074548
NCBI BlastP on this gene
Accession: ASW52403
Location: 2074144-2074548
NCBI BlastP on this gene
A7J09_09920
fatty acid-binding protein DegV
Accession: ASW52402
Location: 2073166-2074017
NCBI BlastP on this gene
Accession: ASW52402
Location: 2073166-2074017
NCBI BlastP on this gene
A7J09_09915
A7J09_09910
A7J09_09905
GntR family transcriptional regulator
Accession: ASW52399
Location: 2070195-2071454
NCBI BlastP on this gene
Accession: ASW52399
Location: 2070195-2071454
NCBI BlastP on this gene
A7J09_09900
A7J09_09895
LytR family transcriptional regulator
Accession: ASW52397
Location: 2067773-2069212
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ASW52397
Location: 2067773-2069212
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
A7J09_09890
capsular biosynthesis protein CpsC
Accession: ASW52396
Location: 2067067-2067756
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: ASW52396
Location: 2067067-2067756
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
A7J09_09885
tyrosine protein kinase
Accession: ASW52395
Location: 2066380-2067057
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 409
Sequence coverage: 97 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: ASW52395
Location: 2066380-2067057
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 409
Sequence coverage: 97 %
E-value: 3e-142
NCBI BlastP on this gene
A7J09_09880
tyrosine protein phosphatase
Accession: ASW52394
Location: 2065611-2066342
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: ASW52394
Location: 2065611-2066342
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
A7J09_09875
A7J09_09870
cell filamentation protein Fic
Accession: ASW52392
Location: 2062955-2063671
NCBI BlastP on this gene
Accession: ASW52392
Location: 2062955-2063671
NCBI BlastP on this gene
A7J09_09865
A7J09_09860
capsular biosynthesis protein
Accession: ASW52390
Location: 2060992-2062221
NCBI BlastP on this gene
Accession: ASW52390
Location: 2060992-2062221
NCBI BlastP on this gene
A7J09_09855
A7J09_09850
A7J09_09845
A7J09_09840
A7J09_09835
A7J09_09830
A7J09_09825
Query: Streptococcus suis 05ZYH33, complete genome.
KU665260 : Streptococcus suis strain YS205 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2080
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP02591
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02591
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02592
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
Accession: AOP02592
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02593
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02593
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02594
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02594
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02595
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02595
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
KT163369 : Streptococcus suis strain YS196 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2080
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
Integral membrane regulatory protein Wzg
Accession: AOP03684
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03684
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03685
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
Accession: AOP03685
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03686
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03686
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03687
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03687
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03688
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03688
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
Query: Streptococcus suis 05ZYH33, complete genome.
JF791163 : Streptococcus suis strain 92-1191 ORFY, ORFX, CpsA, CpsB, CpsC, and CpsD genes Total score: 5.5 Cumulative Blast bit score: 2080
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEP02702
AEP02703
CpsA
Accession: AEP02704
Location: 2914-4353
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 870
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02704
Location: 2914-4353
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 870
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02704
CpsB
Accession: AEP02705
Location: 4370-5059
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AEP02705
Location: 4370-5059
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AEP02705
CpsC
Accession: AEP02706
Location: 5069-5755
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: AEP02706
Location: 5069-5755
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
AEP02706
CpsD
Accession: AEP02707
Location: 5793-6524
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
Accession: AEP02707
Location: 5793-6524
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
AEP02707
Query: Streptococcus suis 05ZYH33, complete genome.
KC537385 : Streptococcus suis strain 14A capsular polysaccharide synthesis locus sequence. Total score: 5.5 Cumulative Blast bit score: 2078
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AGS58356
AGS58357
AGS58358
Cps21A
Accession: AGS58359
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGS58359
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AGS58359
Cps21B
Accession: AGS58360
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AGS58360
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AGS58360
Cps21C
Accession: AGS58361
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: AGS58361
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
AGS58361
Cps21D
Accession: AGS58362
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
Accession: AGS58362
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
AGS58362
AGS58363
AGS58364
AGS58365
AGS58366
AGS58367
AGS58368
AGS58369
AGS58370
AGS58371
AGS58372
Query: Streptococcus suis 05ZYH33, complete genome.
AB737827 : Streptococcus suis DNA, capsular polysaccharide locus, strain: 14A. Total score: 5.5 Cumulative Blast bit score: 2078
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
BAM94809
BAM94810
predicted transcriptional regulator of pyridoxine metabolism
Accession: BAM94811
Location: 1670-2929
NCBI BlastP on this gene
Accession: BAM94811
Location: 1670-2929
NCBI BlastP on this gene
BAM94811
BAM94812
capsular polysaccharide expression regulator
Accession: BAM94813
Location: 3912-5351
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BAM94813
Location: 3912-5351
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps21A
chain length determinant protein/polysaccharide export protein
Accession: BAM94814
Location: 5368-6057
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: BAM94814
Location: 5368-6057
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cps21B
tyrosine-protein kinase
Accession: BAM94815
Location: 6067-6753
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: BAM94815
Location: 6067-6753
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
cps21C
protein-tyrosine phosphatase
Accession: BAM94816
Location: 6791-7522
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
Accession: BAM94816
Location: 6791-7522
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
cps21D
predicted nucleoside-diphosphate sugar epimerase
Accession: BAM94817
Location: 7551-9374
NCBI BlastP on this gene
Accession: BAM94817
Location: 7551-9374
NCBI BlastP on this gene
cps21E
cps21F
cps21G
NAD dependent epimerase/dehydratase family protein
Accession: BAM94820
Location: 10787-11641
NCBI BlastP on this gene
Accession: BAM94820
Location: 10787-11641
NCBI BlastP on this gene
cps21H
cps21I
cps21J
cps21K
cps21L
cps21M
cps21N
Query: Streptococcus suis 05ZYH33, complete genome.
KC537386 : Streptococcus suis strain 92-1191 capsular polysaccharide synthesis locus sequence. Total score: 5.5 Cumulative Blast bit score: 2076
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AGS58378
AGS58379
AGS58380
Cps29A
Accession: AGS58381
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGS58381
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AGS58381
Cps29B
Accession: AGS58382
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AGS58382
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AGS58382
Cps29C
Accession: AGS58383
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: AGS58383
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
AGS58383
Cps29D
Accession: AGS58384
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
Accession: AGS58384
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
AGS58384
AGS58385
AGS58386
AGS58387
AGS58388
AGS58389
AGS58390
AGS58391
AGS58392
AGS58393
Query: Streptococcus suis 05ZYH33, complete genome.
JF791155 : Streptococcus suis strain 93 ORFY, ORFX, CpsA, CpsB, CpsC, and CpsD genes Total score: 5.5 Cumulative Blast bit score: 2076
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEP02663
AEP02664
CpsA
Accession: AEP02665
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02665
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02665
CpsB
Accession: AEP02666
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AEP02666
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AEP02666
CpsC
Accession: AEP02667
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
Accession: AEP02667
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
AEP02667
CpsD
Accession: AEP02668
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AEP02668
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
AEP02668
Query: Streptococcus suis 05ZYH33, complete genome.
JF791154 : Streptococcus suis strain NT77 ORFY, ORFX, CpsA, CpsB, CpsC, and CpsD genes Total score: 5.5 Cumulative Blast bit score: 2076
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEP02657
AEP02658
CpsA
Accession: AEP02659
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02659
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02659
CpsB
Accession: AEP02660
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AEP02660
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AEP02660
CpsC
Accession: AEP02661
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
Accession: AEP02661
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
AEP02661
CpsD
Accession: AEP02662
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AEP02662
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
AEP02662
Query: Streptococcus suis 05ZYH33, complete genome.
JF273654 : Streptococcus suis strain 42A ORF19Z gene Total score: 5.5 Cumulative Blast bit score: 2076
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
SSU05_0577
SSU05_0578
AEH57559
AEH57560
AEH57561
Cps19A
Accession: AEH57562
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57562
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps19A
Cps19B
Accession: AEH57563
Location: 4522-5238
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 396
Sequence coverage: 100 %
E-value: 7e-137
NCBI BlastP on this gene
Accession: AEH57563
Location: 4522-5238
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 396
Sequence coverage: 100 %
E-value: 7e-137
NCBI BlastP on this gene
cps19B
Cps19C
Accession: AEH57564
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
Accession: AEH57564
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
cps19C
Cps19D
Accession: AEH57565
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AEH57565
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
cps19D
cps19E
cps19F
cps19G
cps19H
cps19I
cps19J
cps19K
cps19L
cps19M
cps19N
Query: Streptococcus suis 05ZYH33, complete genome.
