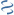MultiGeneBlast hits

Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP041360 : Spirosoma sp. KCTC 42546 chromosome Total score: 1.0 Cumulative Blast bit score: 175
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hxpB
PhzF family phenazine biosynthesis protein
Accession: QDK80659
Location: 4746782-4747627
NCBI BlastP on this gene
Accession: QDK80659
Location: 4746782-4747627
NCBI BlastP on this gene
EXU85_19420
EXU85_19415
GNAT family N-acetyltransferase
Accession: QDK80657
Location: 4744580-4745032
NCBI BlastP on this gene
Accession: QDK80657
Location: 4744580-4745032
NCBI BlastP on this gene
EXU85_19410
radical SAM family heme chaperone HemW
Accession: QDK80656
Location: 4743288-4744424
NCBI BlastP on this gene
Accession: QDK80656
Location: 4743288-4744424
NCBI BlastP on this gene
hemW
monofunctional biosynthetic peptidoglycan transglycosylase
Accession: QDK83781
Location: 4742318-4743025
NCBI BlastP on this gene
Accession: QDK83781
Location: 4742318-4743025
NCBI BlastP on this gene
mtgA
EXU85_19395
polysaccharide lyase
Accession: QDK80654
Location: 4739014-4740657
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 175
Sequence coverage: 92 %
E-value: 4e-45
NCBI BlastP on this gene
Accession: QDK80654
Location: 4739014-4740657
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 175
Sequence coverage: 92 %
E-value: 4e-45
NCBI BlastP on this gene
EXU85_19390
EXU85_19385
EXU85_19380
EXU85_19375
DUF3826 domain-containing protein
Accession: QDK80651
Location: 4731456-4732160
NCBI BlastP on this gene
Accession: QDK80651
Location: 4731456-4732160
NCBI BlastP on this gene
EXU85_19370
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP042913 : Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 174
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Pr1d_41200
N-formylglutamate amidohydrolase
Accession: QEG36783
Location: 4948258-4949055
NCBI BlastP on this gene
Accession: QEG36783
Location: 4948258-4949055
NCBI BlastP on this gene
Pr1d_41190
Pr1d_41180
gshB
autoinducer-2 (AI-2) modifying protein LsrG
Accession: QEG36780
Location: 4944903-4945205
NCBI BlastP on this gene
Accession: QEG36780
Location: 4944903-4945205
NCBI BlastP on this gene
Pr1d_41160
Pr1d_41150
Pr1d_41140
hypothetical protein
Accession: QEG36777
Location: 4941190-4942764
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 174
Sequence coverage: 101 %
E-value: 8e-45
NCBI BlastP on this gene
Accession: QEG36777
Location: 4941190-4942764
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 174
Sequence coverage: 101 %
E-value: 8e-45
NCBI BlastP on this gene
Pr1d_41130
Prolyl tripeptidyl peptidase precursor
Accession: QEG36776
Location: 4939032-4941179
NCBI BlastP on this gene
Accession: QEG36776
Location: 4939032-4941179
NCBI BlastP on this gene
ptpA
ComEC family competence protein
Accession: QEG36775
Location: 4936282-4938795
NCBI BlastP on this gene
Accession: QEG36775
Location: 4936282-4938795
NCBI BlastP on this gene
Pr1d_41110
Pr1d_41100
Pr1d_41090
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP042913 : Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 173
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Pr1d_30760
Type I phosphodiesterase / nucleotide pyrophosphatase
Accession: QEG35769
Location: 3712474-3713841
NCBI BlastP on this gene
Accession: QEG35769
Location: 3712474-3713841
NCBI BlastP on this gene
Pr1d_30750
Pr1d_30740
hypothetical protein
Accession: QEG35767
Location: 3706972-3709233
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 173
Sequence coverage: 92 %
E-value: 7e-44
NCBI BlastP on this gene
Accession: QEG35767
Location: 3706972-3709233
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 173
Sequence coverage: 92 %
E-value: 7e-44
NCBI BlastP on this gene
Pr1d_30730
Pr1d_30720
Pr1d_30710
tktB
1-deoxy-D-xylulose-5-phosphate synthase
Accession: QEG35763
Location: 3701132-3702148
NCBI BlastP on this gene
Accession: QEG35763
Location: 3701132-3702148
NCBI BlastP on this gene
dxs_2
glpK_2
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP041253 : Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 173
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
FKX85_12870
FKX85_12875
carboxylesterase family protein
Accession: QDH79876
Location: 3233256-3234821
NCBI BlastP on this gene
Accession: QDH79876
Location: 3233256-3234821
NCBI BlastP on this gene
FKX85_12880
FKX85_12885
polysaccharide lyase
Accession: QDH79878
Location: 3239102-3240634
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 173
Sequence coverage: 93 %
E-value: 1e-44
NCBI BlastP on this gene
Accession: QDH79878
Location: 3239102-3240634
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 173
Sequence coverage: 93 %
E-value: 1e-44
NCBI BlastP on this gene
FKX85_12890
FKX85_12895
MerC domain-containing protein
Accession: QDH79880
Location: 3242208-3242591
NCBI BlastP on this gene
Accession: QDH79880
Location: 3242208-3242591
NCBI BlastP on this gene
FKX85_12900
FKX85_12905
FKX85_12910
FKX85_12915
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP042913 : Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 172
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hypothetical protein
Accession: QEG37972
Location: 6380709-6382025
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 92 %
E-value: 1e-44
NCBI BlastP on this gene
Accession: QEG37972
Location: 6380709-6382025
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 92 %
E-value: 1e-44
NCBI BlastP on this gene
Pr1d_53200
Pr1d_53190
sglT_14
Pr1d_53170
yhdN_4
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP026605 : Catenovulum sp. CCB-QB4 plasmid unnamed1 Total score: 1.0 Cumulative Blast bit score: 172
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
C2869_22125
C2869_22120
C2869_22115
C2869_22110
hypothetical protein
Accession: AWB69198
Location: 122612-125431
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 98 %
E-value: 7e-43
NCBI BlastP on this gene
Accession: AWB69198
Location: 122612-125431
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 98 %
E-value: 7e-43
NCBI BlastP on this gene
C2869_22105
serine/threonine protein kinase
Accession: AWB69197
Location: 120706-122022
NCBI BlastP on this gene
Accession: AWB69197
Location: 120706-122022
NCBI BlastP on this gene
C2869_22100
C2869_22095
iron ABC transporter substrate-binding protein
Accession: AWB69195
Location: 119031-120044
NCBI BlastP on this gene
Accession: AWB69195
Location: 119031-120044
NCBI BlastP on this gene
C2869_22090
C2869_22085
C2869_22080
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP023344 : Nibricoccus aquaticus strain HZ-65 chromosome Total score: 1.0 Cumulative Blast bit score: 172
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
CMV30_04980
CMV30_04985
CMV30_04990
pectate lyase
Accession: ATC63360
Location: 1174089-1176590
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
Accession: ATC63360
Location: 1174089-1176590
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
CMV30_04995
CMV30_05000
CMV30_05005
CMV30_05010
CMV30_05015
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP013729 : Roseateles depolymerans strain KCTC 42856 Total score: 1.0 Cumulative Blast bit score: 172
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
pectate lyase
Accession: ALV06992
Location: 2963684-2964622
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 7e-43
NCBI BlastP on this gene
Accession: ALV06992
Location: 2963684-2964622
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 7e-43
NCBI BlastP on this gene
RD2015_2525
RD2015_2524
RD2015_2523
RD2015_2522
RD2015_2521
RD2015_2520
RD2015_2519
RD2015_2518
RD2015_2517
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP018439 : Undibacterium sp. KW1 DNA Total score: 1.0 Cumulative Blast bit score: 172
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
UNDKW_3205
UNDKW_3206
glycine dehydrogenase (decarboxylating)
Accession: BBB61480
Location: 3567023-3569911
NCBI BlastP on this gene
Accession: BBB61480
Location: 3567023-3569911
NCBI BlastP on this gene
gcvP
glycine cleavage system H protein
Accession: BBB61481
Location: 3570032-3570409
NCBI BlastP on this gene
Accession: BBB61481
Location: 3570032-3570409
NCBI BlastP on this gene
gcvH
gcvT
hypothetical protein
Accession: BBB61483
Location: 3572377-3573747
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 1e-41
NCBI BlastP on this gene
Accession: BBB61483
Location: 3572377-3573747
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 1e-41
NCBI BlastP on this gene
UNDKW_3210
penicillin acylase family protein
Accession: BBB61484
Location: 3574168-3576588
NCBI BlastP on this gene
Accession: BBB61484
Location: 3574168-3576588
NCBI BlastP on this gene
UNDKW_3211
UNDKW_3212
TetR family transcriptional regulator
Accession: BBB61486
Location: 3578652-3579227
NCBI BlastP on this gene
Accession: BBB61486
Location: 3578652-3579227
NCBI BlastP on this gene
UNDKW_3213
UNDKW_3214
UNDKW_3215
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP017422 : Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 172
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
pectate lyase
Accession: BAV07455
Location: 4215323-4221238
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 90 %
E-value: 2e-42
NCBI BlastP on this gene
Accession: BAV07455
Location: 4215323-4221238
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 90 %
E-value: 2e-42
NCBI BlastP on this gene
FLA_3480
FLA_3479
FLA_3478
FLA_3477
FLA_3476
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP043329 : Pedobacter sp. CJ43 chromosome Total score: 1.0 Cumulative Blast bit score: 170
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
DUF3826 domain-containing protein
Accession: QEK50873
Location: 812440-813123
NCBI BlastP on this gene
Accession: QEK50873
Location: 812440-813123
NCBI BlastP on this gene
FYC62_03690
FYC62_03695
RagB/SusD family nutrient uptake outer membrane protein
Accession: QEK53219
Location: 816621-818501
NCBI BlastP on this gene
Accession: QEK53219
Location: 816621-818501
NCBI BlastP on this gene
FYC62_03700
FYC62_03705
polysaccharide lyase
Accession: QEK50876
Location: 819689-821332
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 170
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
Accession: QEK50876
Location: 819689-821332
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 170
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
FYC62_03710
FYC62_03715
FYC62_03720
FYC62_03725
DUF4038 domain-containing protein
Accession: QEK50879
Location: 826744-828138
NCBI BlastP on this gene
Accession: QEK50879
Location: 826744-828138
NCBI BlastP on this gene
FYC62_03730
FYC62_03735
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP022985 : Mariniflexile sp. TRM1-10 chromosome Total score: 1.0 Cumulative Blast bit score: 170
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Efflux pump membrane transporter BepE
Accession: AXP80800
Location: 1964473-1967607
NCBI BlastP on this gene
Accession: AXP80800
Location: 1964473-1967607
NCBI BlastP on this gene
CJ739_1714
Outer membrane protein OprM precursor
Accession: AXP80801
Location: 1967657-1969027
NCBI BlastP on this gene
Accession: AXP80801
Location: 1967657-1969027
NCBI BlastP on this gene
CJ739_1715
CJ739_1716
CJ739_1717
CJ739_1718
Glycerol-3-phosphate dehydrogenase
Accession: AXP80805
Location: 1971977-1972978
NCBI BlastP on this gene
Accession: AXP80805
Location: 1971977-1972978
NCBI BlastP on this gene
CJ739_1719
CJ739_1720
hypothetical protein
Accession: AXP80807
Location: 1974337-1975854
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 93 %
E-value: 2e-43
NCBI BlastP on this gene
Accession: AXP80807
Location: 1974337-1975854
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 93 %
E-value: 2e-43
