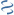MultiGeneBlast hits

Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP013120 : Alteromonas stellipolaris LMG 21856 Total score: 1.0 Cumulative Blast bit score: 165
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
AOR13_381
AOR13_382
AOR13_383
AOR13_384
Protein secretion chaperonin CsaA
Accession: ALM89437
Location: 486893-487228
NCBI BlastP on this gene
Accession: ALM89437
Location: 486893-487228
NCBI BlastP on this gene
AOR13_385
Alginate lyase precursor
Accession: ALM89438
Location: 487733-489268
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 165
Sequence coverage: 99 %
E-value: 2e-42
NCBI BlastP on this gene
Accession: ALM89438
Location: 487733-489268
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 165
Sequence coverage: 99 %
E-value: 2e-42
NCBI BlastP on this gene
AOR13_386
Glyoxalase/bleomycin resistance
Accession: ALM89439
Location: 489371-489706
NCBI BlastP on this gene
Accession: ALM89439
Location: 489371-489706
NCBI BlastP on this gene
AOR13_387
AOR13_388
AOR13_389
AOR13_390
AOR13_391
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
LR136958 : Alteromonas sp. 76-1 genome assembly, chromosome: cAlt76. Total score: 1.0 Cumulative Blast bit score: 164
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
glucose-6-phosphate isomerase
Accession: VEL98201
Location: 3655182-3656831
NCBI BlastP on this gene
Accession: VEL98201
Location: 3655182-3656831
NCBI BlastP on this gene
ALT761_03218
ALT761_03217
ALT761_03216
ALT761_03215
ALT761_03214
alginate lyase
Accession: VEL98196
Location: 3650131-3651666
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 164
Sequence coverage: 99 %
E-value: 4e-42
NCBI BlastP on this gene
Accession: VEL98196
Location: 3650131-3651666
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 164
Sequence coverage: 99 %
E-value: 4e-42
NCBI BlastP on this gene
ALT761_03213
ALT761_03212
ALT761_03211
ALT761_03210
uncharacterized protein DUF4426
Accession: VEL98192
Location: 3645636-3646076
NCBI BlastP on this gene
Accession: VEL98192
Location: 3645636-3646076
NCBI BlastP on this gene
ALT761_03209
ALT761_03208
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP002339 : Alteromonas naphthalenivorans strain SN2 Total score: 1.0 Cumulative Blast bit score: 161
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
pgi
ambt_01980
ambt_01985
putative glutathione S-transferase protein
Accession: AEF01952
Location: 466470-467108
NCBI BlastP on this gene
Accession: AEF01952
Location: 466470-467108
NCBI BlastP on this gene
ambt_01990
ambt_01995
putative alginate lyase
Accession: AEF01954
Location: 468232-469767
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 161
Sequence coverage: 99 %
E-value: 4e-41
NCBI BlastP on this gene
Accession: AEF01954
Location: 468232-469767
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 161
Sequence coverage: 99 %
E-value: 4e-41
NCBI BlastP on this gene
ambt_02000
Glyoxalase/bleomycin resistance
Accession: AEF01955
Location: 469870-470205
NCBI BlastP on this gene
Accession: AEF01955
Location: 469870-470205
NCBI BlastP on this gene
ambt_02005
ambt_02010
ambt_02015
ambt_02020
ambt_02025
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP038493 : Thalassotalea sp. HSM 43 chromosome Total score: 1.0 Cumulative Blast bit score: 160
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: QBY04449
Location: 2010122-2011183
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 103 %
E-value: 5e-42
NCBI BlastP on this gene
Accession: QBY04449
Location: 2010122-2011183
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 103 %
E-value: 5e-42
NCBI BlastP on this gene
E2K93_08615
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP034660 : Colwellia sp. Arc7-635 chromosome Total score: 1.0 Cumulative Blast bit score: 160
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: AZQ84888
Location: 3082695-3083741
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 98 %
E-value: 6e-42
NCBI BlastP on this gene
Accession: AZQ84888
Location: 3082695-3083741
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 98 %
E-value: 6e-42
NCBI BlastP on this gene
EKO29_13345
EKO29_13340
siderophore-interacting protein
Accession: AZQ84886
Location: 3080956-3081687
NCBI BlastP on this gene
Accession: AZQ84886
Location: 3080956-3081687
NCBI BlastP on this gene
EKO29_13335
MarR family transcriptional regulator
Accession: AZQ84885
Location: 3080214-3080630
NCBI BlastP on this gene
Accession: AZQ84885
Location: 3080214-3080630
NCBI BlastP on this gene
EKO29_13330
multidrug efflux SMR transporter
Accession: AZQ84884
Location: 3079573-3079887
NCBI BlastP on this gene
Accession: AZQ84884
Location: 3079573-3079887
NCBI BlastP on this gene
EKO29_13325
ACT domain-containing protein
Accession: AZQ84883
Location: 3079107-3079520
NCBI BlastP on this gene
Accession: AZQ84883
Location: 3079107-3079520
NCBI BlastP on this gene
EKO29_13320
EKO29_13315
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP028924 : Colwellia sp. Arc7-D chromosome Total score: 1.0 Cumulative Blast bit score: 160
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
DBO93_17415
DUF2489 domain-containing protein
Accession: AWB59163
Location: 4012787-4013287
NCBI BlastP on this gene
Accession: AWB59163
Location: 4012787-4013287
NCBI BlastP on this gene
DBO93_17420
oxygen-independent coproporphyrinogen III oxidase
Accession: AWB59164
Location: 4013520-4014890
NCBI BlastP on this gene
Accession: AWB59164
Location: 4013520-4014890
NCBI BlastP on this gene
hemN
DUF1285 domain-containing protein
Accession: AWB59165
Location: 4014898-4015440
NCBI BlastP on this gene
Accession: AWB59165
Location: 4014898-4015440
NCBI BlastP on this gene
DBO93_17430
polysaccharide lyase family 7 protein
Accession: AWB59166
Location: 4016576-4017649
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
Accession: AWB59166
Location: 4016576-4017649
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
DBO93_17440
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP020465 : Colwellia beringensis strain NB097-1 chromosome Total score: 1.0 Cumulative Blast bit score: 159
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: ASP49898
Location: 1218528-1219487
NCBI BlastP on this gene
Accession: ASP49898
Location: 1218528-1219487
NCBI BlastP on this gene
B5D82_05170
B5D82_05165
B5D82_05160
B5D82_05155
polysaccharide lyase family 7 protein
Accession: ASP47198
Location: 1214236-1215312
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 159
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
Accession: ASP47198
Location: 1214236-1215312
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 159
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
B5D82_05150
B5D82_05145
B5D82_05140
B5D82_05135
NAD(P)-dependent oxidoreductase
Accession: ASP47195
Location: 1210757-1211620
NCBI BlastP on this gene
Accession: ASP47195
Location: 1210757-1211620
NCBI BlastP on this gene
B5D82_05130
B5D82_05125
B5D82_05120
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP019419 : Polaribacter reichenbachii strain 6Alg 8T Total score: 1.0 Cumulative Blast bit score: 158
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
BW723_16210
long-chain fatty acid--CoA ligase
Accession: APZ47745
Location: 3791698-3793488
NCBI BlastP on this gene
Accession: APZ47745
Location: 3791698-3793488
NCBI BlastP on this gene
BW723_16215
BW723_16220
BW723_16225
BW723_16230
alginate lyase
Accession: APZ47749
Location: 3795336-3798014
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
Accession: APZ47749
Location: 3795336-3798014
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
BW723_16235
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP019337 : Polaribacter reichenbachii strain KCTC 23969 chromosome. Total score: 1.0 Cumulative Blast bit score: 158
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
BTO17_06630
long-chain fatty acid--CoA ligase
Accession: AUC18380
Location: 1517621-1519411
NCBI BlastP on this gene
Accession: AUC18380
Location: 1517621-1519411
NCBI BlastP on this gene
BTO17_06635
BTO17_06640
BTO17_06645
BTO17_06650
alginate lyase
Accession: AUC18384
Location: 1521259-1523937
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
Accession: AUC18384
Location: 1521259-1523937
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
BTO17_06655
BTO17_06660
BTO17_06665
BTO17_06670
BTO17_06675
BTO17_06680
Arc family DNA binding domain-containing protein
Accession: AUC18389
Location: 1527518-1527682
NCBI BlastP on this gene
Accession: AUC18389
Location: 1527518-1527682
NCBI BlastP on this gene
BTO17_06685
BTO17_06690
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP026604 : Catenovulum sp. CCB-QB4 chromosome Total score: 1.0 Cumulative Blast bit score: 155
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
C2869_09515
cupin domain-containing protein
Accession: AWB66656
Location: 2703613-2703960
NCBI BlastP on this gene
Accession: AWB66656
Location: 2703613-2703960
NCBI BlastP on this gene
C2869_09520
C2869_09525
sugar phosphate isomerase/epimerase
Accession: AWB66658
Location: 2705917-2706837
NCBI BlastP on this gene
Accession: AWB66658
Location: 2705917-2706837
NCBI BlastP on this gene
C2869_09530
AraC family transcriptional regulator
Accession: AWB66659
Location: 2706987-2707739
NCBI BlastP on this gene
Accession: AWB66659
Location: 2706987-2707739
NCBI BlastP on this gene
C2869_09535
antibiotic biosynthesis monooxygenase
Accession: AWB68984
Location: 2707736-2708038
NCBI BlastP on this gene
Accession: AWB68984
Location: 2707736-2708038
NCBI BlastP on this gene
C2869_09540
polysaccharide lyase family 7 protein
Accession: AWB66660