301. : KM972262 Streptococcus suis strain YS27_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2120
SSU05_0564
SSU05_0565
SSU05_0566
SSU05_0567
SSU05_0568
SSU05_0569
SSU05_0570
SSU05_0571
SSU05_0572
SSU05_0573
SSU05_0574
SSU05_0575
SSU05_0576
Membrane protein involved in the export of
Accession: ABP89543.1
Location: 15369-16775
NCBI BlastP on this gene
Accession: ABP89543.1
Location: 15369-16775
NCBI BlastP on this gene
SSU05_0577
SSU05_0578
integral membrane regulatory protein Wzg
Accession: AKE80009
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80009
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 893
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80010
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 384
Sequence coverage: 100 %
E-value: 4e-132
NCBI BlastP on this gene
Accession: AKE80010
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 384
Sequence coverage: 100 %
E-value: 4e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80011
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AKE80011
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80012
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: AKE80012
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80013
Location: 3709-5469
NCBI BlastP on this gene
Accession: AKE80013
Location: 3709-5469
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
302. : KT163371 Streptococcus suis strain YS209 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2119
Integral membrane regulatory protein Wzg
Accession: AOP03744
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 875
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03744
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 875
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03745
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AOP03745
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03746
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP03746
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03747
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
Accession: AOP03747
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 418
Sequence coverage: 100 %
E-value: 3e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03748
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03748
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03754
Location: 10566-11684
NCBI BlastP on this gene
Accession: AOP03754
Location: 10566-11684
NCBI BlastP on this gene
cpsK
cpsL
cpsM
303. : KT163366 Streptococcus suis strain YS189 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2119
Integral membrane regulatory protein Wzg
Accession: AOP03598
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 880
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03598
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 880
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03599
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
Accession: AOP03599
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 1e-135
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03600
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03600
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03601
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03601
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03602
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03602
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03608
Location: 10567-11685
NCBI BlastP on this gene
Accession: AOP03608
Location: 10567-11685
NCBI BlastP on this gene
cpsK
cpsL
cpsM
304. : KM972294 Streptococcus suis strain YS94_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2119
integral membrane regulatory protein Wzg
Accession: AKE80700
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 887
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE80700
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 887
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE80701
Location: 1458-2147
BlastP hit with ABP89531.1
Percentage identity: 85 %
BlastP bit score: 380
Sequence coverage: 100 %
E-value: 6e-131
NCBI BlastP on this gene
Accession: AKE80701
Location: 1458-2147
BlastP hit with ABP89531.1
Percentage identity: 85 %
BlastP bit score: 380
Sequence coverage: 100 %
E-value: 6e-131
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE80702
Location: 2157-2843
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AKE80702
Location: 2157-2843
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE80703
Location: 2882-3613
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 1e-146
NCBI BlastP on this gene
Accession: AKE80703
Location: 2882-3613
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 421
Sequence coverage: 100 %
E-value: 1e-146
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE80704
Location: 3642-5468
NCBI BlastP on this gene
Accession: AKE80704
Location: 3642-5468
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
305. : KJ669337 Streptococcus suis strain CZ130302 hypothetical protein (ChZ00x1), putative transcripti... Total score: 5.5 Cumulative Blast bit score: 2118
ChZ00x1
putative transcriptional regulator of pyridoxine metabolism
Accession: AJA01362
Location: 532-1383
NCBI BlastP on this gene
Accession: AJA01362
Location: 532-1383
NCBI BlastP on this gene
ChZ00x2
putative transcriptional regulator of pyridoxine metabolism
Accession: AJA01363
Location: 1474-2733
NCBI BlastP on this gene
Accession: AJA01363
Location: 1474-2733
NCBI BlastP on this gene
ChZ00x3
ChZ00x4
capsular polysaccharide expression regulator
Accession: AJA01365
Location: 3692-5131
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AJA01365
Location: 3692-5131
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
chzA
chain length determinant protein/polysaccharide export protein
Accession: AJA01366
Location: 5148-5837
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AJA01366
Location: 5148-5837
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
chzB
tyrosine-protein kinase
Accession: AJA01367
Location: 5847-6533
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AJA01367
Location: 5847-6533
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
chzC
protein-tyrosine phosphatase
Accession: AJA01368
Location: 6572-7303
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AJA01368
Location: 6572-7303
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
chzD
putative nucleoside-diphosphate sugar epimerase
Accession: AJA01369
Location: 7332-9158
NCBI BlastP on this gene
Accession: AJA01369
Location: 7332-9158
NCBI BlastP on this gene
chzE
chzF
chzG
NAD dependent epimerase/dehydratase family protein
Accession: AJA01372
Location: 10569-11429
NCBI BlastP on this gene
Accession: AJA01372
Location: 10569-11429
NCBI BlastP on this gene
chzH
putative glycosyltransferase of GT1 family
Accession: AJA01373
Location: 11426-12544
NCBI BlastP on this gene
Accession: AJA01373
Location: 11426-12544
NCBI BlastP on this gene
chzI
chzJ
chzK
chzL
chzM
chzN
306. : CP024974 Streptococcus suis strain CZ130302 chromosome Total score: 5.5 Cumulative Blast bit score: 2118
CVO91_08840
CVO91_08835
CVO91_08830
glutamate-5-semialdehyde dehydrogenase
Accession: ATZ03987
Location: 1734972-1736210
NCBI BlastP on this gene
Accession: ATZ03987
Location: 1734972-1736210
NCBI BlastP on this gene
CVO91_08825
pyrroline-5-carboxylate reductase
Accession: ATZ03986
Location: 1734177-1734962
NCBI BlastP on this gene
Accession: ATZ03986
Location: 1734177-1734962
NCBI BlastP on this gene
proC
CVO91_08815
TetR/AcrR family transcriptional regulator
Accession: ATZ03984
Location: 1732212-1732616
NCBI BlastP on this gene
Accession: ATZ03984
Location: 1732212-1732616
NCBI BlastP on this gene
CVO91_08810
DegV domain-containing protein
Accession: ATZ03983
Location: 1731234-1732085
NCBI BlastP on this gene
Accession: ATZ03983
Location: 1731234-1732085
NCBI BlastP on this gene
CVO91_08805
PLP-dependent aminotransferase family protein
Accession: ATZ03982
Location: 1729884-1731143
NCBI BlastP on this gene
Accession: ATZ03982
Location: 1729884-1731143
NCBI BlastP on this gene
CVO91_08800
CVO91_08795
LytR family transcriptional regulator
Accession: ATZ03980
Location: 1727486-1728925
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ATZ03980
Location: 1727486-1728925
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 873
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
CVO91_08790
capsular biosynthesis protein CpsC
Accession: ATZ03979
Location: 1726780-1727469
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: ATZ03979
Location: 1726780-1727469
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 397
Sequence coverage: 100 %
E-value: 2e-137
NCBI BlastP on this gene
CVO91_08785
tyrosine protein kinase
Accession: ATZ03978
Location: 1726084-1726770
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: ATZ03978
Location: 1726084-1726770
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
CVO91_08780
tyrosine protein phosphatase
Accession: ATZ03977
Location: 1725314-1726045
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: ATZ03977
Location: 1725314-1726045
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
CVO91_08775
polysaccharide biosynthesis protein
Accession: ATZ03976
Location: 1723459-1725285
NCBI BlastP on this gene
Accession: ATZ03976
Location: 1723459-1725285
NCBI BlastP on this gene
CVO91_08770
cell filamentation protein Fic
Accession: ATZ03975
Location: 1722657-1723373
NCBI BlastP on this gene
Accession: ATZ03975
Location: 1722657-1723373
NCBI BlastP on this gene
CVO91_08765
lipid carrier--UDP-N-acetylgalactosaminyltransferase
Accession: ATZ03974
Location: 1722048-1722647
NCBI BlastP on this gene
Accession: ATZ03974
Location: 1722048-1722647
NCBI BlastP on this gene
CVO91_08760
CVO91_08755
CVO91_08750
CVO91_08745
CVO91_08740
glycosyltransferase family 4 protein
Accession: ATZ03970
Location: 1717397-1718476
NCBI BlastP on this gene
Accession: ATZ03970
Location: 1717397-1718476
NCBI BlastP on this gene
CVO91_08735
CVO91_08730
CVO91_08725
307. : KU665275 Streptococcus suis strain YS488 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2117
Integral membrane regulatory protein Wzg
Accession: AOP02953
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02953
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 896
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02954
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 375
Sequence coverage: 100 %
E-value: 1e-128
NCBI BlastP on this gene
Accession: AOP02954
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 375
Sequence coverage: 100 %
E-value: 1e-128
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02955
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
Accession: AOP02955
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 3e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02956
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AOP02956
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02957
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02957
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP02963
Location: 10566-11684
NCBI BlastP on this gene
Accession: AOP02963
Location: 10566-11684