NCBI BlastP on this gene
CJ739_1721
CJ739_1722
putative nicotinate-nucleotide adenylyltransferase
Accession: AXP80809
Location: 1976506-1977084
NCBI BlastP on this gene
Accession: AXP80809
Location: 1976506-1977084
NCBI BlastP on this gene
CJ739_1723
CJ739_1724
CJ739_1725
CJ739_1726
CJ739_1727
Type II secretion system protein D precursor
Accession: AXP80814
Location: 1980558-1982750
NCBI BlastP on this gene
Accession: AXP80814
Location: 1980558-1982750
NCBI BlastP on this gene
CJ739_1728
CJ739_1729
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP002584 : Sphingobacterium sp. 21 Total score: 1.0 Cumulative Blast bit score: 170
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Sph21_2304
Sph21_2305
Sph21_2306
RagB/SusD domain-containing protein
Accession: ADZ78861
Location: 2832004-2833962
NCBI BlastP on this gene
Accession: ADZ78861
Location: 2832004-2833962
NCBI BlastP on this gene
Sph21_2307
Sph21_2308
hypothetical protein
Accession: ADZ78863
Location: 2834797-2836458
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 92 %
E-value: 3e-43
NCBI BlastP on this gene
Accession: ADZ78863
Location: 2834797-2836458
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 92 %
E-value: 3e-43
NCBI BlastP on this gene
Sph21_2309
Sph21_2310
Sph21_2311
Sph21_2312
Sph21_2313
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
LT629740 : Mucilaginibacter mallensis strain MP1X4 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 169
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
SAMN05216490_2653
AraC-type DNA-binding protein
Accession: SDT17347
Location: 3198363-3199262
NCBI BlastP on this gene
Accession: SDT17347
Location: 3198363-3199262
NCBI BlastP on this gene
SAMN05216490_2652
SAMN05216490_2651
TonB-linked outer membrane protein, SusC/RagA family
Accession: SDT17242
Location: 3193928-3197185
NCBI BlastP on this gene
Accession: SDT17242
Location: 3193928-3197185
NCBI BlastP on this gene
SAMN05216490_2650
Starch-binding associating with outer membrane
Accession: SDT17198
Location: 3192027-3193916
NCBI BlastP on this gene
Accession: SDT17198
Location: 3192027-3193916
NCBI BlastP on this gene
SAMN05216490_2649
hypothetical protein
Accession: SDT17153
Location: 3190299-3191966
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 5e-43
NCBI BlastP on this gene
Accession: SDT17153
Location: 3190299-3191966
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 5e-43
NCBI BlastP on this gene
SAMN05216490_2648
SAMN05216490_2647
Putative collagen-binding domain of a collagenase
Accession: SDT17071
Location: 3185749-3187119
NCBI BlastP on this gene
Accession: SDT17071
Location: 3185749-3187119
NCBI BlastP on this gene
SAMN05216490_2646
Glycosyl hydrolases family 43
Accession: SDT17035
Location: 3184792-3185733
NCBI BlastP on this gene
Accession: SDT17035
Location: 3184792-3185733
NCBI BlastP on this gene
SAMN05216490_2645
SAMN05216490_2644
SAMN05216490_2643
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP041253 : Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 169
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
PepSY domain-containing protein
Accession: QDH81091
Location: 5026335-5027594
NCBI BlastP on this gene
Accession: QDH81091
Location: 5026335-5027594
NCBI BlastP on this gene
FKX85_19450
FKX85_19455
efflux RND transporter periplasmic adaptor subunit
Accession: QDH81092
Location: 5029282-5030463
NCBI BlastP on this gene
Accession: QDH81092
Location: 5029282-5030463
NCBI BlastP on this gene
FKX85_19460
efflux RND transporter permease subunit
Accession: QDH81093
Location: 5030466-5033582
NCBI BlastP on this gene
Accession: QDH81093
Location: 5030466-5033582
NCBI BlastP on this gene
FKX85_19465
T9SS C-terminal target domain-containing protein
Accession: FKX85_19470
Location: 5034123-5036591
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 169
Sequence coverage: 91 %
E-value: 4e-42
NCBI BlastP on this gene
Accession: FKX85_19470
Location: 5034123-5036591
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 169
Sequence coverage: 91 %
E-value: 4e-42
NCBI BlastP on this gene
FKX85_19470
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP047647 : Hymenobacter sp. BT182 chromosome Total score: 1.0 Cumulative Blast bit score: 167
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
GUY19_05395
SusC/RagA family TonB-linked outer membrane protein
Accession: QHJ06758
Location: 1216554-1219547
NCBI BlastP on this gene
Accession: QHJ06758
Location: 1216554-1219547
NCBI BlastP on this gene
GUY19_05390
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHJ06757
Location: 1214790-1216538
NCBI BlastP on this gene
Accession: QHJ06757
Location: 1214790-1216538
NCBI BlastP on this gene
GUY19_05385
polysaccharide lyase
Accession: QHJ06756
Location: 1213009-1214673
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 93 %
E-value: 3e-42
NCBI BlastP on this gene
Accession: QHJ06756
Location: 1213009-1214673
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 93 %
E-value: 3e-42
NCBI BlastP on this gene
GUY19_05380
GUY19_05375
DUF4038 domain-containing protein
Accession: QHJ09605
Location: 1208115-1209545
NCBI BlastP on this gene
Accession: QHJ09605
Location: 1208115-1209545
NCBI BlastP on this gene
GUY19_05370
GUY19_05365
GUY19_05360
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP047647 : Hymenobacter sp. BT182 chromosome Total score: 1.0 Cumulative Blast bit score: 167
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
GUY19_05115
PspC domain-containing protein
Accession: QHJ06712
Location: 1134348-1134566
NCBI BlastP on this gene
Accession: QHJ06712
Location: 1134348-1134566
NCBI BlastP on this gene
GUY19_05120
GUY19_05125
GUY19_05130
GUY19_05135
pyruvate dehydrogenase complex E1 component subunit beta
Accession: QHJ06716
Location: 1137438-1138418
NCBI BlastP on this gene
Accession: QHJ06716
Location: 1137438-1138418
NCBI BlastP on this gene
GUY19_05140
GUY19_05145
GUY19_05150
methyltransferase domain-containing protein
Accession: QHJ06718
Location: 1139746-1140483
NCBI BlastP on this gene
Accession: QHJ06718
Location: 1139746-1140483
NCBI BlastP on this gene
GUY19_05155
DUF2256 domain-containing protein
Accession: QHJ06719
Location: 1140589-1140777
NCBI BlastP on this gene
Accession: QHJ06719
Location: 1140589-1140777
NCBI BlastP on this gene
GUY19_05160
GUY19_05165
polysaccharide lyase
Accession: QHJ06721
Location: 1141532-1143169
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 90 %
E-value: 4e-42
NCBI BlastP on this gene
Accession: QHJ06721
Location: 1141532-1143169
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 90 %
E-value: 4e-42
NCBI BlastP on this gene
GUY19_05170
GUY19_05175
GUY19_05180
DUF3826 domain-containing protein
Accession: QHJ06724
Location: 1146119-1146802
NCBI BlastP on this gene
Accession: QHJ06724
Location: 1146119-1146802
NCBI BlastP on this gene
GUY19_05185
SusC/RagA family TonB-linked outer membrane protein
Accession: QHJ06725
Location: 1146833-1150063
NCBI BlastP on this gene
Accession: QHJ06725
Location: 1146833-1150063
NCBI BlastP on this gene
GUY19_05190
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHJ06726
Location: 1150082-1151866
NCBI BlastP on this gene
Accession: QHJ06726
Location: 1150082-1151866
NCBI BlastP on this gene
GUY19_05195
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP041253 : Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 167
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
PepSY domain-containing protein
Accession: QDH78693
Location: 1534448-1535773
NCBI BlastP on this gene
Accession: QDH78693
Location: 1534448-1535773
NCBI BlastP on this gene
FKX85_06445
FKX85_06440
FKX85_06435
FKX85_06430
DUF4919 domain-containing protein
Accession: QDH78689
Location: 1530221-1530916
NCBI BlastP on this gene
Accession: QDH78689
Location: 1530221-1530916
NCBI BlastP on this gene
FKX85_06425
FKX85_06420
glycosyltransferase family 39 protein
Accession: QDH78687
Location: 1527951-1529462
NCBI BlastP on this gene
Accession: QDH78687
Location: 1527951-1529462
NCBI BlastP on this gene
FKX85_06415
polysaccharide lyase
Accession: QDH78686
Location: 1525918-1527618
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 95 %
E-value: 3e-42
NCBI BlastP on this gene
Accession: QDH78686
Location: 1525918-1527618
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 95 %
E-value: 3e-42
NCBI BlastP on this gene
FKX85_06410
FKX85_06405
FKX85_06400
fumC
SPOR domain-containing protein
Accession: QDH78683
Location: 1521582-1522202
NCBI BlastP on this gene
Accession: QDH78683
Location: 1521582-1522202
NCBI BlastP on this gene
FKX85_06390
FKX85_06385
aspS
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP020991 : Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 167
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
NADP-reducing hydrogenase, subunit B
Accession: AUO20297
Location: 2583312-2583692
NCBI BlastP on this gene
Accession: AUO20297
Location: 2583312-2583692
NCBI BlastP on this gene
B9O19_02155
NAD-reducing hydrogenase subunit HoxF
Accession: AUO20298
Location: 2583696-2585492
NCBI BlastP on this gene
Accession: AUO20298
Location: 2583696-2585492
NCBI BlastP on this gene
B9O19_02156
periplasmic [Fe] hydrogenase large subunit
Accession: AUO20299
Location: 2585505-2587247
NCBI BlastP on this gene
Accession: AUO20299
Location: 2585505-2587247
NCBI BlastP on this gene
B9O19_02157
transcriptional regulator, XRE family
Accession: AUO20300
Location: 2587795-2588118
NCBI BlastP on this gene
Accession: AUO20300
Location: 2587795-2588118
NCBI BlastP on this gene
B9O19_02158
B9O19_02159
pectate lyase family 1
Accession: AUO20302
Location: 2589949-2593386
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 98 %
E-value: 7e-41
NCBI BlastP on this gene
Accession: AUO20302
Location: 2589949-2593386
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 98 %
E-value: 7e-41
NCBI BlastP on this gene
B9O19_02160
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP049143 : Roseimicrobium sp. ORNL1 chromosome Total score: 1.0 Cumulative Blast bit score: 166
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
DUF3445 domain-containing protein
Accession: QIF03659
Location: 4868103-4868936
NCBI BlastP on this gene
Accession: QIF03659
Location: 4868103-4868936
NCBI BlastP on this gene
G5S37_19755
G5S37_19750
KamA family radical SAM protein
Accession: QIF03657
Location: 4865099-4866316
NCBI BlastP on this gene
Accession: QIF03657
Location: 4865099-4866316
NCBI BlastP on this gene
G5S37_19745
winged helix-turn-helix transcriptional regulator
Accession: QIF03656
Location: 4863709-4864932
NCBI BlastP on this gene
Accession: QIF03656
Location: 4863709-4864932
NCBI BlastP on this gene
G5S37_19740
G5S37_19735
G5S37_19730
sugar phosphate isomerase/epimerase
Accession: QIF06130
Location: 4860952-4861881
NCBI BlastP on this gene
Accession: QIF06130
Location: 4860952-4861881
NCBI BlastP on this gene
G5S37_19725
pectate lyase
Accession: QIF03655
Location: 4859359-4860723
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 166
Sequence coverage: 74 %
E-value: 3e-42
NCBI BlastP on this gene
Accession: QIF03655
Location: 4859359-4860723
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 166
Sequence coverage: 74 %
E-value: 3e-42
NCBI BlastP on this gene
G5S37_19720
DUF4185 domain-containing protein
Accession: QIF03654
Location: 4858234-4859292
NCBI BlastP on this gene
Accession: QIF03654
Location: 4858234-4859292
NCBI BlastP on this gene
G5S37_19715
G5S37_19710
G5S37_19705
nuclear transport factor 2 family protein
Accession: QIF03651
Location: 4854152-4854622
NCBI BlastP on this gene
Accession: QIF03651
Location: 4854152-4854622
NCBI BlastP on this gene
G5S37_19700
PQQ-binding-like beta-propeller repeat protein
Accession: QIF03650
Location: 4852805-4854076
NCBI BlastP on this gene