Location: 2708351-2709472
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 155
Sequence coverage: 98 %
E-value: 8e-40
NCBI BlastP on this gene
Accession: AWB66660
Location: 2708351-2709472
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 155
Sequence coverage: 98 %
E-value: 8e-40
NCBI BlastP on this gene
C2869_09545
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP010912 : Alteromonas australica strain DE170 Total score: 1.0 Cumulative Blast bit score: 154
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
EP12_15295
EP12_15290
EP12_15285
XRE family transcriptional regulator
Accession: AJP44803
Location: 3350176-3350703
NCBI BlastP on this gene
Accession: AJP44803
Location: 3350176-3350703
NCBI BlastP on this gene
EP12_15280
branched-chain amino acid permease
Accession: AJP44802
Location: 3349457-3350179
NCBI BlastP on this gene
Accession: AJP44802
Location: 3349457-3350179
NCBI BlastP on this gene
EP12_15275
branched-chain amino acid ABC transporter
Accession: AJP44801
Location: 3349120-3349467
NCBI BlastP on this gene
Accession: AJP44801
Location: 3349120-3349467
NCBI BlastP on this gene
EP12_15270
EP12_15265
alginate lyase
Accession: AJP44799
Location: 3346678-3348213
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
Accession: AJP44799
Location: 3346678-3348213
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
EP12_15260
EP12_15255
EP12_15240
EP12_15235
pyrroline-5-carboxylate reductase
Accession: AJP44795
Location: 3341870-3342691
NCBI BlastP on this gene
Accession: AJP44795
Location: 3341870-3342691
NCBI BlastP on this gene
EP12_15230
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP008849 : Alteromonas australica strain H 17 Total score: 1.0 Cumulative Blast bit score: 154
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
EP13_14730
EP13_14725
EP13_14720
XRE family transcriptional regulator
Accession: AIF99835
Location: 3271977-3272504
NCBI BlastP on this gene
Accession: AIF99835
Location: 3271977-3272504
NCBI BlastP on this gene
EP13_14715
branched-chain amino acid permease
Accession: AIF99834
Location: 3271258-3271980
NCBI BlastP on this gene
Accession: AIF99834
Location: 3271258-3271980
NCBI BlastP on this gene
EP13_14710
branched-chain amino acid ABC transporter
Accession: AIF99833
Location: 3270921-3271268
NCBI BlastP on this gene
Accession: AIF99833
Location: 3270921-3271268
NCBI BlastP on this gene
EP13_14705
EP13_14700
alginate lyase
Accession: AIF99831
Location: 3268479-3270014
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
Accession: AIF99831
Location: 3268479-3270014
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
EP13_14695
EP13_14690
EP13_14685
fadH
EP13_14675
EP13_14670
pyrroline-5-carboxylate reductase
Accession: AIF99825
Location: 3263660-3264481
NCBI BlastP on this gene
Accession: AIF99825
Location: 3263660-3264481
NCBI BlastP on this gene
EP13_14665
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP045859 : Vibrio owensii strain SH14 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: QGH47617
Location: 2339593-2340663
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
Accession: QGH47617
Location: 2339593-2340663
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
APZ19_11120
katG
APZ19_11110
uxaC
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP033144 : Vibrio owensii strain V180403 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: AYO20327
Location: 1980872-1981942
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 3e-39
NCBI BlastP on this gene
Accession: AYO20327
Location: 1980872-1981942
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 3e-39
NCBI BlastP on this gene
D0856_09680
katG
D0856_09670
D0856_09665
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP033137 : Vibrio owensii strain 1700302 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
CBS domain-containing protein
Accession: AYO14702
Location: 2057907-2059769
NCBI BlastP on this gene
Accession: AYO14702
Location: 2057907-2059769
NCBI BlastP on this gene
D0812_09910
FadR family transcriptional regulator
Accession: AYO14703
Location: 2059943-2060662
NCBI BlastP on this gene
Accession: AYO14703
Location: 2059943-2060662
NCBI BlastP on this gene
D0812_09915
D0812_09920
FadR family transcriptional regulator
Accession: AYO14705
Location: 2061927-2062643
NCBI BlastP on this gene
Accession: AYO14705
Location: 2061927-2062643
NCBI BlastP on this gene
D0812_09925
MATE family efflux transporter
Accession: AYO14706
Location: 2062758-2064116
NCBI BlastP on this gene
Accession: AYO14706
Location: 2062758-2064116
NCBI BlastP on this gene
D0812_09930
polysaccharide lyase family 7 protein
Accession: AYO14707
Location: 2064377-2065447
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
Accession: AYO14707
Location: 2064377-2065447
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
D0812_09935
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP019959 : Vibrio owensii strain XSBZ03 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
A9237_02610
GntR family transcriptional regulator
Accession: AQW57073
Location: 598004-598723
NCBI BlastP on this gene
Accession: AQW57073
Location: 598004-598723
NCBI BlastP on this gene
A9237_02605
A9237_02600
GntR family transcriptional regulator
Accession: AQW57071
Location: 596023-596739
NCBI BlastP on this gene
Accession: AQW57071
Location: 596023-596739
NCBI BlastP on this gene
A9237_02595
A9237_02590
polysaccharide lyase family 7 protein
Accession: AQW57069
Location: 593219-594289
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
Accession: AQW57069
Location: 593219-594289
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
A9237_02585
A9237_02580
A9237_02575
A9237_02570
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP010912 : Alteromonas australica strain DE170 Total score: 1.0 Cumulative Blast bit score: 152
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
EP12_16265
EP12_16270
EP12_16275
alginate lyase
Accession: AJP44971
Location: 3606913-3608016
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 152
Sequence coverage: 105 %
E-value: 6e-39
NCBI BlastP on this gene
Accession: AJP44971
Location: 3606913-3608016
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 152
Sequence coverage: 105 %
E-value: 6e-39
NCBI BlastP on this gene
EP12_16280
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
HG315671 : Formosa agariphila KMM 3901 Total score: 1.0 Cumulative Blast bit score: 150
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
oxidoreductase of aldo/keto reductase family, subgroup 1
Accession: CDF77716
Location: 2722-3564
NCBI BlastP on this gene
Accession: CDF77716
Location: 2722-3564
NCBI BlastP on this gene
BN863_40
BN863_50
BN863_60
alginate lyase precursor
Accession: CDF77719
Location: 7374-10481
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 53 %
E-value: 5e-34
NCBI BlastP on this gene
Accession: CDF77719
Location: 7374-10481
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 53 %
E-value: 5e-34
NCBI BlastP on this gene
BN863_70
BN863_80
iron-sulfur cluster-binding protein
Accession: CDF77721
Location: 11978-13549
NCBI BlastP on this gene
Accession: CDF77721
Location: 11978-13549
NCBI BlastP on this gene
BN863_90
BN863_100
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP039700 : Vibrio cyclitrophicus strain ECSMB14105 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 150
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
FAZ90_06520
FAZ90_06525
FAZ90_06530
polysaccharide lyase family 7 protein
Accession: QCI70745
Location: 1432843-1433880
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 150
Sequence coverage: 97 %
E-value: 3e-38
NCBI BlastP on this gene
Accession: QCI70745
Location: 1432843-1433880
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 150
Sequence coverage: 97 %
E-value: 3e-38
NCBI BlastP on this gene
FAZ90_06535
MATE family efflux transporter
Accession: QCI70746
Location: 1434249-1435616
NCBI BlastP on this gene
Accession: QCI70746
Location: 1434249-1435616
NCBI BlastP on this gene
FAZ90_06540
FadR family transcriptional regulator
Accession: QCI70747
Location: 1435852-1436574
NCBI BlastP on this gene
Accession: QCI70747
Location: 1435852-1436574
NCBI BlastP on this gene
FAZ90_06545
FAZ90_06550
FadR family transcriptional regulator
Accession: QCI70749
Location: 1437938-1438657
NCBI BlastP on this gene
Accession: QCI70749
Location: 1437938-1438657
NCBI BlastP on this gene
FAZ90_06555
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP019336 : Polaribacter sejongensis strain KCTC 23670 chromosome. Total score: 1.0 Cumulative Blast bit score: 150
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
BTO15_02835
BTO15_02840
BTO15_02845
BTO15_02850
BTO15_02855
BTO15_02860
BTO15_02865
alginate lyase
Accession: AUC21121
Location: 763957-766617
BlastP hit with alyA4
Percentage identity: 32 %
BlastP bit score: 150
Sequence coverage: 51 %
E-value: 6e-34
NCBI BlastP on this gene
Accession: AUC21121
Location: 763957-766617
BlastP hit with alyA4
Percentage identity: 32 %
BlastP bit score: 150
Sequence coverage: 51 %
E-value: 6e-34
NCBI BlastP on this gene
BTO15_02870
BTO15_02875
1-pyrroline-5-carboxylate dehydrogenase
Accession: AUC21123
Location: 767532-769157
NCBI BlastP on this gene
Accession: AUC21123
Location: 767532-769157
NCBI BlastP on this gene
BTO15_02880
BTO15_02885
BTO15_02890
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP016228 : Vibrio crassostreae 9CS106 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 149
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
A134_08070
A134_08075
A134_08080
alginate lyase
Accession: ANP76358
Location: 1774280-1775317