NCBI BlastP on this gene
cpsK
cpsL
cpsM
308. : KT163375 Streptococcus suis strain YS241 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2117
Integral membrane regulatory protein Wzg
Accession: AOP03855
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 882
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03855
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 882
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03856
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
Accession: AOP03856
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 88 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03857
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
Accession: AOP03857
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 428
Sequence coverage: 96 %
E-value: 1e-149
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03858
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03858
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03859
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03859
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03865
Location: 10567-11685
NCBI BlastP on this gene
Accession: AOP03865
Location: 10567-11685
NCBI BlastP on this gene
cpsK
cpsL
cpsM
309. : JF273649 Streptococcus suis strain 8074 ORF7Z gene Total score: 5.5 Cumulative Blast bit score: 2117
AEH57449
AEH57450
AEH57451
Cps7A
Accession: AEH57452
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57452
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps7A
Cps7B
Accession: AEH57453
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
Accession: AEH57453
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
cps7B
Cps7C
Accession: AEH57454
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AEH57454
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
cps7C
Cps7D
Accession: AEH57455
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEH57455
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cps7D
cps7E
cps7F
cps7G
cps7H
cps7I
cps7J
cps7K
cps7L
cps7M
310. : CP002641 Streptococcus suis D9 Total score: 5.5 Cumulative Blast bit score: 2117
SSUD9_0732
proB
gamma-glutamyl phosphate reductase
Accession: AER16959
Location: 728256-729494
NCBI BlastP on this gene
Accession: AER16959
Location: 728256-729494
NCBI BlastP on this gene
proA
pyrroline-5-carboxylate reductase
Accession: AER16960
Location: 729504-730289
NCBI BlastP on this gene
Accession: AER16960
Location: 729504-730289
NCBI BlastP on this gene
proC
SSUD9_0736
SSUD9_0737
SSUD9_0738
SSUD9_0739
SSUD9_0740
integral membrane regulatory protein Wzg
Accession: AER16966
Location: 735552-736991
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AER16966
Location: 735552-736991
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 863
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
wzg
Cps2B
Accession: AER16967
Location: 737007-737696
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
Accession: AER16967
Location: 737007-737696
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 426
Sequence coverage: 100 %
E-value: 9e-149
NCBI BlastP on this gene
SSUD9_0742
tyrosine-protein kinase Wze
Accession: AER16968
Location: 737706-738392
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AER16968
Location: 737706-738392
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 412
Sequence coverage: 96 %
E-value: 2e-143
NCBI BlastP on this gene
wze
protein-tyrosine phosphatase Wzh
Accession: AER16969
Location: 738431-739162
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AER16969
Location: 738431-739162
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
wzh
SSUD9_0745
putative glycosyltransferase Cps7F
Accession: AER16971
Location: 741463-742161
NCBI BlastP on this gene
Accession: AER16971
Location: 741463-742161
NCBI BlastP on this gene
SSUD9_0746
SSUD9_0747
putative glycosyltransferase Cps7H
Accession: AER16973
Location: 743403-744188
NCBI BlastP on this gene
Accession: AER16973
Location: 743403-744188
NCBI BlastP on this gene
SSUD9_0748
SSUD9_0749
SSUD9_0750
polysaccharide polymerase Cps19aI
Accession: AER16976
Location: 746768-748090
NCBI BlastP on this gene
Accession: AER16976
Location: 746768-748090
NCBI BlastP on this gene
SSUD9_0751
SSUD9_0752
311. : KX870065 Streptococcus suis strain 1297150 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2115
Integral membrane regulatory protein Wzg
Accession: APZ79282
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 879
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ79282
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 879
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ79283
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 1e-136
NCBI BlastP on this gene
Accession: APZ79283
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 90 %
BlastP bit score: 395
Sequence coverage: 100 %
E-value: 1e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ79284
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 2e-149
NCBI BlastP on this gene
Accession: APZ79284
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 2e-149
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ79285
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
Accession: APZ79285
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 7e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ79286
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ79286
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
Initial sugar transferase(Glycosyl-1-phosphate transferase)
Accession: APZ79288
Location: 6295-6993
NCBI BlastP on this gene
Accession: APZ79288
Location: 6295-6993
NCBI BlastP on this gene
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: APZ79292
Location: 10559-11677
NCBI BlastP on this gene
Accession: APZ79292
Location: 10559-11677
NCBI BlastP on this gene
cpsK
cpsL
cpsM
312. : KT163373 Streptococcus suis strain YS219 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2115
Integral membrane regulatory protein Wzg
Accession: AOP03804
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 876
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03804
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 876
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03805
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 6e-136
NCBI BlastP on this gene
Accession: AOP03805
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 6e-136
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03806
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 4e-150
NCBI BlastP on this gene
Accession: AOP03806
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 429
Sequence coverage: 96 %
E-value: 4e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03807
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AOP03807
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 7e-145
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03808
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03808
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
Bacterial transferase hexapeptide repeat protein
Accession: AOP03814
Location: 10567-11685
NCBI BlastP on this gene
Accession: AOP03814
Location: 10567-11685
NCBI BlastP on this gene
cpsK
cpsL
cpsM
313. : KU983476 Streptococcus suis strain YS602 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
Integral membrane regulatory protein Wzg
Accession: AOP03455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03455
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03456
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03456
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03457
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03458
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03459
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03459
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03462
Location: 6885-7745
NCBI BlastP on this gene
Accession: AOP03462
Location: 6885-7745
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
314. : KT163377 Streptococcus suis strain YS249 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
Integral membrane regulatory protein Wzg
Accession: AOP03916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03916
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03917
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03917
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03918
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03919
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03920
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03920
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03923
Location: 6885-7745
NCBI BlastP on this gene
Accession: AOP03923
Location: 6885-7745
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
315. : KT163367 Streptococcus suis strain YS190 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
Integral membrane regulatory protein Wzg
Accession: AOP03627
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03627
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03628
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03628
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03629
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03629
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03630
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AOP03630
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03631
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03631
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03634
Location: 6885-7745
NCBI BlastP on this gene
Accession: AOP03634
Location: 6885-7745
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
316. : KM972231 Streptococcus suis strain YS113_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2114
integral membrane regulatory protein Wzg
Accession: AKE79313
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79313
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 91 %
BlastP bit score: 895
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79314
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AKE79314
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 89 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79315
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
Accession: AKE79315
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 89 %
BlastP bit score: 410
Sequence coverage: 96 %
E-value: 1e-142
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79316
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
Accession: AKE79316
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 415
Sequence coverage: 100 %
E-value: 6e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79317
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79317
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
317. : MH763820 Streptococcus suis strain 7619/2012 hypothetical protein gene Total score: 5.5 Cumulative Blast bit score: 2113
QBQ85388
integral membrane regulatory protein Wzg
Accession: QBQ85389
Location: 844-2283
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 856
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: QBQ85389
Location: 844-2283
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 856
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