Accession: QIF03650
Location: 4852805-4854076
NCBI BlastP on this gene
G5S37_19695
G5S37_19690
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP003178 : Niastella koreensis GR20-10 Total score: 1.0 Cumulative Blast bit score: 165
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Niako_2525
Glyoxalase/bleomycin resistance
Accession: AEV98864
Location: 3128007-3128420
NCBI BlastP on this gene
Accession: AEV98864
Location: 3128007-3128420
NCBI BlastP on this gene
Niako_2524
Niako_2523
Peptidoglycan-binding lysin domain protein
Accession: AEV98862
Location: 3126708-3127361
NCBI BlastP on this gene
Accession: AEV98862
Location: 3126708-3127361
NCBI BlastP on this gene
Niako_2522
rifampin ADP-ribosyl transferase
Accession: AEV98861
Location: 3126096-3126521
NCBI BlastP on this gene
Accession: AEV98861
Location: 3126096-3126521
NCBI BlastP on this gene
Niako_2521
Niako_2520
transcriptional regulator, LacI family
Accession: AEV98859
Location: 3124221-3125258
NCBI BlastP on this gene
Accession: AEV98859
Location: 3124221-3125258
NCBI BlastP on this gene
Niako_2519
hypothetical protein
Accession: AEV98858
Location: 3122536-3124167
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 165
Sequence coverage: 92 %
E-value: 2e-41
NCBI BlastP on this gene
Accession: AEV98858
Location: 3122536-3124167
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 165
Sequence coverage: 92 %
E-value: 2e-41
NCBI BlastP on this gene
Niako_2518
putative transcriptional regulator, Crp/Fnr family
Accession: AEV98857
Location: 3121713-3122285
NCBI BlastP on this gene
Accession: AEV98857
Location: 3121713-3122285
NCBI BlastP on this gene
Niako_2517
Niako_2516
Niako_2515
bifunctional deaminase-reductase domain protein
Accession: AEV98854
Location: 3119796-3120359
NCBI BlastP on this gene
Accession: AEV98854
Location: 3119796-3120359
NCBI BlastP on this gene
Niako_2514
Niako_2513
putative RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: AEV98852
Location: 3118067-3119380
NCBI BlastP on this gene
Accession: AEV98852
Location: 3118067-3119380
NCBI BlastP on this gene
Niako_2512
Niako_2511
Niako_2510
Niako_2509
Niako_2508
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP039396 : Muribaculum sp. H5 chromosome. Total score: 1.0 Cumulative Blast bit score: 163
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
E7747_15890
glutamate-5-semialdehyde dehydrogenase
Accession: QCD43605
Location: 3583220-3584470
NCBI BlastP on this gene
Accession: QCD43605
Location: 3583220-3584470
NCBI BlastP on this gene
E7747_15895
proB
pyrroline-5-carboxylate reductase
Accession: QCD43607
Location: 3585676-3586539
NCBI BlastP on this gene
Accession: QCD43607
Location: 3585676-3586539
NCBI BlastP on this gene
E7747_15905
E7747_15910
RagB/SusD family nutrient uptake outer membrane protein
Accession: E7747_15915
Location: 3587122-3589301
NCBI BlastP on this gene
Accession: E7747_15915
Location: 3587122-3589301
NCBI BlastP on this gene
E7747_15915
polysaccharide lyase
Accession: QCD43744
Location: 3589505-3591223
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
Accession: QCD43744
Location: 3589505-3591223
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
E7747_15920
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP039396 : Muribaculum sp. H5 chromosome. Total score: 1.0 Cumulative Blast bit score: 163
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
DUF4957 domain-containing protein
Accession: QCD43458
Location: 3381860-3383332
NCBI BlastP on this gene
Accession: QCD43458
Location: 3381860-3383332
NCBI BlastP on this gene
E7747_15050
E7747_15055
RagB/SusD family nutrient uptake outer membrane protein
Accession: QCD43460
Location: 3386711-3388921
NCBI BlastP on this gene
Accession: QCD43460
Location: 3386711-3388921
NCBI BlastP on this gene
E7747_15060
polysaccharide lyase
Accession: QCD43733
Location: 3389125-3390843
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
Accession: QCD43733
Location: 3389125-3390843
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
E7747_15065
DUF3826 domain-containing protein
Accession: QCD43461
Location: 3390868-3391536
NCBI BlastP on this gene
Accession: QCD43461
Location: 3390868-3391536
NCBI BlastP on this gene
E7747_15070
E7747_15075
DUF1593 domain-containing protein
Accession: QCD43463
Location: 3394946-3396250
NCBI BlastP on this gene
Accession: QCD43463
Location: 3394946-3396250
NCBI BlastP on this gene
E7747_15080
E7747_15085
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP020991 : Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 163
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
S-layer domain-containing protein
Accession: AUO18489
Location: 351097-354123
NCBI BlastP on this gene
Accession: AUO18489
Location: 351097-354123
NCBI BlastP on this gene
B9O19_00305
B9O19_00306
B9O19_00307
B9O19_00308
probabale oxalocrotonate tautomerase
Accession: AUO18493
Location: 356768-356977
NCBI BlastP on this gene
Accession: AUO18493
Location: 356768-356977
NCBI BlastP on this gene
B9O19_00309
B9O19_00310
B9O19_00311
pectate lyase family 1
Accession: AUO18496
Location: 358585-361302
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 97 %
E-value: 5e-40
NCBI BlastP on this gene
Accession: AUO18496
Location: 358585-361302
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 97 %
E-value: 5e-40
NCBI BlastP on this gene
B9O19_00312
B9O19_00313
B9O19_00314
B9O19_00315
B9O19_00316
V-type sodium ATPase catalytic subunit A
Accession: AUO18501
Location: 366216-367982
NCBI BlastP on this gene
Accession: AUO18501
Location: 366216-367982
NCBI BlastP on this gene
B9O19_00317
B9O19_00318
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP002584 : Sphingobacterium sp. 21 Total score: 1.0 Cumulative Blast bit score: 163
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Sph21_2568
Sph21_2569
Sph21_2570
PepSY-associated TM helix domain protein
Accession: ADZ79122
Location: 3168499-3169641
NCBI BlastP on this gene
Accession: ADZ79122
Location: 3168499-3169641
NCBI BlastP on this gene
Sph21_2571
TonB-dependent siderophore receptor
Accession: ADZ79123
Location: 3169644-3171758
NCBI BlastP on this gene
Accession: ADZ79123
Location: 3169644-3171758
NCBI BlastP on this gene
Sph21_2572
Sph21_2573
hypothetical protein
Accession: ADZ79125
Location: 3172491-3174116
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 163
Sequence coverage: 90 %
E-value: 8e-41
NCBI BlastP on this gene
Accession: ADZ79125
Location: 3172491-3174116
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 163
Sequence coverage: 90 %
E-value: 8e-41
NCBI BlastP on this gene
Sph21_2574
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP021382 : Cellvibrio sp. PSBB006 chromosome Total score: 1.0 Cumulative Blast bit score: 162
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
dicarboxylate/amino acid:cation symporter
Accession: ARU28644
Location: 3623654-3624961
NCBI BlastP on this gene
Accession: ARU28644
Location: 3623654-3624961
NCBI BlastP on this gene
CBR65_14995
CBR65_15000
CBR65_15005
4-hydroxybenzoate polyprenyltransferase
Accession: ARU28647
Location: 3626093-3627019
NCBI BlastP on this gene
Accession: ARU28647
Location: 3626093-3627019
NCBI BlastP on this gene
CBR65_15010
phosphate regulon transcriptional regulatory protein PhoB
Accession: ARU28648
Location: 3627143-3627841
NCBI BlastP on this gene
Accession: ARU28648
Location: 3627143-3627841
NCBI BlastP on this gene
CBR65_15015
PAS domain-containing sensor histidine kinase
Accession: ARU28649
Location: 3627987-3629261
NCBI BlastP on this gene
Accession: ARU28649
Location: 3627987-3629261
NCBI BlastP on this gene
CBR65_15020
CBR65_15025
CBR65_15030
hypothetical protein
Accession: ARU28652
Location: 3630869-3633220
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 162
Sequence coverage: 92 %
E-value: 7e-40
NCBI BlastP on this gene
Accession: ARU28652
Location: 3630869-3633220
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 162
Sequence coverage: 92 %
E-value: 7e-40
NCBI BlastP on this gene
CBR65_15035
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
LT629758 : Actinoplanes derwentensis strain DSM 43941 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 161
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
pectate lyase
Accession: SDT74959
Location: 7952377-7953996
BlastP hit with VTO45599.1
Percentage identity: 36 %
BlastP bit score: 161
Sequence coverage: 12 %
E-value: 1e-37
NCBI BlastP on this gene
Accession: SDT74959
Location: 7952377-7953996
BlastP hit with VTO45599.1
Percentage identity: 36 %
BlastP bit score: 161
Sequence coverage: 12 %
E-value: 1e-37
NCBI BlastP on this gene
SAMN04489716_7149
SAMN04489716_7148
SAMN04489716_7147
SAMN04489716_7146
SAMN04489716_7145
SAMN04489716_7144
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP030261 : Flavobacterium sp. HYN0086 chromosome Total score: 1.0 Cumulative Blast bit score: 161
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
HYN86_16520
HYN86_16515
HYN86_16510
SusC/RagA family TonB-linked outer membrane protein
Accession: AXB58103
Location: 3753617-3756997
NCBI BlastP on this gene
Accession: AXB58103
Location: 3753617-3756997
NCBI BlastP on this gene
HYN86_16505
RagB/SusD family nutrient uptake outer membrane protein
Accession: AXB58102
Location: 3752008-3753612
NCBI BlastP on this gene
Accession: AXB58102
Location: 3752008-3753612
NCBI BlastP on this gene
HYN86_16500
polysaccharide lyase
Accession: AXB58101
Location: 3749993-3751696
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 161
Sequence coverage: 90 %
E-value: 4e-40
NCBI BlastP on this gene
Accession: AXB58101
Location: 3749993-3751696
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 161
Sequence coverage: 90 %
E-value: 4e-40
NCBI BlastP on this gene
HYN86_16495
HYN86_16490
HYN86_16485
HYN86_16480
HYN86_16475
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP003346 : Echinicola vietnamensis DSM 17526 Total score: 1.0 Cumulative Blast bit score: 160
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Fe2+-dicitrate sensor, membrane component
Accession: AGA77594
Location: 1528585-1529655
NCBI BlastP on this gene
Accession: AGA77594
Location: 1528585-1529655
NCBI BlastP on this gene
Echvi_1323
TonB-linked outer membrane protein, SusC/RagA family
Accession: AGA77595
Location: 1529773-1533282
NCBI BlastP on this gene
Accession: AGA77595
Location: 1529773-1533282
NCBI BlastP on this gene
Echvi_1324
Echvi_1325
hypothetical protein
Accession: AGA77597
Location: 1535777-1537474
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 93 %
E-value: 8e-40
NCBI BlastP on this gene
Accession: AGA77597
Location: 1535777-1537474
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 93 %
E-value: 8e-40
NCBI BlastP on this gene
Echvi_1326
endoribonuclease L-PSP, putative
Accession: AGA77598
Location: 1537696-1538079
NCBI BlastP on this gene
Accession: AGA77598
Location: 1537696-1538079
NCBI BlastP on this gene
Echvi_1327
Echvi_1328
Echvi_1329
Echvi_1330
peptidyl-prolyl cis-trans isomerase (rotamase) - cyclophilin family
Accession: AGA77602
Location: 1541336-1541920
NCBI BlastP on this gene
Accession: AGA77602
Location: 1541336-1541920
NCBI BlastP on this gene
Echvi_1331
single-stranded-DNA-specific exonuclease RecJ
Accession: AGA77603
Location: 1542293-1544062
NCBI BlastP on this gene
Accession: AGA77603
Location: 1542293-1544062
NCBI BlastP on this gene
Echvi_1332
ABC-type (unclassified) transport system, ATPase component
Accession: AGA77604
Location: 1544065-1544796
NCBI BlastP on this gene
Accession: AGA77604
Location: 1544065-1544796
NCBI BlastP on this gene
Echvi_1333
acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II
Accession: AGA77605
Location: 1544800-1546320
NCBI BlastP on this gene
Accession: AGA77605
Location: 1544800-1546320
NCBI BlastP on this gene