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 96 %
E-value: 1e-37
NCBI BlastP on this gene
Accession: ANP76358
Location: 1774280-1775317
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 96 %
E-value: 1e-37
NCBI BlastP on this gene
A134_08085
A134_08090
GntR family transcriptional regulator
Accession: ANP76360
Location: 1777064-1777786
NCBI BlastP on this gene
Accession: ANP76360
Location: 1777064-1777786
NCBI BlastP on this gene
A134_08095
A134_08100
GntR family transcriptional regulator
Accession: ANP76362
Location: 1779080-1779799
NCBI BlastP on this gene
Accession: ANP76362
Location: 1779080-1779799
NCBI BlastP on this gene
A134_08105
A134_08110
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP031769 : Salinimonas sediminis strain N102 chromosome Total score: 1.0 Cumulative Blast bit score: 144
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
D0Y50_13595
cupin domain-containing protein
Accession: AXR07288
Location: 3061656-3062000
NCBI BlastP on this gene
Accession: AXR07288
Location: 3061656-3062000
NCBI BlastP on this gene
D0Y50_13600
NAD(P)-dependent oxidoreductase
Accession: AXR07289
Location: 3062137-3063015
NCBI BlastP on this gene
Accession: AXR07289
Location: 3062137-3063015
NCBI BlastP on this gene
D0Y50_13605
D0Y50_13610
polysaccharide lyase family 7 protein
Accession: AXR07290
Location: 3064666-3065787
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 110 %
E-value: 1e-35
NCBI BlastP on this gene
Accession: AXR07290
Location: 3064666-3065787
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 110 %
E-value: 1e-35
NCBI BlastP on this gene
D0Y50_13615
LacI family transcriptional regulator
Accession: D0Y50_13620
Location: 3065863-3066899
NCBI BlastP on this gene
Accession: D0Y50_13620
Location: 3065863-3066899
NCBI BlastP on this gene
D0Y50_13620
cysE
trmJ
D0Y50_13635
protein translocase subunit SecF
Accession: AXR07294
Location: 3070519-3071460
NCBI BlastP on this gene
Accession: AXR07294
Location: 3070519-3071460
NCBI BlastP on this gene
secF
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP019650 : Microbulbifer agarilyticus strain GP101 Total score: 1.0 Cumulative Blast bit score: 144
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
Mag101_17585
Mag101_17590
Mag101_17595
Mag101_17600
Mag101_17605
Mag101_17610
polysaccharide lyase family 7 protein
Accession: AQQ69248
Location: 4212960-4214051
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 144
Sequence coverage: 107 %
E-value: 9e-36
NCBI BlastP on this gene
Accession: AQQ69248
Location: 4212960-4214051
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 144
Sequence coverage: 107 %
E-value: 9e-36
NCBI BlastP on this gene
Mag101_17615
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP016177 : Vibrio breoganii strain FF50 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 144
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
A6E01_10035
A6E01_10040
LysR family transcriptional regulator
Accession: ANO33545
Location: 2179265-2180170
NCBI BlastP on this gene
Accession: ANO33545
Location: 2179265-2180170
NCBI BlastP on this gene
A6E01_10050
A6E01_10055
A6E01_10060
A6E01_10065
alginate lyase
Accession: ANO33549
Location: 2182537-2183580
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 144
Sequence coverage: 96 %
E-value: 5e-36
NCBI BlastP on this gene
Accession: ANO33549
Location: 2182537-2183580
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 144
Sequence coverage: 96 %
E-value: 5e-36
NCBI BlastP on this gene
A6E01_10070
2-deoxy-D-gluconate 3-dehydrogenase
Accession: ANO33550
Location: 2183904-2184665
NCBI BlastP on this gene
Accession: ANO33550
Location: 2183904-2184665
NCBI BlastP on this gene
A6E01_10075
A6E01_10080
A6E01_10085
A6E01_10090
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP000282 : Saccharophagus degradans 2-40 Total score: 1.0 Cumulative Blast bit score: 143
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
Sde_2842
ABC transporter, inner membrane protein
Accession: ABD82098
Location: 3588666-3591599
NCBI BlastP on this gene
Accession: ABD82098
Location: 3588666-3591599
NCBI BlastP on this gene
Sde_2841
Sde_2840
putative polysaccharide lyase
Accession: ABD82096
Location: 3586312-3587415
BlastP hit with alyA5
Percentage identity: 36 %
BlastP bit score: 143
Sequence coverage: 91 %
E-value: 1e-35
NCBI BlastP on this gene
Accession: ABD82096
Location: 3586312-3587415
BlastP hit with alyA5
Percentage identity: 36 %
BlastP bit score: 143
Sequence coverage: 91 %
E-value: 1e-35
NCBI BlastP on this gene
ply7E
Sde_2838
Sde_2837
Sde_2836
Sde_2835
gly81A
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP036200 : Shewanella maritima strain D4-2 chromosome Total score: 1.0 Cumulative Blast bit score: 142
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
EXU30_08335
EXU30_08340
EXU30_08345
EXU30_08350
EXU30_08355
polysaccharide lyase family 7 protein
Accession: QBF82700
Location: 1958329-1959390
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 107 %
E-value: 4e-35
NCBI BlastP on this gene
Accession: QBF82700
Location: 1958329-1959390
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 107 %
E-value: 4e-35
NCBI BlastP on this gene
EXU30_08360
ammonia-forming cytochrome c nitrite reductase subunit c552
Accession: QBF82701
Location: 1960113-1961768
NCBI BlastP on this gene
Accession: QBF82701
Location: 1960113-1961768
NCBI BlastP on this gene
EXU30_08365
EXU30_08370
N-acetylneuraminic acid outer membrane channel protein NanC
Accession: QBF84846
Location: 1964039-1964698
NCBI BlastP on this gene
Accession: QBF84846
Location: 1964039-1964698
NCBI BlastP on this gene
EXU30_08375
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP012898 : Algibacter alginicilyticus strain HZ22 chromosome Total score: 1.0 Cumulative Blast bit score: 137
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
APS56_07990
alginate lyase
Accession: ALJ05070
Location: 1924768-1927443
BlastP hit with alyA6
Percentage identity: 33 %
BlastP bit score: 137
Sequence coverage: 39 %
E-value: 7e-30
NCBI BlastP on this gene
Accession: ALJ05070
Location: 1924768-1927443
BlastP hit with alyA6
Percentage identity: 33 %
BlastP bit score: 137
Sequence coverage: 39 %
E-value: 7e-30
NCBI BlastP on this gene
APS56_07995
APS56_08000
APS56_08005
APS56_08010
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP036200 : Shewanella maritima strain D4-2 chromosome Total score: 1.0 Cumulative Blast bit score: 134
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
EXU30_09990
EXU30_09995
polysaccharide lyase family 7 protein
Accession: QBF82988
Location: 2350649-2351587
NCBI BlastP on this gene
Accession: QBF82988
Location: 2350649-2351587
NCBI BlastP on this gene
EXU30_10000
EXU30_10005
polysaccharide lyase family 7 protein
Accession: QBF82989
Location: 2353038-2354108
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 134
Sequence coverage: 109 %
E-value: 4e-32
NCBI BlastP on this gene
Accession: QBF82989
Location: 2353038-2354108
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 134
Sequence coverage: 109 %
E-value: 4e-32
NCBI BlastP on this gene
EXU30_10010
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP033144 : Vibrio owensii strain V180403 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 133
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
lipopolysaccharide assembly protein LapB
Accession: AYO19304
Location: 765644-766819
NCBI BlastP on this gene
Accession: AYO19304
Location: 765644-766819
NCBI BlastP on this gene
lapB
DUF1049 domain-containing protein
Accession: AYO19303
Location: 765332-765625
NCBI BlastP on this gene
Accession: AYO19303
Location: 765332-765625
NCBI BlastP on this gene
D0856_03575
integration host factor subunit beta
Accession: AYO19302
Location: 764907-765191
NCBI BlastP on this gene
Accession: AYO19302
Location: 764907-765191
NCBI BlastP on this gene
ihfB
D0856_03565
D0856_03560
polysaccharide lyase family 7 protein
Accession: AYO19299
Location: 760895-761938
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 133
Sequence coverage: 97 %
E-value: 7e-32
NCBI BlastP on this gene
Accession: AYO19299
Location: 760895-761938
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 133
Sequence coverage: 97 %
E-value: 7e-32
NCBI BlastP on this gene
D0856_03555
sohB
D0856_03545
translesion DNA synthesis-associated protein ImuA
Accession: AYO19296
Location: 757906-758598
NCBI BlastP on this gene
Accession: AYO19296
Location: 757906-758598
NCBI BlastP on this gene
imuA
DNA polymerase Y family protein
Accession: AYO19295
Location: 756498-757901
NCBI BlastP on this gene
Accession: AYO19295
Location: 756498-757901
NCBI BlastP on this gene
D0856_03535
DNA polymerase III subunit alpha
Accession: AYO19294
Location: 753396-756470
NCBI BlastP on this gene
Accession: AYO19294
Location: 753396-756470
NCBI BlastP on this gene
dnaE
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
LT629752 : Polaribacter sp. KT25b genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 132
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
SAMN05216503_2576
uncharacterized radical SAM protein YgiQ
Accession: SDS28860
Location: 2899844-2901805
NCBI BlastP on this gene
Accession: SDS28860
Location: 2899844-2901805
NCBI BlastP on this gene
SAMN05216503_2577
SAMN05216503_2578
Por secretion system C-terminal sorting domain-containing protein
Accession: SDS28918
Location: 2903529-2906090
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 132
Sequence coverage: 42 %
E-value: 3e-28
NCBI BlastP on this gene
Accession: SDS28918
Location: 2903529-2906090
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 132
Sequence coverage: 42 %