QBQ85389
chain length determinant protein Wzd
Accession: QBQ85390
Location: 2299-2988
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: QBQ85390
Location: 2299-2988
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
QBQ85390
tyrosine-protein kinase Wze
Accession: QBQ85391
Location: 2998-3684
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: QBQ85391
Location: 2998-3684
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
QBQ85391
protein-tyrosine phosphatase Wzh
Accession: QBQ85392
Location: 3723-4454
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: QBQ85392
Location: 3723-4454
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
QBQ85392
Nucleoside-diphosphate sugar epimerase
Accession: QBQ85393
Location: 4484-6310
NCBI BlastP on this gene
Accession: QBQ85393
Location: 4484-6310
NCBI BlastP on this gene
QBQ85393
QBQ85394
pyridoxal phosphate (PLP)-dependent aspartate aminotransferase family protein
Accession: QBQ85395
Location: 7462-8679
NCBI BlastP on this gene
Accession: QBQ85395
Location: 7462-8679
NCBI BlastP on this gene
QBQ85395
QBQ85396
QBQ85397
QBQ85398
QBQ85399
QBQ85400
QBQ85401
QBQ85402
318. : LR536835 Streptococcus pneumoniae strain 2245STDY6020240 genome assembly, chromosome: 1. Total score: 5.5 Cumulative Blast bit score: 2113
pbpX
Phospho-N-acetylmuramoyl-pentapeptide- transferase
Accession: VGM74225
Location: 354521-355501
NCBI BlastP on this gene
Accession: VGM74225
Location: 354521-355501
NCBI BlastP on this gene
mraY
SAMEA3175728_00378
Probable ATP-dependent Clp protease ATP-binding subunit
Accession: VGM74227
Location: 356110-358215
NCBI BlastP on this gene
Accession: VGM74227
Location: 356110-358215
NCBI BlastP on this gene
clpC_2
luxS
Uncharacterized protein conserved in bacteria
Accession: VGM74229
Location: 359087-360571
NCBI BlastP on this gene
Accession: VGM74229
Location: 359087-360571
NCBI BlastP on this gene
SAMEA3175728_00381
dexB
Putative transcriptional regulator ywtF
Accession: VGM74231
Location: 363075-364520
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 533
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: VGM74231
Location: 363075-364520
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 533
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
ywtF
Tyrosine-protein phosphatase CpsB
Accession: VGM74232
Location: 364522-365253
BlastP hit with ABP89533.1
Percentage identity: 60 %
BlastP bit score: 303
Sequence coverage: 100 %
E-value: 4e-100
NCBI BlastP on this gene
Accession: VGM74232
Location: 364522-365253
BlastP hit with ABP89533.1
Percentage identity: 60 %
BlastP bit score: 303
Sequence coverage: 100 %
E-value: 4e-100
NCBI BlastP on this gene
cpsB
Capsular polysaccharide type 8 biosynthesis protein cap8A
Accession: VGM74233
Location: 365262-365954
NCBI BlastP on this gene
Accession: VGM74233
Location: 365262-365954
NCBI BlastP on this gene
cap8A
Tyrosine-protein kinase CpsD
Accession: VGM74234
Location: 365964-366653
BlastP hit with ABP89532.1
Percentage identity: 62 %
BlastP bit score: 295
Sequence coverage: 96 %
E-value: 3e-97
NCBI BlastP on this gene
Accession: VGM74234
Location: 365964-366653
BlastP hit with ABP89532.1
Percentage identity: 62 %
BlastP bit score: 295
Sequence coverage: 96 %
E-value: 3e-97
NCBI BlastP on this gene
cpsD
Putative colanic biosynthesis UDP-glucose lipid carrier transferase
Accession: VGM74235
Location: 366669-368036
BlastP hit with ABP89534.1
Percentage identity: 50 %
BlastP bit score: 395
Sequence coverage: 99 %
E-value: 3e-130
NCBI BlastP on this gene
Accession: VGM74235
Location: 366669-368036
BlastP hit with ABP89534.1
Percentage identity: 50 %
BlastP bit score: 395
Sequence coverage: 99 %
E-value: 3e-130
NCBI BlastP on this gene
wcaJ_1
Domain of uncharacterised function (DUF1972)
Accession: VGM74236
Location: 368067-369239
BlastP hit with ABP89535.1
Percentage identity: 71 %
BlastP bit score: 587
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: VGM74236
Location: 368067-369239
BlastP hit with ABP89535.1
Percentage identity: 71 %
BlastP bit score: 587
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
SAMEA3175728_00388
glycogen/starch synthase, ADP-glucose type
Accession: VGM74237
Location: 369236-370519
NCBI BlastP on this gene
Accession: VGM74237
Location: 369236-370519
NCBI BlastP on this gene
SAMEA3175728_00389
Putative acetyltransferase SA2342
Accession: VGM74238
Location: 370497-371186
NCBI BlastP on this gene
Accession: VGM74238
Location: 370497-371186
NCBI BlastP on this gene
SAMEA3175728_00390
ugd_2
D-inositol-3-phosphate glycosyltransferase
Accession: VGM74240
Location: 372473-373597
NCBI BlastP on this gene
Accession: VGM74240
Location: 372473-373597
NCBI BlastP on this gene
mshA_2
SAMEA3175728_00393
SAMEA3175728_00394
SAMEA3175728_00395
rfbX
319. : KM972249 Streptococcus suis strain YS150_seq capsular palysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2113
integral membrane regulatory protein Wzg
Accession: AKE79733
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 884
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AKE79733
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 884
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
chain length determinant protein Wzd
Accession: AKE79734
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
Accession: AKE79734
Location: 1430-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 385
Sequence coverage: 100 %
E-value: 2e-132
NCBI BlastP on this gene
cpsB
tyrosine-protein kinase Wze
Accession: AKE79735
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AKE79735
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 96 %
E-value: 4e-149
NCBI BlastP on this gene
cpsC
protein-tyrosine phosphatase Wzh
Accession: AKE79736
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AKE79736
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 417
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
cpsD
polysaccharide biosynthesis protein
Accession: AKE79737
Location: 3641-5467
NCBI BlastP on this gene
Accession: AKE79737
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
320. : KU983473 Streptococcus suis strain YS494 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2112
Integral membrane regulatory protein Wzg
Accession: AOP03387
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03387
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03388
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03388
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03389
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03389
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03390
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03390
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03391
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03391
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03394
Location: 6886-7746
NCBI BlastP on this gene
Accession: AOP03394
Location: 6886-7746
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
321. : KT163372 Streptococcus suis strain YS210 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2112
Integral membrane regulatory protein Wzg
Accession: AOP03779
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03779
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 885
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03780
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
Accession: AOP03780
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 382
Sequence coverage: 100 %
E-value: 2e-131
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03781
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
Accession: AOP03781
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 431
Sequence coverage: 96 %
E-value: 1e-150
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03782
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
Accession: AOP03782
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 1e-143
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03783
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03783
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
NAD dependent epimerase/dehydratase family
Accession: AOP03786
Location: 6886-7746
NCBI BlastP on this gene
Accession: AOP03786
Location: 6886-7746
NCBI BlastP on this gene
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
322. : JF273655 Streptococcus suis strain 89-2479 ORF23Z gene Total score: 5.5 Cumulative Blast bit score: 2109
AEH57586
AEH57587
AEH57588
Cps23A
Accession: AEH57589
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 855
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57589
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 855
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps23A
Cps23B
Accession: AEH57590
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 3e-148
NCBI BlastP on this gene
Accession: AEH57590
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 92 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 3e-148
NCBI BlastP on this gene
cps23B
Cps23C
Accession: AEH57591
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEH57591
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cps23C
Cps23D
Accession: AEH57592
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEH57592
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cps23D
cps23E
cps23F
cps23G
cps23H
cps23I
cps23J
cps23K
cps23L
cps23M
323. : JF273648 Streptococcus suis strain 11538 ORF5Z gene Total score: 5.5 Cumulative Blast bit score: 2109
AEH57426
AEH57427
AEH57428
Cps5A
Accession: AEH57429
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57429
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps5A
Cps5B
Accession: AEH57430
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AEH57430
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cps5B
Cps5C
Accession: AEH57431
Location: 5093-5770
BlastP hit with ABP89532.1
Percentage identity: 90 %
BlastP bit score: 416
Sequence coverage: 97 %
E-value: 7e-145
NCBI BlastP on this gene
Accession: AEH57431
Location: 5093-5770
BlastP hit with ABP89532.1
Percentage identity: 90 %
BlastP bit score: 416
Sequence coverage: 97 %
E-value: 7e-145
NCBI BlastP on this gene
cps5C
Cps5D
Accession: AEH57432
Location: 5809-6540
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 9e-144
NCBI BlastP on this gene
Accession: AEH57432
Location: 5809-6540
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 9e-144
NCBI BlastP on this gene
cps5D
cps5E
cps5F
cps5G
cps5H
cps5I
cps5J
cps5K
cps5L
cps5M
cps5N
cps5O
324. : JF273646 Streptococcus suis strain 4961 ORF3Z and ORF3Y genes Total score: 5.5 Cumulative Blast bit score: 2109
AEH57389
AEH57390
AEH57391
Cps3A
Accession: AEH57392
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57392
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps3A
Cps3B
Accession: AEH57393
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AEH57393
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cps3B
Cps3C
Accession: AEH57394
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEH57394
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cps3C
Cps3D
Accession: AEH57395
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEH57395