Echvi_1334
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP017422 : Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 160
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
FLA_2726
FLA_2725
FLA_2723
transferase, hexapeptide repeat family
Accession: BAV06703
Location: 3274355-3274663
NCBI BlastP on this gene
Accession: BAV06703
Location: 3274355-3274663
NCBI BlastP on this gene
FLA_2722
transcriptional regulator, AraC family
Accession: BAV06702
Location: 3273879-3274343
NCBI BlastP on this gene
Accession: BAV06702
Location: 3273879-3274343
NCBI BlastP on this gene
FLA_2721
FLA_2720
FLA_2719
FLA_2718
DNA-binding response regulator, AraC family
Accession: BAV06698
Location: 3271207-3271581
NCBI BlastP on this gene
Accession: BAV06698
Location: 3271207-3271581
NCBI BlastP on this gene
FLA_2717
FLA_2716
pectate lyase
Accession: BAV06696
Location: 3268270-3271014
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 87 %
E-value: 5e-39
NCBI BlastP on this gene
Accession: BAV06696
Location: 3268270-3271014
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 87 %
E-value: 5e-39
NCBI BlastP on this gene
FLA_2715
FLA_2714
FLA_2713
endo-1,4-beta-xylanase D precursor
Accession: BAV06693
Location: 3266150-3267754
NCBI BlastP on this gene
Accession: BAV06693
Location: 3266150-3267754
NCBI BlastP on this gene
FLA_2712
rhamnogalacturonan acetylesterase
Accession: BAV06692
Location: 3264418-3266100
NCBI BlastP on this gene
Accession: BAV06692
Location: 3264418-3266100
NCBI BlastP on this gene
FLA_2711
FLA_2710
FLA_2709
FLA_2708
FLA_2707
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP017422 : Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 160
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
FLA_1170
FLA_1171
FLA_1172
outer membrane protein, nutrient binding
Accession: BAV05166
Location: 1385496-1387322
NCBI BlastP on this gene
Accession: BAV05166
Location: 1385496-1387322
NCBI BlastP on this gene
FLA_1173
pectate lyase
Accession: BAV05167
Location: 1387366-1389015
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 91 %
E-value: 5e-40
NCBI BlastP on this gene
Accession: BAV05167
Location: 1387366-1389015
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 91 %
E-value: 5e-40
NCBI BlastP on this gene
FLA_1174
FLA_1175
FLA_1176
FLA_1177
lipase/acylhydrolase family protein
Accession: BAV05171
Location: 1394535-1395293
NCBI BlastP on this gene
Accession: BAV05171
Location: 1394535-1395293
NCBI BlastP on this gene
FLA_1178
FLA_1179
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP043306 : Cellvibrio japonicus strain ADPT1-KOJIBIOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 159
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hypothetical protein
Accession: QEI20559
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QEI20559
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
FY115_15030
FY115_15025
small ribosomal subunit biogenesis GTPase RsgA
Accession: QEI20557
Location: 3702363-3703388
NCBI BlastP on this gene
Accession: QEI20557
Location: 3702363-3703388
NCBI BlastP on this gene
rsgA
FY115_15015
FY115_15010
FY115_15005
FY115_15000
FY115_14995
bla
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP043305 : Cellvibrio japonicus strain ADPT2-NIGEROSE chromosome. Total score: 1.0 Cumulative Blast bit score: 159
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hypothetical protein
Accession: QEI16981
Location: 3704311-3706509
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QEI16981
Location: 3704311-3706509
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
FY116_15035
FY116_15030
small ribosomal subunit biogenesis GTPase RsgA
Accession: QEI16979
Location: 3702358-3703383
NCBI BlastP on this gene
Accession: QEI16979
Location: 3702358-3703383
NCBI BlastP on this gene
rsgA
FY116_15020
FY116_15015
FY116_15010
FY116_15005
FY116_15000
bla
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP043304 : Cellvibrio japonicus strain ADPT3-ISOMALTOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 159
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hypothetical protein
Accession: QEI13407
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QEI13407
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
FY117_15030
FY117_15025
small ribosomal subunit biogenesis GTPase RsgA
Accession: QEI13405
Location: 3702363-3703388
NCBI BlastP on this gene
Accession: QEI13405
Location: 3702363-3703388
NCBI BlastP on this gene
rsgA
FY117_15015
FY117_15010
FY117_15005
FY117_15000
FY117_14995
bla
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP036350 : Planctomycetes bacterium K2D chromosome Total score: 1.0 Cumulative Blast bit score: 159
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
K2D_26110
K2D_26120
K2D_26130
RNA polymerase-binding transcription factor DksA
Accession: QDV79005
Location: 3218170-3218523
NCBI BlastP on this gene
Accession: QDV79005
Location: 3218170-3218523
NCBI BlastP on this gene
dksA
thrC
lacZ_4
hypothetical protein
Accession: QDV79008
Location: 3222879-3225056
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
Accession: QDV79008
Location: 3222879-3225056
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
K2D_26170
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP036349 : Planctomycetes bacterium Spa11 chromosome. Total score: 1.0 Cumulative Blast bit score: 159
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Spa11_26090
Spa11_26100
Spa11_26110
RNA polymerase-binding transcription factor DksA
Accession: QDV74409
Location: 3285998-3286351
NCBI BlastP on this gene
Accession: QDV74409
Location: 3285998-3286351
NCBI BlastP on this gene
dksA
thrC
lacZ_4
hypothetical protein
Accession: QDV74412
Location: 3290707-3292884
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
Accession: QDV74412
Location: 3290707-3292884
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
Spa11_26150
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP000934 : Cellvibrio japonicus Ueda107 Total score: 1.0 Cumulative Blast bit score: 159
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
pectate lyase, putative, pel1G
Accession: ACE83184
Location: 3704298-3706496
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: ACE83184
Location: 3704298-3706496
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
pel1G
rhdA
CJA_3118
orn
pectin methylesterase putative, pme8B
Accession: ACE83435
Location: 3701180-3701401
NCBI BlastP on this gene
Accession: ACE83435
Location: 3701180-3701401
NCBI BlastP on this gene
pme8B
CJA_3115
CJA_3114
CJA_3113
putative dipeptidyl peptidase IV
Accession: ACE86065
Location: 3698344-3699336
NCBI BlastP on this gene
Accession: ACE86065
Location: 3698344-3699336
NCBI BlastP on this gene
CJA_3112
CJA_3111
CJA_3110
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP019371 : Actinoplanes sp. OR16 DNA Total score: 1.0 Cumulative Blast bit score: 158
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
glyceraldehyde 3-phosphate reductase
Accession: BBH65236
Location: 2069915-2070910
NCBI BlastP on this gene
Accession: BBH65236
Location: 2069915-2070910
NCBI BlastP on this gene
ACTI_19210
sulfotransferase family protein
Accession: BBH65237
Location: 2070896-2071813
NCBI BlastP on this gene
Accession: BBH65237
Location: 2070896-2071813
NCBI BlastP on this gene
ACTI_19220
ACTI_19230
ACTI_19240
ACTI_19250
ACTI_19260
ACTI_19270
ACTI_19280
hypothetical protein
Accession: BBH65244
Location: 2077591-2079135
BlastP hit with VTO45599.1
Percentage identity: 37 %
BlastP bit score: 158
Sequence coverage: 12 %
E-value: 1e-36
NCBI BlastP on this gene
Accession: BBH65244
Location: 2077591-2079135
BlastP hit with VTO45599.1
Percentage identity: 37 %
BlastP bit score: 158
Sequence coverage: 12 %
E-value: 1e-36
NCBI BlastP on this gene
ACTI_19290
ACTI_19300
ACTI_19310
ACTI_19320
ACTI_19330
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP020991 : Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 157
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
3-oxoacyl-(acyl-carrier-protein) reductase
Accession: AUO20008
Location: 2212386-2213138
NCBI BlastP on this gene
Accession: AUO20008
Location: 2212386-2213138
NCBI BlastP on this gene
B9O19_01859
B9O19_01860
3-oxoacyl-(acyl-carrier-protein) synthase 2
Accession: AUO20010
Location: 2213664-2214902
NCBI BlastP on this gene
Accession: AUO20010
Location: 2213664-2214902
NCBI BlastP on this gene
B9O19_01861
GCN5-related N-acetyltransferase
Accession: AUO20011
Location: 2214976-2215989
NCBI BlastP on this gene
Accession: AUO20011
Location: 2214976-2215989
NCBI BlastP on this gene
B9O19_01862
B9O19_01863
B9O19_01864
B9O19_01865
copper amine oxidase-like domain-containing protein
Accession: AUO20015
Location: 2218246-2219430
NCBI BlastP on this gene
Accession: AUO20015
Location: 2218246-2219430
NCBI BlastP on this gene
B9O19_01866
pectate lyase family 1
Accession: AUO20016
Location: 2219740-2222430
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 157
Sequence coverage: 90 %
E-value: 5e-38
NCBI BlastP on this gene
Accession: AUO20016
Location: 2219740-2222430
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 157
Sequence coverage: 90 %
E-value: 5e-38
NCBI BlastP on this gene
B9O19_01867
B9O19_01868
B9O19_01869
S-layer domain-containing protein
Accession: AUO20019
Location: 2224870-2230200
NCBI BlastP on this gene
Accession: AUO20019
Location: 2224870-2230200
NCBI BlastP on this gene
B9O19_01870
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP048113 : Chitinophaga sp. H33E-04 chromosome Total score: 1.0 Cumulative Blast bit score: 154
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
T9SS type A sorting domain-containing protein
Accession: QHS62610
Location: 6079946-6081601
NCBI BlastP on this gene
Accession: QHS62610
Location: 6079946-6081601
NCBI BlastP on this gene
GWR21_24420
GWR21_24425
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHS62612
Location: 6085364-6087127
NCBI BlastP on this gene
Accession: QHS62612
Location: 6085364-6087127
NCBI BlastP on this gene
GWR21_24430
DUF3826 domain-containing protein
Accession: QHS62613
Location: 6087189-6089294
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 154
Sequence coverage: 94 %
E-value: 1e-37
NCBI BlastP on this gene
Accession: QHS62613
Location: 6087189-6089294
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 154
Sequence coverage: 94 %
E-value: 1e-37
NCBI BlastP on this gene
GWR21_24435
GWR21_24440
DUF4038 domain-containing protein
Accession: QHS62615
Location: 6092389-6093792
NCBI BlastP on this gene
Accession: QHS62615
Location: 6092389-6093792
NCBI BlastP on this gene
GWR21_24445
glycoside hydrolase family 43 protein
Accession: QHS62616
Location: 6093807-6094709
NCBI BlastP on this gene
Accession: QHS62616
Location: 6093807-6094709
NCBI BlastP on this gene
GWR21_24450
GWR21_24455
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP021904 : Alkalitalea saponilacus strain SC/BZ-SP2 chromosome Total score: 1.0 Cumulative Blast bit score: 154
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
tsaB
CDL62_08555
efflux RND transporter periplasmic adaptor subunit
Accession: ASB49187
Location: 2188471-2189553
NCBI BlastP on this gene
Accession: ASB49187
Location: 2188471-2189553
NCBI BlastP on this gene
CDL62_08560
ABC transporter ATP-binding protein
Accession: ASB49188
Location: 2189622-2190902
NCBI BlastP on this gene
Accession: ASB49188
Location: 2189622-2190902
NCBI BlastP on this gene
CDL62_08565
CDL62_08570
macrolide ABC transporter ATP-binding protein
Accession: ASB51107
Location: 2192321-2193043
NCBI BlastP on this gene
Accession: ASB51107
Location: 2192321-2193043
NCBI BlastP on this gene
CDL62_08575
hypothetical protein
Accession: ASB49190
Location: 2193569-2196439
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 154