E-value: 3e-28
NCBI BlastP on this gene
SAMN05216503_2579
SAMN05216503_2580
SAMN05216503_2581
SAMN05216503_2582
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP019959 : Vibrio owensii strain XSBZ03 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 132
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: AQW59088
Location: 3022601-3023644
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 132
Sequence coverage: 97 %
E-value: 1e-31
NCBI BlastP on this gene
Accession: AQW59088
Location: 3022601-3023644
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 132
Sequence coverage: 97 %
E-value: 1e-31
NCBI BlastP on this gene
A9237_13920
A9237_13915
A9237_13910
A9237_13905
A9237_13900
A9237_13895
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP045859 : Vibrio owensii strain SH14 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 130
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
lipopolysaccharide assembly protein LapB
Accession: QGH46642
Location: 1218862-1220037
NCBI BlastP on this gene
Accession: QGH46642
Location: 1218862-1220037
NCBI BlastP on this gene
lapB
DUF1049 domain-containing protein
Accession: QGH46641
Location: 1218550-1218843
NCBI BlastP on this gene
Accession: QGH46641
Location: 1218550-1218843
NCBI BlastP on this gene
APZ19_05760
integration host factor subunit beta
Accession: QGH46640
Location: 1218125-1218409
NCBI BlastP on this gene
Accession: QGH46640
Location: 1218125-1218409
NCBI BlastP on this gene
ihfB
rpsA
APZ19_05745
polysaccharide lyase family 7 protein
Accession: QGH46637
Location: 1214113-1215156
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
Accession: QGH46637
Location: 1214113-1215156
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
APZ19_05740
sohB
APZ19_05730
translesion DNA synthesis-associated protein ImuA
Accession: QGH46634
Location: 1211126-1211818
NCBI BlastP on this gene
Accession: QGH46634
Location: 1211126-1211818
NCBI BlastP on this gene
imuA
DNA polymerase Y family protein
Accession: QGH46633
Location: 1209718-1211121
NCBI BlastP on this gene
Accession: QGH46633
Location: 1209718-1211121
NCBI BlastP on this gene
APZ19_05720
DNA polymerase III subunit alpha
Accession: QGH46632
Location: 1206616-1209690
NCBI BlastP on this gene
Accession: QGH46632
Location: 1206616-1209690
NCBI BlastP on this gene
dnaE
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP033137 : Vibrio owensii strain 1700302 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 130
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
lipopolysaccharide assembly protein LapB
Accession: AYO13986
Location: 1253812-1254987
NCBI BlastP on this gene
Accession: AYO13986
Location: 1253812-1254987
NCBI BlastP on this gene
lapB
DUF1049 domain-containing protein
Accession: AYO13985
Location: 1253500-1253793
NCBI BlastP on this gene
Accession: AYO13985
Location: 1253500-1253793
NCBI BlastP on this gene
D0812_05955
integration host factor subunit beta
Accession: AYO13984
Location: 1253075-1253359
NCBI BlastP on this gene
Accession: AYO13984
Location: 1253075-1253359
NCBI BlastP on this gene
ihfB
D0812_05945
D0812_05940
polysaccharide lyase family 7 protein
Accession: AYO13981
Location: 1249063-1250106
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
Accession: AYO13981
Location: 1249063-1250106
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
D0812_05935
sohB
D0812_05925
translesion DNA synthesis-associated protein ImuA
Accession: AYO13978
Location: 1246076-1246768
NCBI BlastP on this gene
Accession: AYO13978
Location: 1246076-1246768
NCBI BlastP on this gene
imuA
DNA polymerase Y family protein
Accession: AYO13977
Location: 1244668-1246071
NCBI BlastP on this gene
Accession: AYO13977
Location: 1244668-1246071
NCBI BlastP on this gene
D0812_05915
DNA polymerase III subunit alpha
Accession: AYO13976
Location: 1241566-1244640
NCBI BlastP on this gene
Accession: AYO13976
Location: 1241566-1244640
NCBI BlastP on this gene
dnaE
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP016177 : Vibrio breoganii strain FF50 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 130
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
alginate lyase
Accession: ANO33567
Location: 2204842-2205984
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 130
Sequence coverage: 78 %
E-value: 1e-30
NCBI BlastP on this gene
Accession: ANO33567
Location: 2204842-2205984
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 130
Sequence coverage: 78 %
E-value: 1e-30
NCBI BlastP on this gene
A6E01_10160
A6E01_10155
A6E01_10150
A6E01_10145
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP039700 : Vibrio cyclitrophicus strain ECSMB14105 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 119
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
polysaccharide lyase family 7 protein
Accession: QCI71698
Location: 2580371-2581408
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 120
Sequence coverage: 77 %
E-value: 3e-27
NCBI BlastP on this gene
Accession: QCI71698
Location: 2580371-2581408
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 120
Sequence coverage: 77 %
E-value: 3e-27
NCBI BlastP on this gene
FAZ90_11735
dnaK
dnaJ
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: QCI72380
Location: 2576070-2576501
NCBI BlastP on this gene
Accession: QCI72380
Location: 2576070-2576501
NCBI BlastP on this gene
FAZ90_11720
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: QCI71695
Location: 2575467-2575967
NCBI BlastP on this gene
Accession: QCI71695
Location: 2575467-2575967
NCBI BlastP on this gene
FAZ90_11715
FAZ90_11710
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP016228 : Vibrio crassostreae 9CS106 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 119
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
alginate lyase
Accession: ANP77352
Location: 2960232-2961269
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 119
Sequence coverage: 77 %
E-value: 4e-27
NCBI BlastP on this gene
Accession: ANP77352
Location: 2960232-2961269
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 119
Sequence coverage: 77 %
E-value: 4e-27
NCBI BlastP on this gene
A134_13460
A134_13455
A134_13450
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: ANP77349
Location: 2955937-2956377
NCBI BlastP on this gene
Accession: ANP77349
Location: 2955937-2956377
NCBI BlastP on this gene
A134_13445
A134_13440
A134_13435
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP028924 : Colwellia sp. Arc7-D chromosome Total score: 1.0 Cumulative Blast bit score: 92
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
urease accessory protein UreG
Accession: AWB59485
Location: 1519474-1520085
NCBI BlastP on this gene
Accession: AWB59485
Location: 1519474-1520085
NCBI BlastP on this gene
ureG
DBO93_06595
polysaccharide lyase family 7 protein
Accession: AWB59484
Location: 1517361-1518296
NCBI BlastP on this gene
Accession: AWB59484
Location: 1517361-1518296
NCBI BlastP on this gene
DBO93_06590
DBO93_06585
polysaccharide lyase family 7 protein
Accession: AWB57244
Location: 1513728-1514678
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 92
Sequence coverage: 54 %
E-value: 7e-18
NCBI BlastP on this gene
Accession: AWB57244
Location: 1513728-1514678
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 92
Sequence coverage: 54 %
E-value: 7e-18
NCBI BlastP on this gene
DBO93_06580
DNA-binding response regulator
Accession: DBO93_06575
Location: 1513382-1513513
NCBI BlastP on this gene
Accession: DBO93_06575
Location: 1513382-1513513
NCBI BlastP on this gene
DBO93_06575
DBO93_06570
DNA-binding response regulator
Accession: AWB57242
Location: 1511014-1511673
NCBI BlastP on this gene
Accession: AWB57242
Location: 1511014-1511673
NCBI BlastP on this gene
DBO93_06565
DBO93_06560
DUF3422 domain-containing protein
Accession: AWB57240
Location: 1508066-1509388
NCBI BlastP on this gene
Accession: AWB57240
Location: 1508066-1509388
NCBI BlastP on this gene
DBO93_06555
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP000282 : Saccharophagus degradans 2-40 Total score: 1.0 Cumulative Blast bit score: 78
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
Sde_0030
oligopeptidase A. Metallo peptidase. MEROPS family M03A
Accession: ABD79295
Location: 34108-36165
NCBI BlastP on this gene
Accession: ABD79295
Location: 34108-36165
NCBI BlastP on this gene
Sde_0031
Sde_0032
Sde_0033
polysaccharide lyase-like protein
Accession: ABD79298
Location: 37816-40122
BlastP hit with alyA6
Percentage identity: 34 %
BlastP bit score: 78
Sequence coverage: 18 %
E-value: 1e-11
NCBI BlastP on this gene
Accession: ABD79298
Location: 37816-40122
BlastP hit with alyA6
Percentage identity: 34 %
BlastP bit score: 78
Sequence coverage: 18 %
E-value: 1e-11
NCBI BlastP on this gene
ply6F
Sde_0035
coxB
coxA
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP039451 : Psychroserpens sp. NJDZ02 chromosome Total score: 1.0 Cumulative Blast bit score: 68
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
DUF962 domain-containing protein
Accession: QCE43072
Location: 3885326-3885784
NCBI BlastP on this gene
Accession: QCE43072
Location: 3885326-3885784
NCBI BlastP on this gene
E9099_17155
GIY-YIG nuclease family protein
Accession: QCE43071
Location: 3884323-3884613
NCBI BlastP on this gene
Accession: QCE43071
Location: 3884323-3884613
NCBI BlastP on this gene
E9099_17150
DNA mismatch repair protein MutS
Accession: QCE43070
Location: 3881630-3883804
NCBI BlastP on this gene
Accession: QCE43070
Location: 3881630-3883804
NCBI BlastP on this gene
E9099_17145
polysaccharide lyase family 7 protein
Accession: QCE43568
Location: 3879867-3880772
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 2e-09