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cps3D
cps3E
cps3F
cps3G
cps3H
cps3I
cps3J
cps3K
cps3L
cps3M
cps3N
325. : CP041994 Streptococcus suis strain INT-01 chromosome Total score: 5.5 Cumulative Blast bit score: 2109
FPT06_02825
FPT06_02820
FPT06_02815
glutamate-5-semialdehyde dehydrogenase
Accession: QDS23302
Location: 574772-576010
NCBI BlastP on this gene
Accession: QDS23302
Location: 574772-576010
NCBI BlastP on this gene
FPT06_02810
pyrroline-5-carboxylate reductase
Accession: QDS23301
Location: 573977-574762
NCBI BlastP on this gene
Accession: QDS23301
Location: 573977-574762
NCBI BlastP on this gene
proC
TetR/AcrR family transcriptional regulator
Accession: QDS23300
Location: 573550-573954
NCBI BlastP on this gene
Accession: QDS23300
Location: 573550-573954
NCBI BlastP on this gene
FPT06_02800
FPT06_02795
PLP-dependent aminotransferase family protein
Accession: QDS23298
Location: 571214-572473
NCBI BlastP on this gene
Accession: QDS23298
Location: 571214-572473
NCBI BlastP on this gene
FPT06_02790
yaaA
LytR family transcriptional regulator
Accession: QDS23296
Location: 568788-570227
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: QDS23296
Location: 568788-570227
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
FPT06_02780
capsular biosynthesis protein CpsC
Accession: QDS23295
Location: 568083-568772
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: QDS23295
Location: 568083-568772
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
FPT06_02775
tyrosine-protein kinase
Accession: QDS23294
Location: 567387-568073
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: QDS23294
Location: 567387-568073
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
FPT06_02770
tyrosine protein phosphatase
Accession: QDS23293
Location: 566617-567348
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: QDS23293
Location: 566617-567348
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
FPT06_02765
polysaccharide biosynthesis protein
Accession: QDS23292
Location: 564761-566587
NCBI BlastP on this gene
Accession: QDS23292
Location: 564761-566587
NCBI BlastP on this gene
FPT06_02760
FPT06_02755
aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme
Accession: QDS23290
Location: 562682-563932
NCBI BlastP on this gene
Accession: QDS23290
Location: 562682-563932
NCBI BlastP on this gene
FPT06_02750
FPT06_02745
FPT06_02740
FPT06_02735
FPT06_02730
O-antigen ligase family protein
Accession: QDS23285
Location: 557878-559101
NCBI BlastP on this gene
Accession: QDS23285
Location: 557878-559101
NCBI BlastP on this gene
FPT06_02725
FPT06_02720
FPT06_02715
326. : CP030124 Streptococcus suis strain SH1510 chromosome Total score: 5.5 Cumulative Blast bit score: 2109
DP111_09535
DP111_09530
DP111_09525
gamma-glutamyl-phosphate reductase
Accession: AXI66227
Location: 1879316-1880554
NCBI BlastP on this gene
Accession: AXI66227
Location: 1879316-1880554
NCBI BlastP on this gene
DP111_09520
pyrroline-5-carboxylate reductase
Accession: AXI66226
Location: 1878521-1879306
NCBI BlastP on this gene
Accession: AXI66226
Location: 1878521-1879306
NCBI BlastP on this gene
proC
TetR/AcrR family transcriptional regulator
Accession: AXI66225
Location: 1878094-1878498
NCBI BlastP on this gene
Accession: AXI66225
Location: 1878094-1878498
NCBI BlastP on this gene
DP111_09510
DegV domain-containing protein
Accession: AXI66224
Location: 1877116-1877967
NCBI BlastP on this gene
Accession: AXI66224
Location: 1877116-1877967
NCBI BlastP on this gene
DP111_09505
PLP-dependent aminotransferase family protein
Accession: AXI66223
Location: 1875758-1877017
NCBI BlastP on this gene
Accession: AXI66223
Location: 1875758-1877017
NCBI BlastP on this gene
DP111_09500
DP111_09495
LytR family transcriptional regulator
Accession: AXI66221
Location: 1873332-1874771
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AXI66221
Location: 1873332-1874771
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
DP111_09490
capsular biosynthesis protein CpsC
Accession: AXI66220
Location: 1872627-1873316
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AXI66220
Location: 1872627-1873316
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
DP111_09485
tyrosine protein kinase
Accession: AXI66219
Location: 1871931-1872617
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AXI66219
Location: 1871931-1872617
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
DP111_09480
tyrosine protein phosphatase
Accession: AXI66218
Location: 1871161-1871892
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AXI66218
Location: 1871161-1871892
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
DP111_09475
polysaccharide biosynthesis protein
Accession: AXI66217
Location: 1869305-1871131
NCBI BlastP on this gene
Accession: AXI66217
Location: 1869305-1871131
NCBI BlastP on this gene
DP111_09470
DP111_09465
DegT/DnrJ/EryC1/StrS family aminotransferase
Accession: AXI66215
Location: 1866932-1868152
NCBI BlastP on this gene
Accession: AXI66215
Location: 1866932-1868152
NCBI BlastP on this gene
DP111_09460
DP111_09455
DP111_09450
GNAT family N-acetyltransferase
Accession: AXI66212
Location: 1864544-1865134
NCBI BlastP on this gene
Accession: AXI66212
Location: 1864544-1865134
NCBI BlastP on this gene
DP111_09445
DP111_09440
glycosyltransferase family 2 protein
Accession: AXI66210
Location: 1862491-1863396
NCBI BlastP on this gene
Accession: AXI66210
Location: 1862491-1863396
NCBI BlastP on this gene
DP111_09435
DP111_09430
327. : CP017142 Streptococcus suis strain GZ0565 chromosome Total score: 5.5 Cumulative Blast bit score: 2109
BFP66_06400
BFP66_06395
BFP66_06390
glutamate-5-semialdehyde dehydrogenase
Accession: AOM74869
Location: 1292557-1293795
NCBI BlastP on this gene
Accession: AOM74869
Location: 1292557-1293795
NCBI BlastP on this gene
BFP66_06385
pyrroline-5-carboxylate reductase
Accession: AOM74868
Location: 1291762-1292547
NCBI BlastP on this gene
Accession: AOM74868
Location: 1291762-1292547
NCBI BlastP on this gene
BFP66_06380
TetR family transcriptional regulator
Accession: AOM74867
Location: 1291335-1291739
NCBI BlastP on this gene
Accession: AOM74867
Location: 1291335-1291739
NCBI BlastP on this gene
BFP66_06375
fatty acid-binding protein DegV
Accession: AOM74866
Location: 1290357-1291208
NCBI BlastP on this gene
Accession: AOM74866
Location: 1290357-1291208
NCBI BlastP on this gene
BFP66_06370
GntR family transcriptional regulator
Accession: AOM74865
Location: 1288999-1290258
NCBI BlastP on this gene
Accession: AOM74865
Location: 1288999-1290258
NCBI BlastP on this gene
BFP66_06365
BFP66_06360
LytR family transcriptional regulator
Accession: AOM74863
Location: 1286573-1288012
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOM74863
Location: 1286573-1288012
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
BFP66_06355
capsular biosynthesis protein CpsC
Accession: AOM74862
Location: 1285868-1286557
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AOM74862
Location: 1285868-1286557
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
BFP66_06350
tyrosine protein kinase
Accession: AOM74861
Location: 1285172-1285858
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AOM74861
Location: 1285172-1285858
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
BFP66_06345
tyrosine protein phosphatase
Accession: AOM74860
Location: 1284402-1285133
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOM74860
Location: 1284402-1285133
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
BFP66_06340
BFP66_06335
capsular biosynthesis protein
Accession: AOM74858
Location: 1281780-1282382
NCBI BlastP on this gene
Accession: AOM74858
Location: 1281780-1282382
NCBI BlastP on this gene
BFP66_06330
BFP66_06325
BFP66_06320
BFP66_06315
BFP66_06310
BFP66_06305
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: AOM74853
Location: 1276288-1277007
NCBI BlastP on this gene
Accession: AOM74853
Location: 1276288-1277007
NCBI BlastP on this gene
BFP66_06300
BFP66_06295
BFP66_06290
328. : CP015557 Streptococcus suis strain DN13 Total score: 5.5 Cumulative Blast bit score: 2109
A6M16_06505
A6M16_06500
A6M16_06495
gamma-glutamyl-phosphate reductase
Accession: AND00153
Location: 1315938-1317176
NCBI BlastP on this gene
Accession: AND00153
Location: 1315938-1317176
NCBI BlastP on this gene
A6M16_06490
pyrroline-5-carboxylate reductase
Accession: AND00152
Location: 1315143-1315928
NCBI BlastP on this gene
Accession: AND00152
Location: 1315143-1315928
NCBI BlastP on this gene
A6M16_06485
TetR family transcriptional regulator
Accession: AND00151
Location: 1314716-1315120
NCBI BlastP on this gene
Accession: AND00151
Location: 1314716-1315120
NCBI BlastP on this gene
A6M16_06480
fatty acid-binding protein DegV
Accession: AND00150
Location: 1313738-1314589
NCBI BlastP on this gene
Accession: AND00150
Location: 1313738-1314589
NCBI BlastP on this gene
A6M16_06475
GntR family transcriptional regulator
Accession: AND00149
Location: 1312380-1313639
NCBI BlastP on this gene
Accession: AND00149
Location: 1312380-1313639
NCBI BlastP on this gene
A6M16_06470
A6M16_06465
LytR family transcriptional regulator
Accession: AND00147
Location: 1309954-1311393
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AND00147
Location: 1309954-1311393
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
A6M16_06460
capsular biosynthesis protein CpsC
Accession: AND00146
Location: 1309249-1309938
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AND00146
Location: 1309249-1309938
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
A6M16_06455
tyrosine protein kinase
Accession: AND00145
Location: 1308553-1309239
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AND00145
Location: 1308553-1309239
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
A6M16_06450
tyrosine protein phosphatase
Accession: AND00144
Location: 1307783-1308514
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AND00144
Location: 1307783-1308514
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
A6M16_06445
A6M16_06440
capsular biosynthesis protein
Accession: AND00142
Location: 1305161-1305763
NCBI BlastP on this gene
Accession: AND00142
Location: 1305161-1305763
NCBI BlastP on this gene
A6M16_06435
A6M16_06430
A6M16_06425
A6M16_06420
A6M16_06415
A6M16_06410
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: AND00137
Location: 1299669-1300388
NCBI BlastP on this gene
Accession: AND00137
Location: 1299669-1300388
NCBI BlastP on this gene
A6M16_06405
A6M16_06400
A6M16_06395
329. : CP002633 Streptococcus suis ST3 Total score: 5.5 Cumulative Blast bit score: 2109
Zn-dependent hydrolase, including glyoxylases
Accession: AEB81183
Location: 732324-732962
NCBI BlastP on this gene
Accession: AEB81183
Location: 732324-732962
NCBI BlastP on this gene
SSUST3_0727
SSUST3_0728
SSUST3_0729
gamma-glutamyl phosphate reductase
Accession: AEB81186
Location: 736841-738079
NCBI BlastP on this gene
Accession: AEB81186
Location: 736841-738079
NCBI BlastP on this gene
proA
pyrroline-5-carboxylate reductase
Accession: AEB81187
Location: 738089-738874
NCBI BlastP on this gene
Accession: AEB81187
Location: 738089-738874
NCBI BlastP on this gene
proC
SSUST3_0732
SSUST3_0733
SSUST3_0734
SSUST3_0735
integral membrane regulatory protein Wzg
