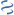MultiGeneBlast hits

Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP001998 : Coraliomargarita akajimensis DSM 45221 Total score: 1.0 Cumulative Blast bit score: 87
ZOBELLIA_4201
ZOBELLIA_4202
GH16
Location: 2758-4377
Location: 2758-4377
ZOBELLIA_4203
Caka_1235
Caka_1234
hypothetical protein
Accession: ADE54253
Location: 1550216-1552576
BlastP hit with agaA
Percentage identity: 32 %
BlastP bit score: 87
Sequence coverage: 31 %
E-value: 7e-15
NCBI BlastP on this gene
Accession: ADE54253
Location: 1550216-1552576
BlastP hit with agaA
Percentage identity: 32 %
BlastP bit score: 87
Sequence coverage: 31 %
E-value: 7e-15
NCBI BlastP on this gene
Caka_1233
protein of unknown function DUF975
Accession: ADE54252
Location: 1548552-1549604
NCBI BlastP on this gene
Accession: ADE54252
Location: 1548552-1549604
NCBI BlastP on this gene
Caka_1232
Caka_1231
Caka_1230
Caka_1229
conserved hypothetical protein
Accession: ADE54248
Location: 1545057-1546769
NCBI BlastP on this gene
Accession: ADE54248
Location: 1545057-1546769
NCBI BlastP on this gene
Caka_1228
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP001998 : Coraliomargarita akajimensis DSM 45221 Total score: 1.0 Cumulative Blast bit score: 85
ZOBELLIA_4201
ZOBELLIA_4202
GH16
Location: 2758-4377
Location: 2758-4377
ZOBELLIA_4203
Beta-agarase
Accession: ADE54273
Location: 1584502-1588413
BlastP hit with agaA
Percentage identity: 39 %
BlastP bit score: 85
Sequence coverage: 24 %
E-value: 8e-14
NCBI BlastP on this gene
Accession: ADE54273
Location: 1584502-1588413
BlastP hit with agaA
Percentage identity: 39 %
BlastP bit score: 85
Sequence coverage: 24 %
E-value: 8e-14
NCBI BlastP on this gene
Caka_1253
Caka_1252
oxidoreductase domain protein
Accession: ADE54271
Location: 1580452-1581771
NCBI BlastP on this gene
Accession: ADE54271
Location: 1580452-1581771
NCBI BlastP on this gene
Caka_1251
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
CP025791 : Flavivirga eckloniae strain ECD14 chromosome Total score: 1.0 Cumulative Blast bit score: 82
ZOBELLIA_4201
ZOBELLIA_4202
GH16
Location: 2758-4377
Location: 2758-4377
ZOBELLIA_4203
hypothetical protein
Accession: AUP80055
Location: 3758544-3761378
BlastP hit with agaA
Percentage identity: 36 %
BlastP bit score: 82
Sequence coverage: 30 %
E-value: 4e-13
NCBI BlastP on this gene
Accession: AUP80055
Location: 3758544-3761378
BlastP hit with agaA
Percentage identity: 36 %
BlastP bit score: 82
Sequence coverage: 30 %
E-value: 4e-13
NCBI BlastP on this gene
C1H87_15620
pyridine nucleotide-disulfide oxidoreductase
Accession: AUP80054
Location: 3757040-3757945
NCBI BlastP on this gene
Accession: AUP80054
Location: 3757040-3757945
NCBI BlastP on this gene
C1H87_15615
C1H87_15610
C1H87_15605
C1H87_15600
Query: Zobellia galactanivorans strain DsiJT chromosome, complete genome.
51. : CP001998 Coraliomargarita akajimensis DSM 45221 Total score: 1.0 Cumulative Blast bit score: 87
ZOBELLIA_4201
ZOBELLIA_4202
GH16
Location: 2758-4377
Location: 2758-4377
ZOBELLIA_4203
Caka_1235
Caka_1234
hypothetical protein
Accession: ADE54253
Location: 1550216-1552576
BlastP hit with agaA
Percentage identity: 32 %
BlastP bit score: 87
Sequence coverage: 31 %
E-value: 7e-15
NCBI BlastP on this gene
Accession: ADE54253
Location: 1550216-1552576
BlastP hit with agaA
Percentage identity: 32 %
BlastP bit score: 87
Sequence coverage: 31 %
E-value: 7e-15
NCBI BlastP on this gene
Caka_1233
protein of unknown function DUF975
Accession: ADE54252
Location: 1548552-1549604
NCBI BlastP on this gene
Accession: ADE54252
Location: 1548552-1549604
NCBI BlastP on this gene
Caka_1232
Caka_1231
Caka_1230
Caka_1229
conserved hypothetical protein
Accession: ADE54248
Location: 1545057-1546769
NCBI BlastP on this gene
Accession: ADE54248
Location: 1545057-1546769
NCBI BlastP on this gene
Caka_1228
52. : CP001998 Coraliomargarita akajimensis DSM 45221 Total score: 1.0 Cumulative Blast bit score: 85
Beta-agarase
Accession: ADE54273
Location: 1584502-1588413
BlastP hit with agaA
Percentage identity: 39 %
BlastP bit score: 85
Sequence coverage: 24 %
E-value: 8e-14
NCBI BlastP on this gene
Accession: ADE54273
Location: 1584502-1588413
BlastP hit with agaA
Percentage identity: 39 %
BlastP bit score: 85
Sequence coverage: 24 %
E-value: 8e-14
NCBI BlastP on this gene
Caka_1253
Caka_1252
oxidoreductase domain protein
Accession: ADE54271
Location: 1580452-1581771
NCBI BlastP on this gene
Accession: ADE54271
Location: 1580452-1581771
NCBI BlastP on this gene
Caka_1251
53. : CP025791 Flavivirga eckloniae strain ECD14 chromosome Total score: 1.0 Cumulative Blast bit score: 82
hypothetical protein
Accession: AUP80055
Location: 3758544-3761378
BlastP hit with agaA
Percentage identity: 36 %
BlastP bit score: 82
Sequence coverage: 30 %
E-value: 4e-13
NCBI BlastP on this gene
Accession: AUP80055
Location: 3758544-3761378
BlastP hit with agaA
Percentage identity: 36 %
BlastP bit score: 82
Sequence coverage: 30 %
E-value: 4e-13
NCBI BlastP on this gene
C1H87_15620
pyridine nucleotide-disulfide oxidoreductase
Accession: AUP80054
Location: 3757040-3757945
NCBI BlastP on this gene
Accession: AUP80054
Location: 3757040-3757945
NCBI BlastP on this gene
C1H87_15615
C1H87_15610
C1H87_15605
C1H87_15600


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.