Sequence coverage: 14 %
E-value: 3e-34
NCBI BlastP on this gene
Accession: ASB49190
Location: 2193569-2196439
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 154
Sequence coverage: 14 %
E-value: 3e-34
NCBI BlastP on this gene
CDL62_08580
pflB
pyruvate formate-lyase 1-activating enzyme
Accession: ASB49192
Location: 2198990-2199805
NCBI BlastP on this gene
Accession: ASB49192
Location: 2198990-2199805
NCBI BlastP on this gene
pflA
CDL62_08595
CDL62_08600
CDL62_08605
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
LT622246 : Bacteroides ovatus V975 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 153
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
BACOV975_03474
BACOV975_03475
two-component system sensor histidine
Accession: SCV09682
Location: 4621311-4625741
NCBI BlastP on this gene
Accession: SCV09682
Location: 4621311-4625741
NCBI BlastP on this gene
BACOV975_03476
hypothetical protein
Accession: SCV09683
Location: 4625800-4627482
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: SCV09683
Location: 4625800-4627482
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
BACOV975_03477
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
FP929033 : Bacteroides xylanisolvens XB1A draft genome. Total score: 1.0 Cumulative Blast bit score: 153
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Beta-galactosidase/beta-glucuronidase
Accession: CBK65668
Location: 490785-493439
NCBI BlastP on this gene
Accession: CBK65668
Location: 490785-493439
NCBI BlastP on this gene
BXY_04110
His Kinase A (phosphoacceptor) domain./Histidine
Accession: CBK65669
Location: 493613-498010
NCBI BlastP on this gene
Accession: CBK65669
Location: 493613-498010
NCBI BlastP on this gene
BXY_04120
hypothetical protein
Accession: CBK65670
Location: 498069-499751
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: CBK65670
Location: 498069-499751
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
BXY_04130
BXY_04140
RNA polymerase sigma factor, sigma-70 family
Accession: CBK65672
Location: 503078-503629
NCBI BlastP on this gene
Accession: CBK65672
Location: 503078-503629
NCBI BlastP on this gene
BXY_04150
BXY_04160
BXY_04170
Uncharacterized conserved protein
Accession: CBK65675
Location: 505413-506180
NCBI BlastP on this gene
Accession: CBK65675
Location: 505413-506180
NCBI BlastP on this gene
BXY_04180
BXY_04190
BXY_04200
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP046397 : Bacteroides ovatus strain FDAARGOS_733 chromosome Total score: 1.0 Cumulative Blast bit score: 153
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
FOC41_03120
FOC41_03125
polysaccharide lyase
Accession: QGT70025
Location: 848992-850674
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: QGT70025
Location: 848992-850674
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
FOC41_03130
FOC41_03135
sigma-70 family RNA polymerase sigma factor
Accession: QGT74087
Location: 854064-854555
NCBI BlastP on this gene
Accession: QGT74087
Location: 854064-854555
NCBI BlastP on this gene
FOC41_03140
FOC41_03145
FOC41_03150
FOC41_03155
FOC41_03160
FOC41_03165
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP041230 : Bacteroides xylanisolvens strain H207 chromosome Total score: 1.0 Cumulative Blast bit score: 153
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
FKZ68_16475
FKZ68_16480
polysaccharide lyase
Accession: QDH55717
Location: 4324837-4326519
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: QDH55717
Location: 4324837-4326519
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
FKZ68_16485
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP012938 : Bacteroides ovatus strain ATCC 8483 Total score: 1.0 Cumulative Blast bit score: 153
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
lacZ_6
tmoS_2
hypothetical protein
Accession: ALJ45347
Location: 858860-860542
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: ALJ45347
Location: 858860-860542
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Bovatus_00681
Bovatus_00682
RNA polymerase sigma factor SigM
Accession: ALJ45349
Location: 863872-864420
NCBI BlastP on this gene
Accession: ALJ45349
Location: 863872-864420
NCBI BlastP on this gene
sigM_1
Bovatus_00684
ybjJ
yjjP_1
Bovatus_00687
Bovatus_00688
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP050831 : Bacteroides sp. CBA7301 chromosome Total score: 1.0 Cumulative Blast bit score: 150
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
BacF7301_17805
BacF7301_17810
BacF7301_17815
polysaccharide lyase
Accession: QIU95892
Location: 4832610-4834292
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 89 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QIU95892
Location: 4832610-4834292
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 89 %
E-value: 2e-36
NCBI BlastP on this gene
BacF7301_17820
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP039396 : Muribaculum sp. H5 chromosome. Total score: 1.0 Cumulative Blast bit score: 150
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
E7747_06865
E7747_06870
DUF5119 domain-containing protein
Accession: QCD42021
Location: 1584612-1585550
NCBI BlastP on this gene
Accession: QCD42021
Location: 1584612-1585550
NCBI BlastP on this gene
E7747_06875
DUF3575 domain-containing protein
Accession: QCD42022
Location: 1585547-1586746
NCBI BlastP on this gene
Accession: QCD42022
Location: 1585547-1586746
NCBI BlastP on this gene
E7747_06880
methylmalonyl Co-A mutase-associated GTPase MeaB
Accession: QCD42023
Location: 1586985-1588070
NCBI BlastP on this gene
Accession: QCD42023
Location: 1586985-1588070
NCBI BlastP on this gene
meaB
DUF1573 domain-containing protein
Accession: QCD42024
Location: 1588096-1589178
NCBI BlastP on this gene
Accession: QCD42024
Location: 1588096-1589178
NCBI BlastP on this gene
E7747_06890
DUF1573 domain-containing protein
Accession: QCD42025
Location: 1589222-1589602
NCBI BlastP on this gene
Accession: QCD42025
Location: 1589222-1589602
NCBI BlastP on this gene
E7747_06895
E7747_06900
hypothetical protein
Accession: E7747_06905
Location: 1590130-1592649
BlastP hit with VTO45597.1
Percentage identity: 39 %
BlastP bit score: 150
Sequence coverage: 61 %
E-value: 1e-35
NCBI BlastP on this gene
Accession: E7747_06905
Location: 1590130-1592649
BlastP hit with VTO45597.1
Percentage identity: 39 %
BlastP bit score: 150
Sequence coverage: 61 %
E-value: 1e-35
NCBI BlastP on this gene
E7747_06905
E7747_06910
leucine-rich repeat domain-containing protein
Accession: QCD42028
Location: 1595441-1597144
NCBI BlastP on this gene
Accession: QCD42028
Location: 1595441-1597144
NCBI BlastP on this gene
E7747_06915
E7747_06920
SPOR domain-containing protein
Accession: QCD42030
Location: 1598420-1598953
NCBI BlastP on this gene
Accession: QCD42030
Location: 1598420-1598953
NCBI BlastP on this gene
E7747_06925
E7747_06930
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP036298 : Planctomycetes bacterium Q31a chromosome Total score: 1.0 Cumulative Blast bit score: 149
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hypothetical protein
Accession: QDV26505
Location: 6324637-6326205
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 69 %
E-value: 4e-36
NCBI BlastP on this gene
Accession: QDV26505
Location: 6324637-6326205
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 69 %
E-value: 4e-36
NCBI BlastP on this gene
Q31a_48790
Q31a_48780
Q31a_48770
Autoinducer 2 sensor kinase/phosphatase LuxQ
Accession: QDV26502
Location: 6317932-6322041
NCBI BlastP on this gene
Accession: QDV26502
Location: 6317932-6322041
NCBI BlastP on this gene
luxQ_2
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP023344 : Nibricoccus aquaticus strain HZ-65 chromosome Total score: 1.0 Cumulative Blast bit score: 149
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
NADH-quinone oxidoreductase subunit C
Accession: ATC64901
Location: 3384889-3385515
NCBI BlastP on this gene
Accession: ATC64901
Location: 3384889-3385515
NCBI BlastP on this gene
CMV30_13510
NADH-quinone oxidoreductase subunit B
Accession: ATC64900
Location: 3384331-3384837
NCBI BlastP on this gene
Accession: ATC64900
Location: 3384331-3384837
NCBI BlastP on this gene
CMV30_13505
CMV30_13500
hybrid sensor histidine kinase/response regulator
Accession: ATC64898
Location: 3380688-3382604
NCBI BlastP on this gene
Accession: ATC64898
Location: 3380688-3382604
NCBI BlastP on this gene
CMV30_13490
DNA-binding response regulator
Accession: ATC64897
Location: 3379793-3380668
NCBI BlastP on this gene
Accession: ATC64897
Location: 3379793-3380668
NCBI BlastP on this gene
CMV30_13485
CMV30_13480
CMV30_13475
Por secretion system protein
Accession: ATC64894
Location: 3375115-3378627
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 83 %
E-value: 5e-35
NCBI BlastP on this gene
Accession: ATC64894
Location: 3375115-3378627
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 83 %
E-value: 5e-35
NCBI BlastP on this gene
CMV30_13470
CMV30_13465
CMV30_13460
CMV30_13455
CMV30_13450
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
251. : CP041360 Spirosoma sp. KCTC 42546 chromosome Total score: 1.0 Cumulative Blast bit score: 175
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
hxpB
PhzF family phenazine biosynthesis protein
Accession: QDK80659
Location: 4746782-4747627
NCBI BlastP on this gene
Accession: QDK80659
Location: 4746782-4747627
NCBI BlastP on this gene
EXU85_19420
EXU85_19415
GNAT family N-acetyltransferase
Accession: QDK80657
Location: 4744580-4745032
NCBI BlastP on this gene
Accession: QDK80657
Location: 4744580-4745032
NCBI BlastP on this gene
EXU85_19410
radical SAM family heme chaperone HemW
Accession: QDK80656
Location: 4743288-4744424
NCBI BlastP on this gene
Accession: QDK80656
Location: 4743288-4744424
NCBI BlastP on this gene
hemW
monofunctional biosynthetic peptidoglycan transglycosylase
Accession: QDK83781
Location: 4742318-4743025
NCBI BlastP on this gene
Accession: QDK83781
Location: 4742318-4743025
NCBI BlastP on this gene
mtgA
EXU85_19395
polysaccharide lyase
Accession: QDK80654
Location: 4739014-4740657
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 175
Sequence coverage: 92 %
E-value: 4e-45
NCBI BlastP on this gene
Accession: QDK80654
Location: 4739014-4740657
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 175
Sequence coverage: 92 %
E-value: 4e-45
NCBI BlastP on this gene
EXU85_19390
EXU85_19385
EXU85_19380
EXU85_19375
DUF3826 domain-containing protein
Accession: QDK80651
Location: 4731456-4732160
NCBI BlastP on this gene
Accession: QDK80651
Location: 4731456-4732160
NCBI BlastP on this gene
EXU85_19370
252. : CP042913 Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 174
Pr1d_41200
N-formylglutamate amidohydrolase
Accession: QEG36783
Location: 4948258-4949055
NCBI BlastP on this gene
Accession: QEG36783
Location: 4948258-4949055
NCBI BlastP on this gene
Pr1d_41190
Pr1d_41180
gshB
autoinducer-2 (AI-2) modifying protein LsrG
Accession: QEG36780
Location: 4944903-4945205
NCBI BlastP on this gene
Accession: QEG36780
Location: 4944903-4945205
NCBI BlastP on this gene
Pr1d_41160
Pr1d_41150
Pr1d_41140
hypothetical protein
Accession: QEG36777
Location: 4941190-4942764
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 174
Sequence coverage: 101 %
E-value: 8e-45
NCBI BlastP on this gene
Accession: QEG36777
Location: 4941190-4942764
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 174
Sequence coverage: 101 %
E-value: 8e-45
NCBI BlastP on this gene
Pr1d_41130
Prolyl tripeptidyl peptidase precursor
Accession: QEG36776
Location: 4939032-4941179
NCBI BlastP on this gene
Accession: QEG36776
Location: 4939032-4941179
NCBI BlastP on this gene
ptpA
ComEC family competence protein
Accession: QEG36775
Location: 4936282-4938795
NCBI BlastP on this gene
Accession: QEG36775
Location: 4936282-4938795