Accession: AEB81192
Location: 742624-744063
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEB81192
Location: 742624-744063
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
wzg
Cps2B
Accession: AEB81193
Location: 744079-744768
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: AEB81193
Location: 744079-744768
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
SSUST3_0737
tyrosine-protein kinase Wze
Accession: AEB81194
Location: 744778-745464
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEB81194
Location: 744778-745464
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
wze
protein-tyrosine phosphatase Wzh
Accession: AEB81195
Location: 745503-746234
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AEB81195
Location: 745503-746234
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
wzh
SSUST3_0740
SSUST3_0741
SSUST3_0742
UDP-galactose phosphate transferase
Accession: AEB81199
Location: 750182-750799
NCBI BlastP on this gene
Accession: AEB81199
Location: 750182-750799
NCBI BlastP on this gene
SSUST3_0743
SSUST3_0744
SSUST3_0745
capsular polysaccharide synthesis protein
Accession: AEB81202
Location: 752575-753753
NCBI BlastP on this gene
Accession: AEB81202
Location: 752575-753753
NCBI BlastP on this gene
SSUST3_0746
SSUST3_0747
SSUST3_0748
330. : KX870051 Streptococcus suis strain 78468-5 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2107
Integral membrane regulatory protein Wzg
Accession: APZ78944
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78944
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ78945
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: APZ78945
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ78946
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: APZ78946
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ78947
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: APZ78947
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ78948
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ78948
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
Nucleoside-diphosphate sugar epimerase
Accession: APZ78955
Location: 10791-11630
NCBI BlastP on this gene
Accession: APZ78955
Location: 10791-11630
NCBI BlastP on this gene
cpsL
cpsM
cpsN
cpsO
331. : KX870050 Streptococcus suis strain 78468-4 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2107
Integral membrane regulatory protein Wzg
Accession: APZ78922
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78922
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ78923
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: APZ78923
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ78924
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: APZ78924
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ78925
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: APZ78925
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ78926
Location: 3641-5467
NCBI BlastP on this gene
Accession: APZ78926
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
Nucleoside-diphosphate sugar epimerase
Accession: APZ78933
Location: 10791-11630
NCBI BlastP on this gene
Accession: APZ78933
Location: 10791-11630
NCBI BlastP on this gene
cpsL
cpsM
cpsN
cpsO
332. : KX870049 Streptococcus suis strain 78468-1 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2107
Integral membrane regulatory protein Wzg
Accession: APZ78900
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: APZ78900
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 850
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: APZ78901
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
Accession: APZ78901
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 427
Sequence coverage: 100 %
E-value: 4e-149
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: APZ78902
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: APZ78902
Location: 2155-2841
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cpsC
Protein-tyrosine phosphatase Wzh
Accession: APZ78903
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: APZ78903
Location: 2880-3611
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: APZ78904
Location: 4007-5467
NCBI BlastP on this gene
Accession: APZ78904
Location: 4007-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
Nucleoside-diphosphate sugar epimerase
Accession: APZ78911
Location: 10791-11630
NCBI BlastP on this gene
Accession: APZ78911
Location: 10791-11630
NCBI BlastP on this gene
cpsL
cpsM
cpsN
cpsO
333. : KM576773 Streptococcus suis strain 43640 Cps31A (cps31A), Cps31B (cps31B), Cps31C (cps31C), Cps3... Total score: 5.5 Cumulative Blast bit score: 2107
Cps31A
Accession: AIZ73034
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 871
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIZ73034
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 90 %
BlastP bit score: 871
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps31A
Cps31B
Accession: AIZ73035
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 82 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
Accession: AIZ73035
Location: 1456-2145
BlastP hit with ABP89531.1
Percentage identity: 82 %
BlastP bit score: 393
Sequence coverage: 100 %
E-value: 7e-136
NCBI BlastP on this gene
cps31B
Cps31C
Accession: AIZ73036
Location: 2155-2838
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 423
Sequence coverage: 95 %
E-value: 1e-147
NCBI BlastP on this gene
Accession: AIZ73036
Location: 2155-2838
BlastP hit with ABP89532.1
Percentage identity: 93 %
BlastP bit score: 423
Sequence coverage: 95 %
E-value: 1e-147
NCBI BlastP on this gene
cps31C
Cps31D
Accession: AIZ73037
Location: 2879-3610
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 420
Sequence coverage: 100 %
E-value: 5e-146
NCBI BlastP on this gene
Accession: AIZ73037
Location: 2879-3610
BlastP hit with ABP89533.1
Percentage identity: 87 %
BlastP bit score: 420
Sequence coverage: 100 %
E-value: 5e-146
NCBI BlastP on this gene
cps31D
cps31E
cps31F
cps31G
cps31H
cps31I
cps31J
cps31K
cps31L
cps31M
334. : JF273647 Streptococcus suis strain 6407 ORF4Z gene Total score: 5.5 Cumulative Blast bit score: 2107
AEH57407
AEH57408
AEH57409
Cps4A
Accession: AEH57410
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57410
Location: 2939-4378
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps4A
Cps4B
Accession: AEH57411
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
Accession: AEH57411
Location: 4394-5083
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
cps4B
Cps4C
Accession: AEH57412
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AEH57412
Location: 5093-5779
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
cps4C
Cps4D
Accession: AEH57413
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
Accession: AEH57413
Location: 5818-6549
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
cps4D
cps4E
cps4F
cps4G
cps4H
cps4I
cps4J
cps4K
cps4L
cps4M
335. : CP008921 Streptococcus suis 6407 Total score: 5.5 Cumulative Blast bit score: 2107
ID09_07160
ID09_07155
ID09_07145
gamma-glutamyl phosphate reductase
Accession: AIG43812
Location: 1433256-1434494
NCBI BlastP on this gene
Accession: AIG43812
Location: 1433256-1434494
NCBI BlastP on this gene
ID09_07140
pyrroline-5-carboxylate reductase
Accession: AIG43811
Location: 1432461-1433246
NCBI BlastP on this gene
Accession: AIG43811
Location: 1432461-1433246
NCBI BlastP on this gene
ID09_07135
TetR family transcriptional regulator
Accession: AIG43810
Location: 1432034-1432438
NCBI BlastP on this gene
Accession: AIG43810
Location: 1432034-1432438
NCBI BlastP on this gene
ID09_07130
ID09_07125
GntR family transcriptional regulator
Accession: AIG43808
Location: 1429698-1430957
NCBI BlastP on this gene
Accession: AIG43808
Location: 1429698-1430957
NCBI BlastP on this gene
ID09_07120
ID09_07115
LytR family transcriptional regulator
Accession: AIG43806
Location: 1427272-1428711
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AIG43806
Location: 1427272-1428711
BlastP hit with ABP89530.1
Percentage identity: 87 %
BlastP bit score: 852
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
ID09_07110
capsular biosynthesis protein CpsC
Accession: AIG43805
Location: 1426567-1427256
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
Accession: AIG43805
Location: 1426567-1427256
BlastP hit with ABP89531.1
Percentage identity: 93 %
BlastP bit score: 428
Sequence coverage: 100 %
E-value: 9e-150
NCBI BlastP on this gene
ID09_07105
tyrosine protein kinase
Accession: AIG43804
Location: 1425871-1426557
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
Accession: AIG43804
Location: 1425871-1426557
BlastP hit with ABP89532.1
Percentage identity: 91 %
BlastP bit score: 414
Sequence coverage: 96 %
E-value: 5e-144
NCBI BlastP on this gene
ID09_07100
tyrosine protein phosphatase
Accession: AIG43803
Location: 1425101-1425832
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
Accession: AIG43803
Location: 1425101-1425832
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 413
Sequence coverage: 100 %
E-value: 3e-143
NCBI BlastP on this gene
ID09_07095
ID09_07090
ID09_07085
capsular biosynthesis protein
Accession: AIG43800
Location: 1420872-1422092
NCBI BlastP on this gene
Accession: AIG43800
Location: 1420872-1422092
NCBI BlastP on this gene
ID09_07080
amylovoran biosynthesis protein AmsE
Accession: AIG43799
Location: 1420049-1420879
NCBI BlastP on this gene
Accession: AIG43799
Location: 1420049-1420879
NCBI BlastP on this gene
ID09_07075
ID09_07070
ID09_07065
ID09_07060
ID09_07055
ID09_07050
336. : KY574603 Streptococcus suis strain 1388970_seq cps gene cluster Total score: 5.5 Cumulative Blast bit score: 2099
integral membrane regulatory protein Wzg
Accession: ARO73588
Location: 1-1338
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 833
Sequence coverage: 91 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ARO73588
Location: 1-1338
BlastP hit with ABP89530.1
Percentage identity: 92 %
BlastP bit score: 833
Sequence coverage: 91 %
E-value: 0.0
NCBI BlastP on this gene
cps9A
chain length determinant protein Wzd
Accession: ARO73589
Location: 1354-2043
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 7e-148
NCBI BlastP on this gene
Accession: ARO73589
Location: 1354-2043
BlastP hit with ABP89531.1
Percentage identity: 91 %
BlastP bit score: 424
Sequence coverage: 100 %
E-value: 7e-148
NCBI BlastP on this gene
cps9B
tyrosine-protein kinase Wze
Accession: ARO73590
Location: 2053-2739
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 426
Sequence coverage: 96 %
E-value: 1e-148
NCBI BlastP on this gene
Accession: ARO73590
Location: 2053-2739
BlastP hit with ABP89532.1
Percentage identity: 94 %
BlastP bit score: 426
Sequence coverage: 96 %
E-value: 1e-148
NCBI BlastP on this gene
cps9C
protein-tyrosine phosphatase Wzh
Accession: ARO73591
Location: 2778-3509
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: ARO73591
Location: 2778-3509
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 2e-144
NCBI BlastP on this gene
cps9D
nucleoside-diphosphate sugar epimerase
Accession: ARO73592
Location: 3539-5365
NCBI BlastP on this gene
Accession: ARO73592
Location: 3539-5365
NCBI BlastP on this gene