NCBI BlastP on this gene
Accession: QCE43568
Location: 3879867-3880772
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 2e-09
NCBI BlastP on this gene
E9099_17140
E9099_17135
E9099_17130
E9099_17125
E9099_17120
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP031612 : Olleya aquimaris strain DAU311 chromosome Total score: 1.0 Cumulative Blast bit score: 68
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
DZC78_02395
DZC78_02400
DZC78_02405
DZC78_02410
DZC78_02415
translation initiation factor SUI1
Accession: AXO79277
Location: 509761-511614
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 3e-09
NCBI BlastP on this gene
Accession: AXO79277
Location: 509761-511614
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 3e-09
NCBI BlastP on this gene
DZC78_02420
ABC transporter substrate-binding protein
Accession: AXO79278
Location: 511695-512549
NCBI BlastP on this gene
Accession: AXO79278
Location: 511695-512549
NCBI BlastP on this gene
DZC78_02425
DZC78_02430
DUF1835 domain-containing protein
Accession: AXO79280
Location: 513420-514349
NCBI BlastP on this gene
Accession: AXO79280
Location: 513420-514349
NCBI BlastP on this gene
DZC78_02435
DZC78_02440
isopenicillin N synthase family oxygenase
Accession: AXO79282
Location: 514757-515704
NCBI BlastP on this gene
Accession: AXO79282
Location: 514757-515704
NCBI BlastP on this gene
DZC78_02445
DZC78_02450
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
201. : CP013120 Alteromonas stellipolaris LMG 21856 Total score: 1.0 Cumulative Blast bit score: 165
PL6|PL6 1
Location: 1-2352
Location: 1-2352
ZOBELLIA_4130
PL7 5|PL7
Location: 2369-3427
Location: 2369-3427
ZOBELLIA_4131
PL6|PL6 1
Location: 3577-5910
Location: 3577-5910
ZOBELLIA_4132
AOR13_381
AOR13_382
AOR13_383
AOR13_384
Protein secretion chaperonin CsaA
Accession: ALM89437
Location: 486893-487228
NCBI BlastP on this gene
Accession: ALM89437
Location: 486893-487228
NCBI BlastP on this gene
AOR13_385
Alginate lyase precursor
Accession: ALM89438
Location: 487733-489268
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 165
Sequence coverage: 99 %
E-value: 2e-42
NCBI BlastP on this gene
Accession: ALM89438
Location: 487733-489268
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 165
Sequence coverage: 99 %
E-value: 2e-42
NCBI BlastP on this gene
AOR13_386
Glyoxalase/bleomycin resistance
Accession: ALM89439
Location: 489371-489706
NCBI BlastP on this gene
Accession: ALM89439
Location: 489371-489706
NCBI BlastP on this gene
AOR13_387
AOR13_388
AOR13_389
AOR13_390
AOR13_391
202. : LR136958 Alteromonas sp. 76-1 genome assembly, chromosome: cAlt76. Total score: 1.0 Cumulative Blast bit score: 164
glucose-6-phosphate isomerase
Accession: VEL98201
Location: 3655182-3656831
NCBI BlastP on this gene
Accession: VEL98201
Location: 3655182-3656831
NCBI BlastP on this gene
ALT761_03218
ALT761_03217
ALT761_03216
ALT761_03215
ALT761_03214
alginate lyase
Accession: VEL98196
Location: 3650131-3651666
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 164
Sequence coverage: 99 %
E-value: 4e-42
NCBI BlastP on this gene
Accession: VEL98196
Location: 3650131-3651666
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 164
Sequence coverage: 99 %
E-value: 4e-42
NCBI BlastP on this gene
ALT761_03213
ALT761_03212
ALT761_03211
ALT761_03210
uncharacterized protein DUF4426
Accession: VEL98192
Location: 3645636-3646076
NCBI BlastP on this gene
Accession: VEL98192
Location: 3645636-3646076
NCBI BlastP on this gene
ALT761_03209
ALT761_03208
203. : CP002339 Alteromonas naphthalenivorans strain SN2 Total score: 1.0 Cumulative Blast bit score: 161
pgi
ambt_01980
ambt_01985
putative glutathione S-transferase protein
Accession: AEF01952
Location: 466470-467108
NCBI BlastP on this gene
Accession: AEF01952
Location: 466470-467108
NCBI BlastP on this gene
ambt_01990
ambt_01995
putative alginate lyase
Accession: AEF01954
Location: 468232-469767
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 161
Sequence coverage: 99 %
E-value: 4e-41
NCBI BlastP on this gene
Accession: AEF01954
Location: 468232-469767
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 161
Sequence coverage: 99 %
E-value: 4e-41
NCBI BlastP on this gene
ambt_02000
Glyoxalase/bleomycin resistance
Accession: AEF01955
Location: 469870-470205
NCBI BlastP on this gene
Accession: AEF01955
Location: 469870-470205
NCBI BlastP on this gene
ambt_02005
ambt_02010
ambt_02015
ambt_02020
ambt_02025
204. : CP038493 Thalassotalea sp. HSM 43 chromosome Total score: 1.0 Cumulative Blast bit score: 160
polysaccharide lyase family 7 protein
Accession: QBY04449
Location: 2010122-2011183
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 103 %
E-value: 5e-42
NCBI BlastP on this gene
Accession: QBY04449
Location: 2010122-2011183
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 103 %
E-value: 5e-42
NCBI BlastP on this gene
E2K93_08615
205. : CP034660 Colwellia sp. Arc7-635 chromosome Total score: 1.0 Cumulative Blast bit score: 160
polysaccharide lyase family 7 protein
Accession: AZQ84888
Location: 3082695-3083741
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 98 %
E-value: 6e-42
NCBI BlastP on this gene
Accession: AZQ84888
Location: 3082695-3083741
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 160
Sequence coverage: 98 %
E-value: 6e-42
NCBI BlastP on this gene
EKO29_13345
EKO29_13340
siderophore-interacting protein
Accession: AZQ84886
Location: 3080956-3081687
NCBI BlastP on this gene
Accession: AZQ84886
Location: 3080956-3081687
NCBI BlastP on this gene
EKO29_13335
MarR family transcriptional regulator
Accession: AZQ84885
Location: 3080214-3080630
NCBI BlastP on this gene
Accession: AZQ84885
Location: 3080214-3080630
NCBI BlastP on this gene
EKO29_13330
multidrug efflux SMR transporter
Accession: AZQ84884
Location: 3079573-3079887
NCBI BlastP on this gene
Accession: AZQ84884
Location: 3079573-3079887
NCBI BlastP on this gene
EKO29_13325
ACT domain-containing protein
Accession: AZQ84883
Location: 3079107-3079520
NCBI BlastP on this gene
Accession: AZQ84883
Location: 3079107-3079520
NCBI BlastP on this gene
EKO29_13320
EKO29_13315
206. : CP028924 Colwellia sp. Arc7-D chromosome Total score: 1.0 Cumulative Blast bit score: 160
DBO93_17415
DUF2489 domain-containing protein
Accession: AWB59163
Location: 4012787-4013287
NCBI BlastP on this gene
Accession: AWB59163
Location: 4012787-4013287
NCBI BlastP on this gene
DBO93_17420
oxygen-independent coproporphyrinogen III oxidase
Accession: AWB59164
Location: 4013520-4014890
NCBI BlastP on this gene
Accession: AWB59164
Location: 4013520-4014890
NCBI BlastP on this gene
hemN
DUF1285 domain-containing protein
Accession: AWB59165
Location: 4014898-4015440
NCBI BlastP on this gene
Accession: AWB59165
Location: 4014898-4015440
NCBI BlastP on this gene
DBO93_17430
polysaccharide lyase family 7 protein
Accession: AWB59166
Location: 4016576-4017649
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
Accession: AWB59166
Location: 4016576-4017649
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
DBO93_17440
207. : CP020465 Colwellia beringensis strain NB097-1 chromosome Total score: 1.0 Cumulative Blast bit score: 159
polysaccharide lyase family 7 protein
Accession: ASP49898
Location: 1218528-1219487
NCBI BlastP on this gene
Accession: ASP49898
Location: 1218528-1219487
NCBI BlastP on this gene
B5D82_05170
B5D82_05165
B5D82_05160
B5D82_05155
polysaccharide lyase family 7 protein
Accession: ASP47198
Location: 1214236-1215312
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 159
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
Accession: ASP47198
Location: 1214236-1215312
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 159
Sequence coverage: 96 %
E-value: 1e-41
NCBI BlastP on this gene
B5D82_05150
B5D82_05145
B5D82_05140
B5D82_05135
NAD(P)-dependent oxidoreductase
Accession: ASP47195
Location: 1210757-1211620
NCBI BlastP on this gene
Accession: ASP47195
Location: 1210757-1211620
NCBI BlastP on this gene
B5D82_05130
B5D82_05125
B5D82_05120
208. : CP019419 Polaribacter reichenbachii strain 6Alg 8T Total score: 1.0 Cumulative Blast bit score: 158
BW723_16210
long-chain fatty acid--CoA ligase
Accession: APZ47745
Location: 3791698-3793488
NCBI BlastP on this gene
Accession: APZ47745
Location: 3791698-3793488
NCBI BlastP on this gene
BW723_16215
BW723_16220
BW723_16225
BW723_16230
alginate lyase
Accession: APZ47749
Location: 3795336-3798014
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
Accession: APZ47749
Location: 3795336-3798014
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
BW723_16235
209. : CP019337 Polaribacter reichenbachii strain KCTC 23969 chromosome. Total score: 1.0 Cumulative Blast bit score: 158
BTO17_06630
long-chain fatty acid--CoA ligase
Accession: AUC18380
Location: 1517621-1519411
NCBI BlastP on this gene
Accession: AUC18380
Location: 1517621-1519411
NCBI BlastP on this gene
BTO17_06635
BTO17_06640
BTO17_06645
BTO17_06650
alginate lyase
Accession: AUC18384
Location: 1521259-1523937
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
Accession: AUC18384
Location: 1521259-1523937
BlastP hit with alyA4
Percentage identity: 31 %
BlastP bit score: 158
Sequence coverage: 53 %
E-value: 1e-36
NCBI BlastP on this gene
BTO17_06655
BTO17_06660
BTO17_06665
BTO17_06670
BTO17_06675
BTO17_06680
Arc family DNA binding domain-containing protein
Accession: AUC18389
Location: 1527518-1527682
NCBI BlastP on this gene
Accession: AUC18389
Location: 1527518-1527682
NCBI BlastP on this gene
BTO17_06685
BTO17_06690
210. : CP026604 Catenovulum sp. CCB-QB4 chromosome Total score: 1.0 Cumulative Blast bit score: 155