NCBI BlastP on this gene
Pr1d_41110
Pr1d_41100
Pr1d_41090
253. : CP042913 Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 173
Pr1d_30760
Type I phosphodiesterase / nucleotide pyrophosphatase
Accession: QEG35769
Location: 3712474-3713841
NCBI BlastP on this gene
Accession: QEG35769
Location: 3712474-3713841
NCBI BlastP on this gene
Pr1d_30750
Pr1d_30740
hypothetical protein
Accession: QEG35767
Location: 3706972-3709233
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 173
Sequence coverage: 92 %
E-value: 7e-44
NCBI BlastP on this gene
Accession: QEG35767
Location: 3706972-3709233
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 173
Sequence coverage: 92 %
E-value: 7e-44
NCBI BlastP on this gene
Pr1d_30730
Pr1d_30720
Pr1d_30710
tktB
1-deoxy-D-xylulose-5-phosphate synthase
Accession: QEG35763
Location: 3701132-3702148
NCBI BlastP on this gene
Accession: QEG35763
Location: 3701132-3702148
NCBI BlastP on this gene
dxs_2
glpK_2
254. : CP041253 Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 173
FKX85_12870
FKX85_12875
carboxylesterase family protein
Accession: QDH79876
Location: 3233256-3234821
NCBI BlastP on this gene
Accession: QDH79876
Location: 3233256-3234821
NCBI BlastP on this gene
FKX85_12880
FKX85_12885
polysaccharide lyase
Accession: QDH79878
Location: 3239102-3240634
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 173
Sequence coverage: 93 %
E-value: 1e-44
NCBI BlastP on this gene
Accession: QDH79878
Location: 3239102-3240634
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 173
Sequence coverage: 93 %
E-value: 1e-44
NCBI BlastP on this gene
FKX85_12890
FKX85_12895
MerC domain-containing protein
Accession: QDH79880
Location: 3242208-3242591
NCBI BlastP on this gene
Accession: QDH79880
Location: 3242208-3242591
NCBI BlastP on this gene
FKX85_12900
FKX85_12905
FKX85_12910
FKX85_12915
255. : CP042913 Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 172
hypothetical protein
Accession: QEG37972
Location: 6380709-6382025
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 92 %
E-value: 1e-44
NCBI BlastP on this gene
Accession: QEG37972
Location: 6380709-6382025
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 92 %
E-value: 1e-44
NCBI BlastP on this gene
Pr1d_53200
Pr1d_53190
sglT_14
Pr1d_53170
yhdN_4
256. : CP026605 Catenovulum sp. CCB-QB4 plasmid unnamed1 Total score: 1.0 Cumulative Blast bit score: 172
C2869_22125
C2869_22120
C2869_22115
C2869_22110
hypothetical protein
Accession: AWB69198
Location: 122612-125431
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 98 %
E-value: 7e-43
NCBI BlastP on this gene
Accession: AWB69198
Location: 122612-125431
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 98 %
E-value: 7e-43
NCBI BlastP on this gene
C2869_22105
serine/threonine protein kinase
Accession: AWB69197
Location: 120706-122022
NCBI BlastP on this gene
Accession: AWB69197
Location: 120706-122022
NCBI BlastP on this gene
C2869_22100
C2869_22095
iron ABC transporter substrate-binding protein
Accession: AWB69195
Location: 119031-120044
NCBI BlastP on this gene
Accession: AWB69195
Location: 119031-120044
NCBI BlastP on this gene
C2869_22090
C2869_22085
C2869_22080
257. : CP023344 Nibricoccus aquaticus strain HZ-65 chromosome Total score: 1.0 Cumulative Blast bit score: 172
CMV30_04980
CMV30_04985
CMV30_04990
pectate lyase
Accession: ATC63360
Location: 1174089-1176590
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
Accession: ATC63360
Location: 1174089-1176590
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 172
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
CMV30_04995
CMV30_05000
CMV30_05005
CMV30_05010
CMV30_05015
258. : CP013729 Roseateles depolymerans strain KCTC 42856 Total score: 1.0 Cumulative Blast bit score: 172
pectate lyase
Accession: ALV06992
Location: 2963684-2964622
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 7e-43
NCBI BlastP on this gene
Accession: ALV06992
Location: 2963684-2964622
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 7e-43
NCBI BlastP on this gene
RD2015_2525
RD2015_2524
RD2015_2523
RD2015_2522
RD2015_2521
RD2015_2520
RD2015_2519
RD2015_2518
RD2015_2517
259. : AP018439 Undibacterium sp. KW1 DNA Total score: 1.0 Cumulative Blast bit score: 172
UNDKW_3205
UNDKW_3206
glycine dehydrogenase (decarboxylating)
Accession: BBB61480
Location: 3567023-3569911
NCBI BlastP on this gene
Accession: BBB61480
Location: 3567023-3569911
NCBI BlastP on this gene
gcvP
glycine cleavage system H protein
Accession: BBB61481
Location: 3570032-3570409
NCBI BlastP on this gene
Accession: BBB61481
Location: 3570032-3570409
NCBI BlastP on this gene
gcvH
gcvT
hypothetical protein
Accession: BBB61483
Location: 3572377-3573747
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 1e-41
NCBI BlastP on this gene
Accession: BBB61483
Location: 3572377-3573747
BlastP hit with VTO45599.1
Percentage identity: 38 %
BlastP bit score: 172
Sequence coverage: 15 %
E-value: 1e-41
NCBI BlastP on this gene
UNDKW_3210
penicillin acylase family protein
Accession: BBB61484
Location: 3574168-3576588
NCBI BlastP on this gene
Accession: BBB61484
Location: 3574168-3576588
NCBI BlastP on this gene
UNDKW_3211
UNDKW_3212
TetR family transcriptional regulator
Accession: BBB61486
Location: 3578652-3579227
NCBI BlastP on this gene
Accession: BBB61486
Location: 3578652-3579227
NCBI BlastP on this gene
UNDKW_3213
UNDKW_3214
UNDKW_3215
260. : AP017422 Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 172
pectate lyase
Accession: BAV07455
Location: 4215323-4221238
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 90 %
E-value: 2e-42
NCBI BlastP on this gene
Accession: BAV07455
Location: 4215323-4221238
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 172
Sequence coverage: 90 %
E-value: 2e-42
NCBI BlastP on this gene
FLA_3480
FLA_3479
FLA_3478
FLA_3477
FLA_3476
261. : CP043329 Pedobacter sp. CJ43 chromosome Total score: 1.0 Cumulative Blast bit score: 170
DUF3826 domain-containing protein
Accession: QEK50873
Location: 812440-813123
NCBI BlastP on this gene
Accession: QEK50873
Location: 812440-813123
NCBI BlastP on this gene
FYC62_03690
FYC62_03695
RagB/SusD family nutrient uptake outer membrane protein
Accession: QEK53219
Location: 816621-818501
NCBI BlastP on this gene
Accession: QEK53219
Location: 816621-818501
NCBI BlastP on this gene
FYC62_03700
FYC62_03705
polysaccharide lyase
Accession: QEK50876
Location: 819689-821332
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 170
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
Accession: QEK50876
Location: 819689-821332
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 170
Sequence coverage: 91 %
E-value: 3e-43
NCBI BlastP on this gene
FYC62_03710
FYC62_03715
FYC62_03720
FYC62_03725
DUF4038 domain-containing protein
Accession: QEK50879
Location: 826744-828138
NCBI BlastP on this gene
Accession: QEK50879
Location: 826744-828138
NCBI BlastP on this gene
FYC62_03730
FYC62_03735
262. : CP022985 Mariniflexile sp. TRM1-10 chromosome Total score: 1.0 Cumulative Blast bit score: 170
Efflux pump membrane transporter BepE
Accession: AXP80800
Location: 1964473-1967607
NCBI BlastP on this gene
Accession: AXP80800
Location: 1964473-1967607
NCBI BlastP on this gene
CJ739_1714
Outer membrane protein OprM precursor
Accession: AXP80801
Location: 1967657-1969027
NCBI BlastP on this gene
Accession: AXP80801
Location: 1967657-1969027
NCBI BlastP on this gene
CJ739_1715
CJ739_1716
CJ739_1717
CJ739_1718
Glycerol-3-phosphate dehydrogenase
Accession: AXP80805
Location: 1971977-1972978
NCBI BlastP on this gene
Accession: AXP80805
Location: 1971977-1972978
NCBI BlastP on this gene
CJ739_1719
CJ739_1720
hypothetical protein
Accession: AXP80807
Location: 1974337-1975854
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 93 %
E-value: 2e-43
NCBI BlastP on this gene
Accession: AXP80807
Location: 1974337-1975854
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 93 %
E-value: 2e-43
NCBI BlastP on this gene
CJ739_1721
CJ739_1722
putative nicotinate-nucleotide adenylyltransferase
Accession: AXP80809
Location: 1976506-1977084
NCBI BlastP on this gene
Accession: AXP80809
Location: 1976506-1977084
NCBI BlastP on this gene
CJ739_1723
CJ739_1724
CJ739_1725
CJ739_1726
CJ739_1727
Type II secretion system protein D precursor
Accession: AXP80814
Location: 1980558-1982750
NCBI BlastP on this gene
Accession: AXP80814
Location: 1980558-1982750
NCBI BlastP on this gene
CJ739_1728
CJ739_1729
263. : CP002584 Sphingobacterium sp. 21 Total score: 1.0 Cumulative Blast bit score: 170
Sph21_2304
Sph21_2305
Sph21_2306
RagB/SusD domain-containing protein
Accession: ADZ78861
Location: 2832004-2833962
NCBI BlastP on this gene
Accession: ADZ78861
Location: 2832004-2833962
NCBI BlastP on this gene
Sph21_2307
Sph21_2308
hypothetical protein
Accession: ADZ78863
Location: 2834797-2836458
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 92 %
E-value: 3e-43
NCBI BlastP on this gene
Accession: ADZ78863
Location: 2834797-2836458
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 170
Sequence coverage: 92 %
E-value: 3e-43
NCBI BlastP on this gene
Sph21_2309
Sph21_2310
Sph21_2311
Sph21_2312
Sph21_2313
264. : LT629740 Mucilaginibacter mallensis strain MP1X4 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 169
SAMN05216490_2653
AraC-type DNA-binding protein
Accession: SDT17347
Location: 3198363-3199262
NCBI BlastP on this gene
Accession: SDT17347
Location: 3198363-3199262
NCBI BlastP on this gene
SAMN05216490_2652
SAMN05216490_2651
TonB-linked outer membrane protein, SusC/RagA family
Accession: SDT17242
Location: 3193928-3197185
NCBI BlastP on this gene
Accession: SDT17242
Location: 3193928-3197185
NCBI BlastP on this gene
SAMN05216490_2650
Starch-binding associating with outer membrane
Accession: SDT17198
Location: 3192027-3193916
NCBI BlastP on this gene
Accession: SDT17198
Location: 3192027-3193916
NCBI BlastP on this gene
SAMN05216490_2649
hypothetical protein
Accession: SDT17153
Location: 3190299-3191966
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 5e-43
NCBI BlastP on this gene
Accession: SDT17153
Location: 3190299-3191966
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 169
Sequence coverage: 94 %
E-value: 5e-43
NCBI BlastP on this gene
SAMN05216490_2648
SAMN05216490_2647
Putative collagen-binding domain of a collagenase
Accession: SDT17071
Location: 3185749-3187119
NCBI BlastP on this gene
Accession: SDT17071
Location: 3185749-3187119
NCBI BlastP on this gene
SAMN05216490_2646
Glycosyl hydrolases family 43
Accession: SDT17035
Location: 3184792-3185733
NCBI BlastP on this gene
Accession: SDT17035
Location: 3184792-3185733
NCBI BlastP on this gene
SAMN05216490_2645
SAMN05216490_2644
SAMN05216490_2643
265. : CP041253 Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 169
PepSY domain-containing protein
Accession: QDH81091
Location: 5026335-5027594
NCBI BlastP on this gene
Accession: QDH81091
Location: 5026335-5027594
NCBI BlastP on this gene
FKX85_19450
FKX85_19455
efflux RND transporter periplasmic adaptor subunit
Accession: QDH81092
Location: 5029282-5030463
NCBI BlastP on this gene
Accession: QDH81092
Location: 5029282-5030463
NCBI BlastP on this gene
FKX85_19460
efflux RND transporter permease subunit
Accession: QDH81093
Location: 5030466-5033582
NCBI BlastP on this gene
Accession: QDH81093
Location: 5030466-5033582
NCBI BlastP on this gene
FKX85_19465
T9SS C-terminal target domain-containing protein
Accession: FKX85_19470
Location: 5034123-5036591
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 169
Sequence coverage: 91 %
E-value: 4e-42
NCBI BlastP on this gene
Accession: FKX85_19470
Location: 5034123-5036591
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 169
Sequence coverage: 91 %
E-value: 4e-42
NCBI BlastP on this gene