cps9E
cps9F
cps9G
cps9H
cps9I
cps9J
capsular polysaccharide synthesis protein
Accession: ARO73598
Location: 9925-10884
NCBI BlastP on this gene
Accession: ARO73598
Location: 9925-10884
NCBI BlastP on this gene
cps9K
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
Accession: ARO73599
Location: 10903-11622
NCBI BlastP on this gene
Accession: ARO73599
Location: 10903-11622
NCBI BlastP on this gene
cps9L
NAD-dependent epimerase/dehydratase
Accession: ARO73600
Location: 11631-12686
NCBI BlastP on this gene
Accession: ARO73600
Location: 11631-12686
NCBI BlastP on this gene
cps9M
cps9N
337. : CR931684 Streptococcus pneumoniae strain 1039/41 (serotype 23b). Total score: 5.5 Cumulative Blast bit score: 2097
dexB
integral membrane regulatory protein Wzg
Accession: CAI33834
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 543
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33834
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 543
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wzg
wzh
capsular polysaccharide biosynthesis protein Wzd
Accession: CAI33836
Location: 4774-5466
NCBI BlastP on this gene
Accession: CAI33836
Location: 4774-5466
NCBI BlastP on this gene
wzd
wze
undecaprenylphosphate glucosephosphotransferase WchA (initial sugar transferase)
Accession: CAI33838
Location: 6180-7541
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
Accession: CAI33838
Location: 6180-7541
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
wchA
putative rhamnosyl transferase WchF
Accession: CAI33839
Location: 7528-8751
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33839
Location: 7528-8751
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wchF
oligosaccharide repeat unit polymerase Wzy
Accession: CAI33840
Location: 9052-10239
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
Accession: CAI33840
Location: 9052-10239
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
wzy
putative glycosyl transferase
Accession: CAI33841
Location: 10249-11217
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 178
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
Accession: CAI33841
Location: 10249-11217
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 178
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
wchV
wchW
wzx
putative glycerol phosphotransferase
Accession: CAI33844
Location: 13840-14994
NCBI BlastP on this gene
Accession: CAI33844
Location: 13840-14994
NCBI BlastP on this gene
wchX
putative glycerol-2-phosphate dehydrogenase
Accession: CAI33845
Location: 15030-16058
NCBI BlastP on this gene
Accession: CAI33845
Location: 15030-16058
NCBI BlastP on this gene
gtp1
gtp2
gtp3
glucose-1-phosphate thymidylyltransferase RmlA
Accession: CAI33848
Location: 17786-18655
NCBI BlastP on this gene
Accession: CAI33848
Location: 17786-18655
NCBI BlastP on this gene
rmlA
338. : CR931674 Streptococcus pneumoniae strain Gethens (serotype 18f). Total score: 5.5 Cumulative Blast bit score: 2094
dexB
integral membrane regulatory protein Wzg
Accession: CAI33599
Location: 1560-3005
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 536
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33599
Location: 1560-3005
BlastP hit with ABP89530.1
Percentage identity: 58 %
BlastP bit score: 536
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wzg
protein-tyrosine phosphatase Wzh
Accession: CAI33600
Location: 3007-3738
BlastP hit with ABP89533.1
Percentage identity: 59 %
BlastP bit score: 297
Sequence coverage: 100 %
E-value: 1e-97
NCBI BlastP on this gene
Accession: CAI33600
Location: 3007-3738
BlastP hit with ABP89533.1
Percentage identity: 59 %
BlastP bit score: 297
Sequence coverage: 100 %
E-value: 1e-97
NCBI BlastP on this gene
wzh
capsular polysaccharide biosynthesis protein Wzd
Accession: CAI33601
Location: 3747-4439
NCBI BlastP on this gene
Accession: CAI33601
Location: 3747-4439
NCBI BlastP on this gene
wzd
tyrosine-protein kinase Wze
Accession: CAI33602
Location: 4449-5132
BlastP hit with ABP89532.1
Percentage identity: 61 %
BlastP bit score: 294
Sequence coverage: 96 %
E-value: 6e-97
NCBI BlastP on this gene
Accession: CAI33602
Location: 4449-5132
BlastP hit with ABP89532.1
Percentage identity: 61 %
BlastP bit score: 294
Sequence coverage: 96 %
E-value: 6e-97
NCBI BlastP on this gene
wze
undecaprenylphosphate glucosephosphotransferase WchA (initial sugar transferase)
Accession: CAI33603
Location: 5148-6515
BlastP hit with ABP89534.1
Percentage identity: 48 %
BlastP bit score: 385
Sequence coverage: 98 %
E-value: 2e-126
NCBI BlastP on this gene
Accession: CAI33603
Location: 5148-6515
BlastP hit with ABP89534.1
Percentage identity: 48 %
BlastP bit score: 385
Sequence coverage: 98 %
E-value: 2e-126
NCBI BlastP on this gene
wchA
putative rhamnosyl transferase WchF
Accession: CAI33604
Location: 6546-7718
BlastP hit with ABP89535.1
Percentage identity: 69 %
BlastP bit score: 582
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: CAI33604
Location: 6546-7718
BlastP hit with ABP89535.1
Percentage identity: 69 %
BlastP bit score: 582
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wchF
wciU
wcxM
wciV
wciW
wzx
oligosaccharide repeat unit polymerase Wzy
Accession: CAI33610
Location: 13129-14403
NCBI BlastP on this gene
Accession: CAI33610
Location: 13129-14403
NCBI BlastP on this gene
wzy
wciX
putative glycerol phosphotransferase
Accession: CAI33612
Location: 15455-16615
NCBI BlastP on this gene
Accession: CAI33612
Location: 15455-16615
NCBI BlastP on this gene
wciY
339. : LT594598 Streptococcus pneumoniae capsular locus (cps) operon, strain 23B1. Total score: 5.5 Cumulative Blast bit score: 2091
tnp
tnp
tnp
integral membrane regulatory protein Wzg
Accession: SBT85355
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 538
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: SBT85355
Location: 2578-4032
BlastP hit with ABP89530.1
Percentage identity: 57 %
BlastP bit score: 538
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wzg
wzh
capsular polysaccharide biosynthesis protein Wzd
Accession: SBT85357
Location: 4774-5466
NCBI BlastP on this gene
Accession: SBT85357
Location: 4774-5466
NCBI BlastP on this gene
wzd
wze
galactosyl trasnferase WchA
Accession: SBT85359
Location: 6171-7532
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
Accession: SBT85359
Location: 6171-7532
BlastP hit with ABP89534.1
Percentage identity: 49 %
BlastP bit score: 389
Sequence coverage: 99 %
E-value: 7e-128
NCBI BlastP on this gene
wchA
putative rhamnosyl transferase WchF
Accession: SBT85360
Location: 7519-8742
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: SBT85360
Location: 7519-8742
BlastP hit with ABP89535.1
Percentage identity: 74 %
BlastP bit score: 612
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
wchF
oligosaccharide repeat unit polymerase Wzy
Accession: SBT85361
Location: 9043-10230
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
Accession: SBT85361
Location: 9043-10230
BlastP hit with ABP89538.1
Percentage identity: 48 %
BlastP bit score: 375
Sequence coverage: 97 %
E-value: 2e-123
NCBI BlastP on this gene
wzy
glycosyl transferase wchV
Accession: SBT85362
Location: 10240-11208
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 177
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
Accession: SBT85362
Location: 10240-11208
BlastP hit with ABP89540.1
Percentage identity: 41 %
BlastP bit score: 177
Sequence coverage: 72 %
E-value: 5e-49
NCBI BlastP on this gene
wchV
wchW
capsule biosynthesis repeating unit flippase Wzx
Accession: SBT85364
Location: 12446-13834
NCBI BlastP on this gene
Accession: SBT85364
Location: 12446-13834
NCBI BlastP on this gene
wzx
wchX
glycerol-2-phosphate dehydrogenase Gtp1
Accession: SBT85366
Location: 15021-16049
NCBI BlastP on this gene
Accession: SBT85366
Location: 15021-16049
NCBI BlastP on this gene
gtp1
gtp2
gtp3
glucose-1-phosphate thymidylyltransferase RmlA
Accession: SBT85369
Location: 17758-18627
NCBI BlastP on this gene
Accession: SBT85369
Location: 17758-18627
NCBI BlastP on this gene
rmlA
340. : JF791167 Streptococcus suis strain 88-1861 ORFY gene Total score: 5.5 Cumulative Blast bit score: 2085
AEP02726
CpsA
Accession: AEP02727
Location: 3092-4531
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 861
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02727
Location: 3092-4531
BlastP hit with ABP89530.1
Percentage identity: 88 %
BlastP bit score: 861
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02727
CpsB
Accession: AEP02728
Location: 4548-5237
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 408
Sequence coverage: 100 %
E-value: 1e-141
NCBI BlastP on this gene
Accession: AEP02728
Location: 4548-5237
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 408
Sequence coverage: 100 %
E-value: 1e-141
NCBI BlastP on this gene
AEP02728
CpsC
Accession: AEP02729
Location: 5247-5933
BlastP hit with ABP89532.1
Percentage identity: 86 %
BlastP bit score: 397
Sequence coverage: 96 %
E-value: 2e-137
NCBI BlastP on this gene
Accession: AEP02729
Location: 5247-5933
BlastP hit with ABP89532.1
Percentage identity: 86 %
BlastP bit score: 397
Sequence coverage: 96 %
E-value: 2e-137
NCBI BlastP on this gene
AEP02729
CpsD
Accession: AEP02730
Location: 5971-6702
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 2e-145
NCBI BlastP on this gene
Accession: AEP02730
Location: 5971-6702
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 419
Sequence coverage: 100 %
E-value: 2e-145
NCBI BlastP on this gene
AEP02730
341. : CP017092 Streptococcus suis strain ISU2812 chromosome Total score: 5.5 Cumulative Blast bit score: 2085
glutamate-5-semialdehyde dehydrogenase
Accession: ASW52408
Location: 2078301-2079539
NCBI BlastP on this gene
Accession: ASW52408
Location: 2078301-2079539
NCBI BlastP on this gene
A7J09_09945
pyrroline-5-carboxylate reductase
Accession: ASW52407
Location: 2077506-2078291
NCBI BlastP on this gene
Accession: ASW52407
Location: 2077506-2078291
NCBI BlastP on this gene
A7J09_09940
A7J09_09935
A7J09_09930
A7J09_09925
TetR family transcriptional regulator
Accession: ASW52403
Location: 2074144-2074548
NCBI BlastP on this gene
Accession: ASW52403
Location: 2074144-2074548
NCBI BlastP on this gene
A7J09_09920
fatty acid-binding protein DegV
Accession: ASW52402
Location: 2073166-2074017
NCBI BlastP on this gene
Accession: ASW52402
Location: 2073166-2074017
NCBI BlastP on this gene
A7J09_09915
A7J09_09910
A7J09_09905
GntR family transcriptional regulator
Accession: ASW52399
Location: 2070195-2071454
NCBI BlastP on this gene
Accession: ASW52399
Location: 2070195-2071454
NCBI BlastP on this gene
A7J09_09900
A7J09_09895
LytR family transcriptional regulator
Accession: ASW52397
Location: 2067773-2069212
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: ASW52397
Location: 2067773-2069212
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
A7J09_09890
capsular biosynthesis protein CpsC
Accession: ASW52396
Location: 2067067-2067756
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: ASW52396
Location: 2067067-2067756
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
A7J09_09885
tyrosine protein kinase
Accession: ASW52395
Location: 2066380-2067057
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 409