C2869_09515
cupin domain-containing protein
Accession: AWB66656
Location: 2703613-2703960
NCBI BlastP on this gene
Accession: AWB66656
Location: 2703613-2703960
NCBI BlastP on this gene
C2869_09520
C2869_09525
sugar phosphate isomerase/epimerase
Accession: AWB66658
Location: 2705917-2706837
NCBI BlastP on this gene
Accession: AWB66658
Location: 2705917-2706837
NCBI BlastP on this gene
C2869_09530
AraC family transcriptional regulator
Accession: AWB66659
Location: 2706987-2707739
NCBI BlastP on this gene
Accession: AWB66659
Location: 2706987-2707739
NCBI BlastP on this gene
C2869_09535
antibiotic biosynthesis monooxygenase
Accession: AWB68984
Location: 2707736-2708038
NCBI BlastP on this gene
Accession: AWB68984
Location: 2707736-2708038
NCBI BlastP on this gene
C2869_09540
polysaccharide lyase family 7 protein
Accession: AWB66660
Location: 2708351-2709472
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 155
Sequence coverage: 98 %
E-value: 8e-40
NCBI BlastP on this gene
Accession: AWB66660
Location: 2708351-2709472
BlastP hit with alyA5
Percentage identity: 35 %
BlastP bit score: 155
Sequence coverage: 98 %
E-value: 8e-40
NCBI BlastP on this gene
C2869_09545
211. : CP010912 Alteromonas australica strain DE170 Total score: 1.0 Cumulative Blast bit score: 154
EP12_15295
EP12_15290
EP12_15285
XRE family transcriptional regulator
Accession: AJP44803
Location: 3350176-3350703
NCBI BlastP on this gene
Accession: AJP44803
Location: 3350176-3350703
NCBI BlastP on this gene
EP12_15280
branched-chain amino acid permease
Accession: AJP44802
Location: 3349457-3350179
NCBI BlastP on this gene
Accession: AJP44802
Location: 3349457-3350179
NCBI BlastP on this gene
EP12_15275
branched-chain amino acid ABC transporter
Accession: AJP44801
Location: 3349120-3349467
NCBI BlastP on this gene
Accession: AJP44801
Location: 3349120-3349467
NCBI BlastP on this gene
EP12_15270
EP12_15265
alginate lyase
Accession: AJP44799
Location: 3346678-3348213
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
Accession: AJP44799
Location: 3346678-3348213
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
EP12_15260
EP12_15255
EP12_15240
EP12_15235
pyrroline-5-carboxylate reductase
Accession: AJP44795
Location: 3341870-3342691
NCBI BlastP on this gene
Accession: AJP44795
Location: 3341870-3342691
NCBI BlastP on this gene
EP12_15230
212. : CP008849 Alteromonas australica strain H 17 Total score: 1.0 Cumulative Blast bit score: 154
EP13_14730
EP13_14725
EP13_14720
XRE family transcriptional regulator
Accession: AIF99835
Location: 3271977-3272504
NCBI BlastP on this gene
Accession: AIF99835
Location: 3271977-3272504
NCBI BlastP on this gene
EP13_14715
branched-chain amino acid permease
Accession: AIF99834
Location: 3271258-3271980
NCBI BlastP on this gene
Accession: AIF99834
Location: 3271258-3271980
NCBI BlastP on this gene
EP13_14710
branched-chain amino acid ABC transporter
Accession: AIF99833
Location: 3270921-3271268
NCBI BlastP on this gene
Accession: AIF99833
Location: 3270921-3271268
NCBI BlastP on this gene
EP13_14705
EP13_14700
alginate lyase
Accession: AIF99831
Location: 3268479-3270014
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
Accession: AIF99831
Location: 3268479-3270014
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 154
Sequence coverage: 99 %
E-value: 8e-39
NCBI BlastP on this gene
EP13_14695
EP13_14690
EP13_14685
fadH
EP13_14675
EP13_14670
pyrroline-5-carboxylate reductase
Accession: AIF99825
Location: 3263660-3264481
NCBI BlastP on this gene
Accession: AIF99825
Location: 3263660-3264481
NCBI BlastP on this gene
EP13_14665
213. : CP045859 Vibrio owensii strain SH14 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
polysaccharide lyase family 7 protein
Accession: QGH47617
Location: 2339593-2340663
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
Accession: QGH47617
Location: 2339593-2340663
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
APZ19_11120
katG
APZ19_11110
uxaC
214. : CP033144 Vibrio owensii strain V180403 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
polysaccharide lyase family 7 protein
Accession: AYO20327
Location: 1980872-1981942
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 3e-39
NCBI BlastP on this gene
Accession: AYO20327
Location: 1980872-1981942
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 3e-39
NCBI BlastP on this gene
D0856_09680
katG
D0856_09670
D0856_09665
215. : CP033137 Vibrio owensii strain 1700302 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
CBS domain-containing protein
Accession: AYO14702
Location: 2057907-2059769
NCBI BlastP on this gene
Accession: AYO14702
Location: 2057907-2059769
NCBI BlastP on this gene
D0812_09910
FadR family transcriptional regulator
Accession: AYO14703
Location: 2059943-2060662
NCBI BlastP on this gene
Accession: AYO14703
Location: 2059943-2060662
NCBI BlastP on this gene
D0812_09915
D0812_09920
FadR family transcriptional regulator
Accession: AYO14705
Location: 2061927-2062643
NCBI BlastP on this gene
Accession: AYO14705
Location: 2061927-2062643
NCBI BlastP on this gene
D0812_09925
MATE family efflux transporter
Accession: AYO14706
Location: 2062758-2064116
NCBI BlastP on this gene
Accession: AYO14706
Location: 2062758-2064116
NCBI BlastP on this gene
D0812_09930
polysaccharide lyase family 7 protein
Accession: AYO14707
Location: 2064377-2065447
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
Accession: AYO14707
Location: 2064377-2065447
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
D0812_09935
216. : CP019959 Vibrio owensii strain XSBZ03 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 153
A9237_02610
GntR family transcriptional regulator
Accession: AQW57073
Location: 598004-598723
NCBI BlastP on this gene
Accession: AQW57073
Location: 598004-598723
NCBI BlastP on this gene
A9237_02605
A9237_02600
GntR family transcriptional regulator
Accession: AQW57071
Location: 596023-596739
NCBI BlastP on this gene
Accession: AQW57071
Location: 596023-596739
NCBI BlastP on this gene
A9237_02595
A9237_02590
polysaccharide lyase family 7 protein
Accession: AQW57069
Location: 593219-594289
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
Accession: AQW57069
Location: 593219-594289
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 153
Sequence coverage: 95 %
E-value: 4e-39
NCBI BlastP on this gene
A9237_02585
A9237_02580
A9237_02575
A9237_02570
217. : CP010912 Alteromonas australica strain DE170 Total score: 1.0 Cumulative Blast bit score: 152
EP12_16265
EP12_16270
EP12_16275
alginate lyase
Accession: AJP44971
Location: 3606913-3608016
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 152
Sequence coverage: 105 %
E-value: 6e-39
NCBI BlastP on this gene
Accession: AJP44971
Location: 3606913-3608016
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 152
Sequence coverage: 105 %
E-value: 6e-39
NCBI BlastP on this gene
EP12_16280
218. : HG315671 Formosa agariphila KMM 3901 Total score: 1.0 Cumulative Blast bit score: 150
oxidoreductase of aldo/keto reductase family, subgroup 1
Accession: CDF77716
Location: 2722-3564
NCBI BlastP on this gene
Accession: CDF77716
Location: 2722-3564
NCBI BlastP on this gene
BN863_40
BN863_50
BN863_60
alginate lyase precursor
Accession: CDF77719
Location: 7374-10481
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 53 %
E-value: 5e-34
NCBI BlastP on this gene
Accession: CDF77719
Location: 7374-10481
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 53 %
E-value: 5e-34
NCBI BlastP on this gene
BN863_70
BN863_80
iron-sulfur cluster-binding protein
Accession: CDF77721
Location: 11978-13549
NCBI BlastP on this gene
Accession: CDF77721
Location: 11978-13549
NCBI BlastP on this gene
BN863_90
BN863_100
219. : CP039700 Vibrio cyclitrophicus strain ECSMB14105 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 150
FAZ90_06520
FAZ90_06525
FAZ90_06530
polysaccharide lyase family 7 protein
Accession: QCI70745
Location: 1432843-1433880
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 150
Sequence coverage: 97 %
E-value: 3e-38
NCBI BlastP on this gene
Accession: QCI70745
Location: 1432843-1433880
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 150
Sequence coverage: 97 %
E-value: 3e-38
NCBI BlastP on this gene
FAZ90_06535
MATE family efflux transporter
Accession: QCI70746
Location: 1434249-1435616
NCBI BlastP on this gene
Accession: QCI70746
Location: 1434249-1435616
NCBI BlastP on this gene
FAZ90_06540
FadR family transcriptional regulator
Accession: QCI70747
Location: 1435852-1436574
NCBI BlastP on this gene
Accession: QCI70747
Location: 1435852-1436574
NCBI BlastP on this gene
FAZ90_06545
FAZ90_06550
FadR family transcriptional regulator
Accession: QCI70749
Location: 1437938-1438657
NCBI BlastP on this gene
Accession: QCI70749
Location: 1437938-1438657
NCBI BlastP on this gene
FAZ90_06555
220. : CP019336 Polaribacter sejongensis strain KCTC 23670 chromosome. Total score: 1.0 Cumulative Blast bit score: 150
BTO15_02835
BTO15_02840
BTO15_02845
BTO15_02850
BTO15_02855
BTO15_02860
BTO15_02865
alginate lyase
Accession: AUC21121
Location: 763957-766617
BlastP hit with alyA4
Percentage identity: 32 %
BlastP bit score: 150
Sequence coverage: 51 %
E-value: 6e-34
NCBI BlastP on this gene
Accession: AUC21121
Location: 763957-766617
BlastP hit with alyA4
Percentage identity: 32 %
BlastP bit score: 150
Sequence coverage: 51 %
E-value: 6e-34
NCBI BlastP on this gene
BTO15_02870
BTO15_02875
1-pyrroline-5-carboxylate dehydrogenase
Accession: AUC21123
Location: 767532-769157
NCBI BlastP on this gene
Accession: AUC21123
Location: 767532-769157
NCBI BlastP on this gene
BTO15_02880
BTO15_02885
BTO15_02890