FKX85_19470
266. : CP047647 Hymenobacter sp. BT182 chromosome Total score: 1.0 Cumulative Blast bit score: 167
GUY19_05395
SusC/RagA family TonB-linked outer membrane protein
Accession: QHJ06758
Location: 1216554-1219547
NCBI BlastP on this gene
Accession: QHJ06758
Location: 1216554-1219547
NCBI BlastP on this gene
GUY19_05390
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHJ06757
Location: 1214790-1216538
NCBI BlastP on this gene
Accession: QHJ06757
Location: 1214790-1216538
NCBI BlastP on this gene
GUY19_05385
polysaccharide lyase
Accession: QHJ06756
Location: 1213009-1214673
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 93 %
E-value: 3e-42
NCBI BlastP on this gene
Accession: QHJ06756
Location: 1213009-1214673
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 93 %
E-value: 3e-42
NCBI BlastP on this gene
GUY19_05380
GUY19_05375
DUF4038 domain-containing protein
Accession: QHJ09605
Location: 1208115-1209545
NCBI BlastP on this gene
Accession: QHJ09605
Location: 1208115-1209545
NCBI BlastP on this gene
GUY19_05370
GUY19_05365
GUY19_05360
267. : CP047647 Hymenobacter sp. BT182 chromosome Total score: 1.0 Cumulative Blast bit score: 167
GUY19_05115
PspC domain-containing protein
Accession: QHJ06712
Location: 1134348-1134566
NCBI BlastP on this gene
Accession: QHJ06712
Location: 1134348-1134566
NCBI BlastP on this gene
GUY19_05120
GUY19_05125
GUY19_05130
GUY19_05135
pyruvate dehydrogenase complex E1 component subunit beta
Accession: QHJ06716
Location: 1137438-1138418
NCBI BlastP on this gene
Accession: QHJ06716
Location: 1137438-1138418
NCBI BlastP on this gene
GUY19_05140
GUY19_05145
GUY19_05150
methyltransferase domain-containing protein
Accession: QHJ06718
Location: 1139746-1140483
NCBI BlastP on this gene
Accession: QHJ06718
Location: 1139746-1140483
NCBI BlastP on this gene
GUY19_05155
DUF2256 domain-containing protein
Accession: QHJ06719
Location: 1140589-1140777
NCBI BlastP on this gene
Accession: QHJ06719
Location: 1140589-1140777
NCBI BlastP on this gene
GUY19_05160
GUY19_05165
polysaccharide lyase
Accession: QHJ06721
Location: 1141532-1143169
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 90 %
E-value: 4e-42
NCBI BlastP on this gene
Accession: QHJ06721
Location: 1141532-1143169
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 90 %
E-value: 4e-42
NCBI BlastP on this gene
GUY19_05170
GUY19_05175
GUY19_05180
DUF3826 domain-containing protein
Accession: QHJ06724
Location: 1146119-1146802
NCBI BlastP on this gene
Accession: QHJ06724
Location: 1146119-1146802
NCBI BlastP on this gene
GUY19_05185
SusC/RagA family TonB-linked outer membrane protein
Accession: QHJ06725
Location: 1146833-1150063
NCBI BlastP on this gene
Accession: QHJ06725
Location: 1146833-1150063
NCBI BlastP on this gene
GUY19_05190
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHJ06726
Location: 1150082-1151866
NCBI BlastP on this gene
Accession: QHJ06726
Location: 1150082-1151866
NCBI BlastP on this gene
GUY19_05195
268. : CP041253 Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 167
PepSY domain-containing protein
Accession: QDH78693
Location: 1534448-1535773
NCBI BlastP on this gene
Accession: QDH78693
Location: 1534448-1535773
NCBI BlastP on this gene
FKX85_06445
FKX85_06440
FKX85_06435
FKX85_06430
DUF4919 domain-containing protein
Accession: QDH78689
Location: 1530221-1530916
NCBI BlastP on this gene
Accession: QDH78689
Location: 1530221-1530916
NCBI BlastP on this gene
FKX85_06425
FKX85_06420
glycosyltransferase family 39 protein
Accession: QDH78687
Location: 1527951-1529462
NCBI BlastP on this gene
Accession: QDH78687
Location: 1527951-1529462
NCBI BlastP on this gene
FKX85_06415
polysaccharide lyase
Accession: QDH78686
Location: 1525918-1527618
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 95 %
E-value: 3e-42
NCBI BlastP on this gene
Accession: QDH78686
Location: 1525918-1527618
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 95 %
E-value: 3e-42
NCBI BlastP on this gene
FKX85_06410
FKX85_06405
FKX85_06400
fumC
SPOR domain-containing protein
Accession: QDH78683
Location: 1521582-1522202
NCBI BlastP on this gene
Accession: QDH78683
Location: 1521582-1522202
NCBI BlastP on this gene
FKX85_06390
FKX85_06385
aspS
269. : CP020991 Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 167
NADP-reducing hydrogenase, subunit B
Accession: AUO20297
Location: 2583312-2583692
NCBI BlastP on this gene
Accession: AUO20297
Location: 2583312-2583692
NCBI BlastP on this gene
B9O19_02155
NAD-reducing hydrogenase subunit HoxF
Accession: AUO20298
Location: 2583696-2585492
NCBI BlastP on this gene
Accession: AUO20298
Location: 2583696-2585492
NCBI BlastP on this gene
B9O19_02156
periplasmic [Fe] hydrogenase large subunit
Accession: AUO20299
Location: 2585505-2587247
NCBI BlastP on this gene
Accession: AUO20299
Location: 2585505-2587247
NCBI BlastP on this gene
B9O19_02157
transcriptional regulator, XRE family
Accession: AUO20300
Location: 2587795-2588118
NCBI BlastP on this gene
Accession: AUO20300
Location: 2587795-2588118
NCBI BlastP on this gene
B9O19_02158
B9O19_02159
pectate lyase family 1
Accession: AUO20302
Location: 2589949-2593386
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 98 %
E-value: 7e-41
NCBI BlastP on this gene
Accession: AUO20302
Location: 2589949-2593386
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 167
Sequence coverage: 98 %
E-value: 7e-41
NCBI BlastP on this gene
B9O19_02160
270. : CP049143 Roseimicrobium sp. ORNL1 chromosome Total score: 1.0 Cumulative Blast bit score: 166
DUF3445 domain-containing protein
Accession: QIF03659
Location: 4868103-4868936
NCBI BlastP on this gene
Accession: QIF03659
Location: 4868103-4868936
NCBI BlastP on this gene
G5S37_19755
G5S37_19750
KamA family radical SAM protein
Accession: QIF03657
Location: 4865099-4866316
NCBI BlastP on this gene
Accession: QIF03657
Location: 4865099-4866316
NCBI BlastP on this gene
G5S37_19745
winged helix-turn-helix transcriptional regulator
Accession: QIF03656
Location: 4863709-4864932
NCBI BlastP on this gene
Accession: QIF03656
Location: 4863709-4864932
NCBI BlastP on this gene
G5S37_19740
G5S37_19735
G5S37_19730
sugar phosphate isomerase/epimerase
Accession: QIF06130
Location: 4860952-4861881
NCBI BlastP on this gene
Accession: QIF06130
Location: 4860952-4861881
NCBI BlastP on this gene
G5S37_19725
pectate lyase
Accession: QIF03655
Location: 4859359-4860723
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 166
Sequence coverage: 74 %
E-value: 3e-42
NCBI BlastP on this gene
Accession: QIF03655
Location: 4859359-4860723
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 166
Sequence coverage: 74 %
E-value: 3e-42
NCBI BlastP on this gene
G5S37_19720
DUF4185 domain-containing protein
Accession: QIF03654
Location: 4858234-4859292
NCBI BlastP on this gene
Accession: QIF03654
Location: 4858234-4859292
NCBI BlastP on this gene
G5S37_19715
G5S37_19710
G5S37_19705
nuclear transport factor 2 family protein
Accession: QIF03651
Location: 4854152-4854622
NCBI BlastP on this gene
Accession: QIF03651
Location: 4854152-4854622
NCBI BlastP on this gene
G5S37_19700
PQQ-binding-like beta-propeller repeat protein
Accession: QIF03650
Location: 4852805-4854076
NCBI BlastP on this gene
Accession: QIF03650
Location: 4852805-4854076
NCBI BlastP on this gene
G5S37_19695
G5S37_19690
271. : CP003178 Niastella koreensis GR20-10 Total score: 1.0 Cumulative Blast bit score: 165
Niako_2525
Glyoxalase/bleomycin resistance
Accession: AEV98864
Location: 3128007-3128420
NCBI BlastP on this gene
Accession: AEV98864
Location: 3128007-3128420
NCBI BlastP on this gene
Niako_2524
Niako_2523
Peptidoglycan-binding lysin domain protein
Accession: AEV98862
Location: 3126708-3127361
NCBI BlastP on this gene
Accession: AEV98862
Location: 3126708-3127361
NCBI BlastP on this gene
Niako_2522
rifampin ADP-ribosyl transferase
Accession: AEV98861
Location: 3126096-3126521
NCBI BlastP on this gene
Accession: AEV98861
Location: 3126096-3126521
NCBI BlastP on this gene
Niako_2521
Niako_2520
transcriptional regulator, LacI family
Accession: AEV98859
Location: 3124221-3125258
NCBI BlastP on this gene
Accession: AEV98859
Location: 3124221-3125258
NCBI BlastP on this gene
Niako_2519
hypothetical protein
Accession: AEV98858
Location: 3122536-3124167
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 165
Sequence coverage: 92 %
E-value: 2e-41
NCBI BlastP on this gene
Accession: AEV98858
Location: 3122536-3124167
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 165
Sequence coverage: 92 %
E-value: 2e-41
NCBI BlastP on this gene
Niako_2518
putative transcriptional regulator, Crp/Fnr family
Accession: AEV98857
Location: 3121713-3122285
NCBI BlastP on this gene
Accession: AEV98857
Location: 3121713-3122285
NCBI BlastP on this gene
Niako_2517
Niako_2516
Niako_2515
bifunctional deaminase-reductase domain protein
Accession: AEV98854
Location: 3119796-3120359
NCBI BlastP on this gene
Accession: AEV98854
Location: 3119796-3120359
NCBI BlastP on this gene
Niako_2514
Niako_2513
putative RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: AEV98852
Location: 3118067-3119380
NCBI BlastP on this gene
Accession: AEV98852
Location: 3118067-3119380
NCBI BlastP on this gene
Niako_2512
Niako_2511
Niako_2510
Niako_2509
Niako_2508
272. : CP039396 Muribaculum sp. H5 chromosome. Total score: 1.0 Cumulative Blast bit score: 163
E7747_15890
glutamate-5-semialdehyde dehydrogenase
Accession: QCD43605
Location: 3583220-3584470
NCBI BlastP on this gene
Accession: QCD43605
Location: 3583220-3584470
NCBI BlastP on this gene
E7747_15895
proB
pyrroline-5-carboxylate reductase
Accession: QCD43607
Location: 3585676-3586539
NCBI BlastP on this gene
Accession: QCD43607
Location: 3585676-3586539
NCBI BlastP on this gene
E7747_15905
E7747_15910
RagB/SusD family nutrient uptake outer membrane protein
Accession: E7747_15915
Location: 3587122-3589301
NCBI BlastP on this gene
Accession: E7747_15915
Location: 3587122-3589301
NCBI BlastP on this gene
E7747_15915
polysaccharide lyase
Accession: QCD43744
Location: 3589505-3591223
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
Accession: QCD43744
Location: 3589505-3591223
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
E7747_15920
273. : CP039396 Muribaculum sp. H5 chromosome. Total score: 1.0 Cumulative Blast bit score: 163
DUF4957 domain-containing protein
Accession: QCD43458
Location: 3381860-3383332
NCBI BlastP on this gene
Accession: QCD43458
Location: 3381860-3383332
NCBI BlastP on this gene
E7747_15050
E7747_15055
RagB/SusD family nutrient uptake outer membrane protein
Accession: QCD43460
Location: 3386711-3388921
NCBI BlastP on this gene
Accession: QCD43460
Location: 3386711-3388921
NCBI BlastP on this gene
E7747_15060
polysaccharide lyase
Accession: QCD43733
Location: 3389125-3390843
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
Accession: QCD43733
Location: 3389125-3390843
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 91 %
E-value: 9e-41
NCBI BlastP on this gene
E7747_15065
DUF3826 domain-containing protein
Accession: QCD43461
Location: 3390868-3391536
NCBI BlastP on this gene
Accession: QCD43461
Location: 3390868-3391536
NCBI BlastP on this gene
E7747_15070
E7747_15075
DUF1593 domain-containing protein
Accession: QCD43463
Location: 3394946-3396250
NCBI BlastP on this gene
Accession: QCD43463
Location: 3394946-3396250
NCBI BlastP on this gene
E7747_15080
E7747_15085
274. : CP020991 Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 163
S-layer domain-containing protein
Accession: AUO18489
Location: 351097-354123
NCBI BlastP on this gene
Accession: AUO18489
Location: 351097-354123
NCBI BlastP on this gene
B9O19_00305
B9O19_00306
B9O19_00307
B9O19_00308
probabale oxalocrotonate tautomerase
Accession: AUO18493
Location: 356768-356977