Sequence coverage: 97 %
E-value: 3e-142
NCBI BlastP on this gene
Accession: ASW52395
Location: 2066380-2067057
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 409
Sequence coverage: 97 %
E-value: 3e-142
NCBI BlastP on this gene
A7J09_09880
tyrosine protein phosphatase
Accession: ASW52394
Location: 2065611-2066342
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: ASW52394
Location: 2065611-2066342
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
A7J09_09875
A7J09_09870
cell filamentation protein Fic
Accession: ASW52392
Location: 2062955-2063671
NCBI BlastP on this gene
Accession: ASW52392
Location: 2062955-2063671
NCBI BlastP on this gene
A7J09_09865
A7J09_09860
capsular biosynthesis protein
Accession: ASW52390
Location: 2060992-2062221
NCBI BlastP on this gene
Accession: ASW52390
Location: 2060992-2062221
NCBI BlastP on this gene
A7J09_09855
A7J09_09850
A7J09_09845
A7J09_09840
A7J09_09835
A7J09_09830
A7J09_09825
342. : KU665260 Streptococcus suis strain YS205 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2080
Integral membrane regulatory protein Wzg
Accession: AOP02591
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP02591
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP02592
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
Accession: AOP02592
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP02593
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP02593
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP02594
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP02594
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP02595
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP02595
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
343. : KT163369 Streptococcus suis strain YS196 capsular polysaccharide synthesis gene cluster Total score: 5.5 Cumulative Blast bit score: 2080
Integral membrane regulatory protein Wzg
Accession: AOP03684
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AOP03684
Location: 1-1440
BlastP hit with ABP89530.1
Percentage identity: 85 %
BlastP bit score: 845
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cpsA
Chain length determinant protein Wzd
Accession: AOP03685
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
Accession: AOP03685
Location: 1457-2146
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 386
Sequence coverage: 100 %
E-value: 4e-133
NCBI BlastP on this gene
cpsB
Tyrosine-protein kinase Wze
Accession: AOP03686
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
Accession: AOP03686
Location: 2156-2842
BlastP hit with ABP89532.1
Percentage identity: 95 %
BlastP bit score: 433
Sequence coverage: 96 %
E-value: 2e-151
NCBI BlastP on this gene
cpsC
Protein-Tyrosine phosphatase Wzh
Accession: AOP03687
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
Accession: AOP03687
Location: 2881-3612
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 3e-144
NCBI BlastP on this gene
cpsD
Polysaccharide biosynthesis protein
Accession: AOP03688
Location: 3641-5467
NCBI BlastP on this gene
Accession: AOP03688
Location: 3641-5467
NCBI BlastP on this gene
cpsE
cpsF
cpsG
cpsH
cpsI
cpsJ
cpsK
cpsL
cpsM
cpsN
344. : JF791163 Streptococcus suis strain 92-1191 ORFY, ORFX, CpsA, CpsB, CpsC, and CpsD genes Total score: 5.5 Cumulative Blast bit score: 2080
AEP02702
AEP02703
CpsA
Accession: AEP02704
Location: 2914-4353
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 870
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02704
Location: 2914-4353
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 870
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02704
CpsB
Accession: AEP02705
Location: 4370-5059
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AEP02705
Location: 4370-5059
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AEP02705
CpsC
Accession: AEP02706
Location: 5069-5755
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: AEP02706
Location: 5069-5755
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
AEP02706
CpsD
Accession: AEP02707
Location: 5793-6524
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
Accession: AEP02707
Location: 5793-6524
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
AEP02707
345. : KC537385 Streptococcus suis strain 14A capsular polysaccharide synthesis locus sequence. Total score: 5.5 Cumulative Blast bit score: 2078
AGS58356
AGS58357
AGS58358
Cps21A
Accession: AGS58359
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGS58359
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AGS58359
Cps21B
Accession: AGS58360
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AGS58360
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AGS58360
Cps21C
Accession: AGS58361
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: AGS58361
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
AGS58361
Cps21D
Accession: AGS58362
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
Accession: AGS58362
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
AGS58362
AGS58363
AGS58364
AGS58365
AGS58366
AGS58367
AGS58368
AGS58369
AGS58370
AGS58371
AGS58372
346. : AB737827 Streptococcus suis DNA, capsular polysaccharide locus, strain: 14A. Total score: 5.5 Cumulative Blast bit score: 2078
BAM94809
BAM94810
predicted transcriptional regulator of pyridoxine metabolism
Accession: BAM94811
Location: 1670-2929
NCBI BlastP on this gene
Accession: BAM94811
Location: 1670-2929
NCBI BlastP on this gene
BAM94811
BAM94812
capsular polysaccharide expression regulator
Accession: BAM94813
Location: 3912-5351
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BAM94813
Location: 3912-5351
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps21A
chain length determinant protein/polysaccharide export protein
Accession: BAM94814
Location: 5368-6057
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: BAM94814
Location: 5368-6057
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
cps21B
tyrosine-protein kinase
Accession: BAM94815
Location: 6067-6753
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: BAM94815
Location: 6067-6753
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
cps21C
protein-tyrosine phosphatase
Accession: BAM94816
Location: 6791-7522
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
Accession: BAM94816
Location: 6791-7522
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 412
Sequence coverage: 100 %
E-value: 7e-143
NCBI BlastP on this gene
cps21D
predicted nucleoside-diphosphate sugar epimerase
Accession: BAM94817
Location: 7551-9374
NCBI BlastP on this gene
Accession: BAM94817
Location: 7551-9374
NCBI BlastP on this gene
cps21E
cps21F
cps21G
NAD dependent epimerase/dehydratase family protein
Accession: BAM94820
Location: 10787-11641
NCBI BlastP on this gene
Accession: BAM94820
Location: 10787-11641
NCBI BlastP on this gene
cps21H
cps21I
cps21J
cps21K
cps21L
cps21M
cps21N
347. : KC537386 Streptococcus suis strain 92-1191 capsular polysaccharide synthesis locus sequence. Total score: 5.5 Cumulative Blast bit score: 2076
AGS58378
AGS58379
AGS58380
Cps29A
Accession: AGS58381
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AGS58381
Location: 3351-4790
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AGS58381
Cps29B
Accession: AGS58382
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AGS58382
Location: 4807-5496
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AGS58382
Cps29C
Accession: AGS58383
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
Accession: AGS58383
Location: 5506-6192
BlastP hit with ABP89532.1
Percentage identity: 88 %
BlastP bit score: 406
Sequence coverage: 96 %
E-value: 5e-141
NCBI BlastP on this gene
AGS58383
Cps29D
Accession: AGS58384
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
Accession: AGS58384
Location: 6230-6961
BlastP hit with ABP89533.1
Percentage identity: 84 %
BlastP bit score: 410
Sequence coverage: 100 %
E-value: 4e-142
NCBI BlastP on this gene
AGS58384
AGS58385
AGS58386
AGS58387
AGS58388
AGS58389
AGS58390
AGS58391
AGS58392
AGS58393
348. : JF791155 Streptococcus suis strain 93 ORFY, ORFX, CpsA, CpsB, CpsC, and CpsD genes Total score: 5.5 Cumulative Blast bit score: 2076
AEP02663
AEP02664
CpsA
Accession: AEP02665
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02665
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02665
CpsB
Accession: AEP02666
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AEP02666
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AEP02666
CpsC
Accession: AEP02667
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
Accession: AEP02667
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
AEP02667
CpsD
Accession: AEP02668
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AEP02668
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
AEP02668
349. : JF791154 Streptococcus suis strain NT77 ORFY, ORFX, CpsA, CpsB, CpsC, and CpsD genes Total score: 5.5 Cumulative Blast bit score: 2076
AEP02657
AEP02658
CpsA
Accession: AEP02659
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEP02659
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
AEP02659
CpsB
Accession: AEP02660
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
Accession: AEP02660
Location: 4549-5238
BlastP hit with ABP89531.1
Percentage identity: 86 %
BlastP bit score: 394
Sequence coverage: 100 %
E-value: 4e-136
NCBI BlastP on this gene
AEP02660
CpsC
Accession: AEP02661
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
Accession: AEP02661
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
AEP02661
CpsD
Accession: AEP02662
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
Accession: AEP02662
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 86 %
BlastP bit score: 416
Sequence coverage: 100 %
E-value: 1e-144
NCBI BlastP on this gene
AEP02662
350. : JF273654 Streptococcus suis strain 42A ORF19Z gene Total score: 5.5 Cumulative Blast bit score: 2076
AEH57559
AEH57560
AEH57561
Cps19A
Accession: AEH57562
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
Accession: AEH57562
Location: 3093-4532
BlastP hit with ABP89530.1
Percentage identity: 89 %
BlastP bit score: 866
Sequence coverage: 98 %
E-value: 0.0
NCBI BlastP on this gene
cps19A
Cps19B
Accession: AEH57563
Location: 4522-5238
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 396
Sequence coverage: 100 %
E-value: 7e-137
NCBI BlastP on this gene
Accession: AEH57563
Location: 4522-5238
BlastP hit with ABP89531.1
Percentage identity: 87 %
BlastP bit score: 396
Sequence coverage: 100 %
E-value: 7e-137
NCBI BlastP on this gene
cps19B
Cps19C
Accession: AEH57564
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
Accession: AEH57564
Location: 5248-5934
BlastP hit with ABP89532.1
Percentage identity: 87 %
BlastP bit score: 400
Sequence coverage: 96 %
E-value: 1e-138
NCBI BlastP on this gene
cps19C
Cps19D
Accession: AEH57565
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
Accession: AEH57565
Location: 5972-6703
BlastP hit with ABP89533.1
Percentage identity: 85 %
BlastP bit score: 414
Sequence coverage: 100 %
E-value: 2e-143
NCBI BlastP on this gene
cps19D
cps19E
cps19F
cps19G
cps19H
cps19I
cps19J
cps19K
cps19L
cps19M
cps19N


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.