221. : CP016228 Vibrio crassostreae 9CS106 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 149
A134_08070
A134_08075
A134_08080
alginate lyase
Accession: ANP76358
Location: 1774280-1775317
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 96 %
E-value: 1e-37
NCBI BlastP on this gene
Accession: ANP76358
Location: 1774280-1775317
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 96 %
E-value: 1e-37
NCBI BlastP on this gene
A134_08085
A134_08090
GntR family transcriptional regulator
Accession: ANP76360
Location: 1777064-1777786
NCBI BlastP on this gene
Accession: ANP76360
Location: 1777064-1777786
NCBI BlastP on this gene
A134_08095
A134_08100
GntR family transcriptional regulator
Accession: ANP76362
Location: 1779080-1779799
NCBI BlastP on this gene
Accession: ANP76362
Location: 1779080-1779799
NCBI BlastP on this gene
A134_08105
A134_08110
222. : CP031769 Salinimonas sediminis strain N102 chromosome Total score: 1.0 Cumulative Blast bit score: 144
D0Y50_13595
cupin domain-containing protein
Accession: AXR07288
Location: 3061656-3062000
NCBI BlastP on this gene
Accession: AXR07288
Location: 3061656-3062000
NCBI BlastP on this gene
D0Y50_13600
NAD(P)-dependent oxidoreductase
Accession: AXR07289
Location: 3062137-3063015
NCBI BlastP on this gene
Accession: AXR07289
Location: 3062137-3063015
NCBI BlastP on this gene
D0Y50_13605
D0Y50_13610
polysaccharide lyase family 7 protein
Accession: AXR07290
Location: 3064666-3065787
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 110 %
E-value: 1e-35
NCBI BlastP on this gene
Accession: AXR07290
Location: 3064666-3065787
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 144
Sequence coverage: 110 %
E-value: 1e-35
NCBI BlastP on this gene
D0Y50_13615
LacI family transcriptional regulator
Accession: D0Y50_13620
Location: 3065863-3066899
NCBI BlastP on this gene
Accession: D0Y50_13620
Location: 3065863-3066899
NCBI BlastP on this gene
D0Y50_13620
cysE
trmJ
D0Y50_13635
protein translocase subunit SecF
Accession: AXR07294
Location: 3070519-3071460
NCBI BlastP on this gene
Accession: AXR07294
Location: 3070519-3071460
NCBI BlastP on this gene
secF
223. : CP019650 Microbulbifer agarilyticus strain GP101 Total score: 1.0 Cumulative Blast bit score: 144
Mag101_17585
Mag101_17590
Mag101_17595
Mag101_17600
Mag101_17605
Mag101_17610
polysaccharide lyase family 7 protein
Accession: AQQ69248
Location: 4212960-4214051
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 144
Sequence coverage: 107 %
E-value: 9e-36
NCBI BlastP on this gene
Accession: AQQ69248
Location: 4212960-4214051
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 144
Sequence coverage: 107 %
E-value: 9e-36
NCBI BlastP on this gene
Mag101_17615
224. : CP016177 Vibrio breoganii strain FF50 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 144
A6E01_10035
A6E01_10040
LysR family transcriptional regulator
Accession: ANO33545
Location: 2179265-2180170
NCBI BlastP on this gene
Accession: ANO33545
Location: 2179265-2180170
NCBI BlastP on this gene
A6E01_10050
A6E01_10055
A6E01_10060
A6E01_10065
alginate lyase
Accession: ANO33549
Location: 2182537-2183580
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 144
Sequence coverage: 96 %
E-value: 5e-36
NCBI BlastP on this gene
Accession: ANO33549
Location: 2182537-2183580
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 144
Sequence coverage: 96 %
E-value: 5e-36
NCBI BlastP on this gene
A6E01_10070
2-deoxy-D-gluconate 3-dehydrogenase
Accession: ANO33550
Location: 2183904-2184665
NCBI BlastP on this gene
Accession: ANO33550
Location: 2183904-2184665
NCBI BlastP on this gene
A6E01_10075
A6E01_10080
A6E01_10085
A6E01_10090
225. : CP000282 Saccharophagus degradans 2-40 Total score: 1.0 Cumulative Blast bit score: 143
Sde_2842
ABC transporter, inner membrane protein
Accession: ABD82098
Location: 3588666-3591599
NCBI BlastP on this gene
Accession: ABD82098
Location: 3588666-3591599
NCBI BlastP on this gene
Sde_2841
Sde_2840
putative polysaccharide lyase
Accession: ABD82096
Location: 3586312-3587415
BlastP hit with alyA5
Percentage identity: 36 %
BlastP bit score: 143
Sequence coverage: 91 %
E-value: 1e-35
NCBI BlastP on this gene
Accession: ABD82096
Location: 3586312-3587415
BlastP hit with alyA5
Percentage identity: 36 %
BlastP bit score: 143
Sequence coverage: 91 %
E-value: 1e-35
NCBI BlastP on this gene
ply7E
Sde_2838
Sde_2837
Sde_2836
Sde_2835
gly81A
226. : CP036200 Shewanella maritima strain D4-2 chromosome Total score: 1.0 Cumulative Blast bit score: 142
EXU30_08335
EXU30_08340
EXU30_08345
EXU30_08350
EXU30_08355
polysaccharide lyase family 7 protein
Accession: QBF82700
Location: 1958329-1959390
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 107 %
E-value: 4e-35
NCBI BlastP on this gene
Accession: QBF82700
Location: 1958329-1959390
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 107 %
E-value: 4e-35
NCBI BlastP on this gene
EXU30_08360
ammonia-forming cytochrome c nitrite reductase subunit c552
Accession: QBF82701
Location: 1960113-1961768
NCBI BlastP on this gene
Accession: QBF82701
Location: 1960113-1961768
NCBI BlastP on this gene
EXU30_08365
EXU30_08370
N-acetylneuraminic acid outer membrane channel protein NanC
Accession: QBF84846
Location: 1964039-1964698
NCBI BlastP on this gene
Accession: QBF84846
Location: 1964039-1964698
NCBI BlastP on this gene
EXU30_08375
227. : CP012898 Algibacter alginicilyticus strain HZ22 chromosome Total score: 1.0 Cumulative Blast bit score: 137
APS56_07990
alginate lyase
Accession: ALJ05070
Location: 1924768-1927443
BlastP hit with alyA6
Percentage identity: 33 %
BlastP bit score: 137
Sequence coverage: 39 %
E-value: 7e-30
NCBI BlastP on this gene
Accession: ALJ05070
Location: 1924768-1927443
BlastP hit with alyA6
Percentage identity: 33 %
BlastP bit score: 137
Sequence coverage: 39 %
E-value: 7e-30
NCBI BlastP on this gene
APS56_07995
APS56_08000
APS56_08005
APS56_08010
228. : CP036200 Shewanella maritima strain D4-2 chromosome Total score: 1.0 Cumulative Blast bit score: 134
EXU30_09990
EXU30_09995
polysaccharide lyase family 7 protein
Accession: QBF82988
Location: 2350649-2351587
NCBI BlastP on this gene
Accession: QBF82988
Location: 2350649-2351587
NCBI BlastP on this gene
EXU30_10000
EXU30_10005
polysaccharide lyase family 7 protein
Accession: QBF82989
Location: 2353038-2354108
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 134
Sequence coverage: 109 %
E-value: 4e-32
NCBI BlastP on this gene
Accession: QBF82989
Location: 2353038-2354108
BlastP hit with alyA5
Percentage identity: 31 %
BlastP bit score: 134
Sequence coverage: 109 %
E-value: 4e-32
NCBI BlastP on this gene
EXU30_10010
229. : CP033144 Vibrio owensii strain V180403 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 133
lipopolysaccharide assembly protein LapB
Accession: AYO19304
Location: 765644-766819
NCBI BlastP on this gene
Accession: AYO19304
Location: 765644-766819
NCBI BlastP on this gene
lapB
DUF1049 domain-containing protein
Accession: AYO19303
Location: 765332-765625
NCBI BlastP on this gene
Accession: AYO19303
Location: 765332-765625
NCBI BlastP on this gene
D0856_03575
integration host factor subunit beta
Accession: AYO19302
Location: 764907-765191
NCBI BlastP on this gene
Accession: AYO19302
Location: 764907-765191
NCBI BlastP on this gene
ihfB
D0856_03565
D0856_03560
polysaccharide lyase family 7 protein
Accession: AYO19299
Location: 760895-761938
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 133
Sequence coverage: 97 %
E-value: 7e-32
NCBI BlastP on this gene
Accession: AYO19299
Location: 760895-761938
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 133
Sequence coverage: 97 %
E-value: 7e-32
NCBI BlastP on this gene
D0856_03555
sohB
D0856_03545
translesion DNA synthesis-associated protein ImuA
Accession: AYO19296
Location: 757906-758598
NCBI BlastP on this gene
Accession: AYO19296
Location: 757906-758598
NCBI BlastP on this gene
imuA
DNA polymerase Y family protein
Accession: AYO19295
Location: 756498-757901
NCBI BlastP on this gene
Accession: AYO19295
Location: 756498-757901
NCBI BlastP on this gene
D0856_03535
DNA polymerase III subunit alpha
Accession: AYO19294
Location: 753396-756470
NCBI BlastP on this gene
Accession: AYO19294
Location: 753396-756470
NCBI BlastP on this gene
dnaE
230. : LT629752 Polaribacter sp. KT25b genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 132
SAMN05216503_2576
uncharacterized radical SAM protein YgiQ
Accession: SDS28860
Location: 2899844-2901805
NCBI BlastP on this gene
Accession: SDS28860
Location: 2899844-2901805
NCBI BlastP on this gene
SAMN05216503_2577
SAMN05216503_2578
Por secretion system C-terminal sorting domain-containing protein
Accession: SDS28918
Location: 2903529-2906090
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 132
Sequence coverage: 42 %
E-value: 3e-28
NCBI BlastP on this gene
Accession: SDS28918
Location: 2903529-2906090
BlastP hit with alyA6
Percentage identity: 31 %
BlastP bit score: 132
Sequence coverage: 42 %
E-value: 3e-28
NCBI BlastP on this gene
SAMN05216503_2579
SAMN05216503_2580
SAMN05216503_2581
SAMN05216503_2582
231. : CP019959 Vibrio owensii strain XSBZ03 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 132
polysaccharide lyase family 7 protein
Accession: AQW59088
Location: 3022601-3023644
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 132
Sequence coverage: 97 %
E-value: 1e-31
NCBI BlastP on this gene
Accession: AQW59088
Location: 3022601-3023644
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 132
Sequence coverage: 97 %
E-value: 1e-31
NCBI BlastP on this gene
A9237_13920
A9237_13915
A9237_13910
A9237_13905
A9237_13900
A9237_13895
232. : CP045859 Vibrio owensii strain SH14 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 130
lipopolysaccharide assembly protein LapB
Accession: QGH46642
Location: 1218862-1220037
NCBI BlastP on this gene
Accession: QGH46642
Location: 1218862-1220037
NCBI BlastP on this gene
lapB
DUF1049 domain-containing protein
Accession: QGH46641
Location: 1218550-1218843
NCBI BlastP on this gene
Accession: QGH46641
Location: 1218550-1218843
NCBI BlastP on this gene
APZ19_05760
integration host factor subunit beta
Accession: QGH46640
Location: 1218125-1218409
NCBI BlastP on this gene
Accession: QGH46640
Location: 1218125-1218409
NCBI BlastP on this gene
ihfB
rpsA
APZ19_05745
polysaccharide lyase family 7 protein
Accession: QGH46637
Location: 1214113-1215156
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
Accession: QGH46637
Location: 1214113-1215156
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
APZ19_05740
sohB
APZ19_05730
translesion DNA synthesis-associated protein ImuA
Accession: QGH46634
Location: 1211126-1211818
NCBI BlastP on this gene
Accession: QGH46634
Location: 1211126-1211818
NCBI BlastP on this gene
imuA
DNA polymerase Y family protein
Accession: QGH46633
Location: 1209718-1211121
NCBI BlastP on this gene
Accession: QGH46633
Location: 1209718-1211121
NCBI BlastP on this gene
APZ19_05720
DNA polymerase III subunit alpha
Accession: QGH46632
Location: 1206616-1209690
NCBI BlastP on this gene
Accession: QGH46632
Location: 1206616-1209690
NCBI BlastP on this gene
dnaE
233. : CP033137 Vibrio owensii strain 1700302 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 130
lipopolysaccharide assembly protein LapB
Accession: AYO13986
Location: 1253812-1254987
NCBI BlastP on this gene
Accession: AYO13986
Location: 1253812-1254987
NCBI BlastP on this gene
lapB
DUF1049 domain-containing protein
Accession: AYO13985
Location: 1253500-1253793
NCBI BlastP on this gene
Accession: AYO13985
Location: 1253500-1253793
NCBI BlastP on this gene
D0812_05955
integration host factor subunit beta
Accession: AYO13984
Location: 1253075-1253359
NCBI BlastP on this gene
Accession: AYO13984
Location: 1253075-1253359
NCBI BlastP on this gene
ihfB
D0812_05945
D0812_05940
polysaccharide lyase family 7 protein
Accession: AYO13981
Location: 1249063-1250106
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
Accession: AYO13981
Location: 1249063-1250106
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 130
Sequence coverage: 97 %
E-value: 4e-31
NCBI BlastP on this gene
D0812_05935
sohB
D0812_05925
translesion DNA synthesis-associated protein ImuA
Accession: AYO13978
Location: 1246076-1246768
NCBI BlastP on this gene
Accession: AYO13978
Location: 1246076-1246768
NCBI BlastP on this gene
imuA
DNA polymerase Y family protein
Accession: AYO13977
Location: 1244668-1246071
NCBI BlastP on this gene
Accession: AYO13977
Location: 1244668-1246071
NCBI BlastP on this gene
D0812_05915
DNA polymerase III subunit alpha
Accession: AYO13976
Location: 1241566-1244640
NCBI BlastP on this gene
Accession: AYO13976
Location: 1241566-1244640
NCBI BlastP on this gene
dnaE
234. : CP016177 Vibrio breoganii strain FF50 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 130
alginate lyase
Accession: ANO33567
Location: 2204842-2205984
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 130
Sequence coverage: 78 %
E-value: 1e-30
NCBI BlastP on this gene
Accession: ANO33567
Location: 2204842-2205984
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 130
Sequence coverage: 78 %
E-value: 1e-30
NCBI BlastP on this gene
A6E01_10160
A6E01_10155
A6E01_10150
A6E01_10145
235. : CP039700 Vibrio cyclitrophicus strain ECSMB14105 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 119
polysaccharide lyase family 7 protein
Accession: QCI71698
Location: 2580371-2581408
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 120
Sequence coverage: 77 %
E-value: 3e-27
NCBI BlastP on this gene
Accession: QCI71698
Location: 2580371-2581408
BlastP hit with alyA5
Percentage identity: 34 %
BlastP bit score: 120
Sequence coverage: 77 %
E-value: 3e-27
NCBI BlastP on this gene
FAZ90_11735
dnaK
dnaJ
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: QCI72380
Location: 2576070-2576501
NCBI BlastP on this gene
Accession: QCI72380
Location: 2576070-2576501
NCBI BlastP on this gene
FAZ90_11720
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: QCI71695
Location: 2575467-2575967
NCBI BlastP on this gene
Accession: QCI71695
Location: 2575467-2575967
NCBI BlastP on this gene
FAZ90_11715
FAZ90_11710
236. : CP016228 Vibrio crassostreae 9CS106 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 119
alginate lyase
Accession: ANP77352
Location: 2960232-2961269
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 119
Sequence coverage: 77 %
E-value: 4e-27
NCBI BlastP on this gene
Accession: ANP77352
Location: 2960232-2961269
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 119
Sequence coverage: 77 %
E-value: 4e-27
NCBI BlastP on this gene
A134_13460
A134_13455
A134_13450
prepilin-type N-terminal cleavage/methylation domain-containing protein
Accession: ANP77349
Location: 2955937-2956377
NCBI BlastP on this gene
Accession: ANP77349
Location: 2955937-2956377
NCBI BlastP on this gene
A134_13445
A134_13440
A134_13435
237. : CP028924 Colwellia sp. Arc7-D chromosome Total score: 1.0 Cumulative Blast bit score: 92
urease accessory protein UreG
Accession: AWB59485
Location: 1519474-1520085
NCBI BlastP on this gene
Accession: AWB59485
Location: 1519474-1520085
NCBI BlastP on this gene
ureG
DBO93_06595
polysaccharide lyase family 7 protein
Accession: AWB59484
Location: 1517361-1518296
NCBI BlastP on this gene
Accession: AWB59484
Location: 1517361-1518296
NCBI BlastP on this gene
DBO93_06590
DBO93_06585
polysaccharide lyase family 7 protein
Accession: AWB57244
Location: 1513728-1514678
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 92
Sequence coverage: 54 %
E-value: 7e-18
NCBI BlastP on this gene
Accession: AWB57244
Location: 1513728-1514678
BlastP hit with alyA5
Percentage identity: 32 %
BlastP bit score: 92
Sequence coverage: 54 %
E-value: 7e-18
NCBI BlastP on this gene
DBO93_06580
DNA-binding response regulator
Accession: DBO93_06575
Location: 1513382-1513513
NCBI BlastP on this gene
Accession: DBO93_06575
Location: 1513382-1513513
NCBI BlastP on this gene
DBO93_06575
DBO93_06570
DNA-binding response regulator
Accession: AWB57242
Location: 1511014-1511673
NCBI BlastP on this gene
Accession: AWB57242
Location: 1511014-1511673
NCBI BlastP on this gene
DBO93_06565
DBO93_06560
DUF3422 domain-containing protein
Accession: AWB57240
Location: 1508066-1509388
NCBI BlastP on this gene
Accession: AWB57240
Location: 1508066-1509388
NCBI BlastP on this gene
DBO93_06555
238. : CP000282 Saccharophagus degradans 2-40 Total score: 1.0 Cumulative Blast bit score: 78
Sde_0030
oligopeptidase A. Metallo peptidase. MEROPS family M03A
Accession: ABD79295
Location: 34108-36165
NCBI BlastP on this gene
Accession: ABD79295
Location: 34108-36165
NCBI BlastP on this gene
Sde_0031
Sde_0032
Sde_0033
polysaccharide lyase-like protein
Accession: ABD79298
Location: 37816-40122
BlastP hit with alyA6
Percentage identity: 34 %
BlastP bit score: 78
Sequence coverage: 18 %
E-value: 1e-11
NCBI BlastP on this gene
Accession: ABD79298
Location: 37816-40122
BlastP hit with alyA6
Percentage identity: 34 %
BlastP bit score: 78
Sequence coverage: 18 %
E-value: 1e-11
NCBI BlastP on this gene
ply6F
Sde_0035
coxB
coxA
239. : CP039451 Psychroserpens sp. NJDZ02 chromosome Total score: 1.0 Cumulative Blast bit score: 68
DUF962 domain-containing protein
Accession: QCE43072
Location: 3885326-3885784
NCBI BlastP on this gene
Accession: QCE43072
Location: 3885326-3885784
NCBI BlastP on this gene
E9099_17155
GIY-YIG nuclease family protein
Accession: QCE43071
Location: 3884323-3884613
NCBI BlastP on this gene
Accession: QCE43071
Location: 3884323-3884613
NCBI BlastP on this gene
E9099_17150
DNA mismatch repair protein MutS
Accession: QCE43070
Location: 3881630-3883804
NCBI BlastP on this gene
Accession: QCE43070
Location: 3881630-3883804
NCBI BlastP on this gene
E9099_17145
polysaccharide lyase family 7 protein
Accession: QCE43568
Location: 3879867-3880772
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 2e-09
NCBI BlastP on this gene
Accession: QCE43568
Location: 3879867-3880772
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 2e-09
NCBI BlastP on this gene
E9099_17140
E9099_17135
E9099_17130
E9099_17125
E9099_17120
240. : CP031612 Olleya aquimaris strain DAU311 chromosome Total score: 1.0 Cumulative Blast bit score: 68
DZC78_02395
DZC78_02400
DZC78_02405
DZC78_02410
DZC78_02415
translation initiation factor SUI1
Accession: AXO79277
Location: 509761-511614
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 3e-09
NCBI BlastP on this gene
Accession: AXO79277
Location: 509761-511614
BlastP hit with alyA5
Percentage identity: 33 %
BlastP bit score: 68
Sequence coverage: 41 %
E-value: 3e-09
NCBI BlastP on this gene
DZC78_02420
ABC transporter substrate-binding protein
Accession: AXO79278
Location: 511695-512549
NCBI BlastP on this gene
Accession: AXO79278
Location: 511695-512549
NCBI BlastP on this gene
DZC78_02425
DZC78_02430
DUF1835 domain-containing protein
Accession: AXO79280
Location: 513420-514349
NCBI BlastP on this gene
Accession: AXO79280
Location: 513420-514349
NCBI BlastP on this gene
DZC78_02435
DZC78_02440
isopenicillin N synthase family oxygenase
Accession: AXO79282
Location: 514757-515704
NCBI BlastP on this gene
Accession: AXO79282
Location: 514757-515704
NCBI BlastP on this gene
DZC78_02445
DZC78_02450


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.