NCBI BlastP on this gene
Accession: AUO18493
Location: 356768-356977
NCBI BlastP on this gene
B9O19_00309
B9O19_00310
B9O19_00311
pectate lyase family 1
Accession: AUO18496
Location: 358585-361302
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 97 %
E-value: 5e-40
NCBI BlastP on this gene
Accession: AUO18496
Location: 358585-361302
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 163
Sequence coverage: 97 %
E-value: 5e-40
NCBI BlastP on this gene
B9O19_00312
B9O19_00313
B9O19_00314
B9O19_00315
B9O19_00316
V-type sodium ATPase catalytic subunit A
Accession: AUO18501
Location: 366216-367982
NCBI BlastP on this gene
Accession: AUO18501
Location: 366216-367982
NCBI BlastP on this gene
B9O19_00317
B9O19_00318
275. : CP002584 Sphingobacterium sp. 21 Total score: 1.0 Cumulative Blast bit score: 163
Sph21_2568
Sph21_2569
Sph21_2570
PepSY-associated TM helix domain protein
Accession: ADZ79122
Location: 3168499-3169641
NCBI BlastP on this gene
Accession: ADZ79122
Location: 3168499-3169641
NCBI BlastP on this gene
Sph21_2571
TonB-dependent siderophore receptor
Accession: ADZ79123
Location: 3169644-3171758
NCBI BlastP on this gene
Accession: ADZ79123
Location: 3169644-3171758
NCBI BlastP on this gene
Sph21_2572
Sph21_2573
hypothetical protein
Accession: ADZ79125
Location: 3172491-3174116
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 163
Sequence coverage: 90 %
E-value: 8e-41
NCBI BlastP on this gene
Accession: ADZ79125
Location: 3172491-3174116
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 163
Sequence coverage: 90 %
E-value: 8e-41
NCBI BlastP on this gene
Sph21_2574
276. : CP021382 Cellvibrio sp. PSBB006 chromosome Total score: 1.0 Cumulative Blast bit score: 162
dicarboxylate/amino acid:cation symporter
Accession: ARU28644
Location: 3623654-3624961
NCBI BlastP on this gene
Accession: ARU28644
Location: 3623654-3624961
NCBI BlastP on this gene
CBR65_14995
CBR65_15000
CBR65_15005
4-hydroxybenzoate polyprenyltransferase
Accession: ARU28647
Location: 3626093-3627019
NCBI BlastP on this gene
Accession: ARU28647
Location: 3626093-3627019
NCBI BlastP on this gene
CBR65_15010
phosphate regulon transcriptional regulatory protein PhoB
Accession: ARU28648
Location: 3627143-3627841
NCBI BlastP on this gene
Accession: ARU28648
Location: 3627143-3627841
NCBI BlastP on this gene
CBR65_15015
PAS domain-containing sensor histidine kinase
Accession: ARU28649
Location: 3627987-3629261
NCBI BlastP on this gene
Accession: ARU28649
Location: 3627987-3629261
NCBI BlastP on this gene
CBR65_15020
CBR65_15025
CBR65_15030
hypothetical protein
Accession: ARU28652
Location: 3630869-3633220
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 162
Sequence coverage: 92 %
E-value: 7e-40
NCBI BlastP on this gene
Accession: ARU28652
Location: 3630869-3633220
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 162
Sequence coverage: 92 %
E-value: 7e-40
NCBI BlastP on this gene
CBR65_15035
277. : LT629758 Actinoplanes derwentensis strain DSM 43941 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 161
pectate lyase
Accession: SDT74959
Location: 7952377-7953996
BlastP hit with VTO45599.1
Percentage identity: 36 %
BlastP bit score: 161
Sequence coverage: 12 %
E-value: 1e-37
NCBI BlastP on this gene
Accession: SDT74959
Location: 7952377-7953996
BlastP hit with VTO45599.1
Percentage identity: 36 %
BlastP bit score: 161
Sequence coverage: 12 %
E-value: 1e-37
NCBI BlastP on this gene
SAMN04489716_7149
SAMN04489716_7148
SAMN04489716_7147
SAMN04489716_7146
SAMN04489716_7145
SAMN04489716_7144
278. : CP030261 Flavobacterium sp. HYN0086 chromosome Total score: 1.0 Cumulative Blast bit score: 161
HYN86_16520
HYN86_16515
HYN86_16510
SusC/RagA family TonB-linked outer membrane protein
Accession: AXB58103
Location: 3753617-3756997
NCBI BlastP on this gene
Accession: AXB58103
Location: 3753617-3756997
NCBI BlastP on this gene
HYN86_16505
RagB/SusD family nutrient uptake outer membrane protein
Accession: AXB58102
Location: 3752008-3753612
NCBI BlastP on this gene
Accession: AXB58102
Location: 3752008-3753612
NCBI BlastP on this gene
HYN86_16500
polysaccharide lyase
Accession: AXB58101
Location: 3749993-3751696
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 161
Sequence coverage: 90 %
E-value: 4e-40
NCBI BlastP on this gene
Accession: AXB58101
Location: 3749993-3751696
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 161
Sequence coverage: 90 %
E-value: 4e-40
NCBI BlastP on this gene
HYN86_16495
HYN86_16490
HYN86_16485
HYN86_16480
HYN86_16475
279. : CP003346 Echinicola vietnamensis DSM 17526 Total score: 1.0 Cumulative Blast bit score: 160
Fe2+-dicitrate sensor, membrane component
Accession: AGA77594
Location: 1528585-1529655
NCBI BlastP on this gene
Accession: AGA77594
Location: 1528585-1529655
NCBI BlastP on this gene
Echvi_1323
TonB-linked outer membrane protein, SusC/RagA family
Accession: AGA77595
Location: 1529773-1533282
NCBI BlastP on this gene
Accession: AGA77595
Location: 1529773-1533282
NCBI BlastP on this gene
Echvi_1324
Echvi_1325
hypothetical protein
Accession: AGA77597
Location: 1535777-1537474
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 93 %
E-value: 8e-40
NCBI BlastP on this gene
Accession: AGA77597
Location: 1535777-1537474
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 93 %
E-value: 8e-40
NCBI BlastP on this gene
Echvi_1326
endoribonuclease L-PSP, putative
Accession: AGA77598
Location: 1537696-1538079
NCBI BlastP on this gene
Accession: AGA77598
Location: 1537696-1538079
NCBI BlastP on this gene
Echvi_1327
Echvi_1328
Echvi_1329
Echvi_1330
peptidyl-prolyl cis-trans isomerase (rotamase) - cyclophilin family
Accession: AGA77602
Location: 1541336-1541920
NCBI BlastP on this gene
Accession: AGA77602
Location: 1541336-1541920
NCBI BlastP on this gene
Echvi_1331
single-stranded-DNA-specific exonuclease RecJ
Accession: AGA77603
Location: 1542293-1544062
NCBI BlastP on this gene
Accession: AGA77603
Location: 1542293-1544062
NCBI BlastP on this gene
Echvi_1332
ABC-type (unclassified) transport system, ATPase component
Accession: AGA77604
Location: 1544065-1544796
NCBI BlastP on this gene
Accession: AGA77604
Location: 1544065-1544796
NCBI BlastP on this gene
Echvi_1333
acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II
Accession: AGA77605
Location: 1544800-1546320
NCBI BlastP on this gene
Accession: AGA77605
Location: 1544800-1546320
NCBI BlastP on this gene
Echvi_1334
280. : AP017422 Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 160
FLA_2726
FLA_2725
FLA_2723
transferase, hexapeptide repeat family
Accession: BAV06703
Location: 3274355-3274663
NCBI BlastP on this gene
Accession: BAV06703
Location: 3274355-3274663
NCBI BlastP on this gene
FLA_2722
transcriptional regulator, AraC family
Accession: BAV06702
Location: 3273879-3274343
NCBI BlastP on this gene
Accession: BAV06702
Location: 3273879-3274343
NCBI BlastP on this gene
FLA_2721
FLA_2720
FLA_2719
FLA_2718
DNA-binding response regulator, AraC family
Accession: BAV06698
Location: 3271207-3271581
NCBI BlastP on this gene
Accession: BAV06698
Location: 3271207-3271581
NCBI BlastP on this gene
FLA_2717
FLA_2716
pectate lyase
Accession: BAV06696
Location: 3268270-3271014
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 87 %
E-value: 5e-39
NCBI BlastP on this gene
Accession: BAV06696
Location: 3268270-3271014
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 160
Sequence coverage: 87 %
E-value: 5e-39
NCBI BlastP on this gene
FLA_2715
FLA_2714
FLA_2713
endo-1,4-beta-xylanase D precursor
Accession: BAV06693
Location: 3266150-3267754
NCBI BlastP on this gene
Accession: BAV06693
Location: 3266150-3267754
NCBI BlastP on this gene
FLA_2712
rhamnogalacturonan acetylesterase
Accession: BAV06692
Location: 3264418-3266100
NCBI BlastP on this gene
Accession: BAV06692
Location: 3264418-3266100
NCBI BlastP on this gene
FLA_2711
FLA_2710
FLA_2709
FLA_2708
FLA_2707
281. : AP017422 Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 160
FLA_1170
FLA_1171
FLA_1172
outer membrane protein, nutrient binding
Accession: BAV05166
Location: 1385496-1387322
NCBI BlastP on this gene
Accession: BAV05166
Location: 1385496-1387322
NCBI BlastP on this gene
FLA_1173
pectate lyase
Accession: BAV05167
Location: 1387366-1389015
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 91 %
E-value: 5e-40
NCBI BlastP on this gene
Accession: BAV05167
Location: 1387366-1389015
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 160
Sequence coverage: 91 %
E-value: 5e-40
NCBI BlastP on this gene
FLA_1174
FLA_1175
FLA_1176
FLA_1177
lipase/acylhydrolase family protein
Accession: BAV05171
Location: 1394535-1395293
NCBI BlastP on this gene
Accession: BAV05171
Location: 1394535-1395293
NCBI BlastP on this gene
FLA_1178
FLA_1179
282. : CP043306 Cellvibrio japonicus strain ADPT1-KOJIBIOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 159
hypothetical protein
Accession: QEI20559
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QEI20559
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
FY115_15030
FY115_15025
small ribosomal subunit biogenesis GTPase RsgA
Accession: QEI20557
Location: 3702363-3703388
NCBI BlastP on this gene
Accession: QEI20557
Location: 3702363-3703388
NCBI BlastP on this gene
rsgA
FY115_15015
FY115_15010
FY115_15005
FY115_15000
FY115_14995
bla
283. : CP043305 Cellvibrio japonicus strain ADPT2-NIGEROSE chromosome. Total score: 1.0 Cumulative Blast bit score: 159
hypothetical protein
Accession: QEI16981
Location: 3704311-3706509
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QEI16981
Location: 3704311-3706509
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
FY116_15035
FY116_15030
small ribosomal subunit biogenesis GTPase RsgA
Accession: QEI16979
Location: 3702358-3703383
NCBI BlastP on this gene
Accession: QEI16979
Location: 3702358-3703383
NCBI BlastP on this gene
rsgA
FY116_15020
FY116_15015
FY116_15010
FY116_15005
FY116_15000
bla
284. : CP043304 Cellvibrio japonicus strain ADPT3-ISOMALTOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 159
hypothetical protein
Accession: QEI13407
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QEI13407
Location: 3704316-3706514
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
FY117_15030
FY117_15025
small ribosomal subunit biogenesis GTPase RsgA
Accession: QEI13405
Location: 3702363-3703388
NCBI BlastP on this gene
Accession: QEI13405
Location: 3702363-3703388
NCBI BlastP on this gene
rsgA
FY117_15015
FY117_15010
FY117_15005
FY117_15000
FY117_14995
bla
285. : CP036350 Planctomycetes bacterium K2D chromosome Total score: 1.0 Cumulative Blast bit score: 159
K2D_26110
K2D_26120
K2D_26130
RNA polymerase-binding transcription factor DksA
Accession: QDV79005
Location: 3218170-3218523
NCBI BlastP on this gene
Accession: QDV79005
Location: 3218170-3218523
NCBI BlastP on this gene
dksA
thrC
lacZ_4
hypothetical protein
Accession: QDV79008
Location: 3222879-3225056
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
Accession: QDV79008
Location: 3222879-3225056
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
K2D_26170
286. : CP036349 Planctomycetes bacterium Spa11 chromosome. Total score: 1.0 Cumulative Blast bit score: 159
Spa11_26090
Spa11_26100
Spa11_26110
RNA polymerase-binding transcription factor DksA
Accession: QDV74409
Location: 3285998-3286351
NCBI BlastP on this gene
Accession: QDV74409
Location: 3285998-3286351
NCBI BlastP on this gene
dksA
thrC
lacZ_4
hypothetical protein
Accession: QDV74412
Location: 3290707-3292884
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
Accession: QDV74412
Location: 3290707-3292884
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 159
Sequence coverage: 92 %
E-value: 7e-39
NCBI BlastP on this gene
Spa11_26150
287. : CP000934 Cellvibrio japonicus Ueda107 Total score: 1.0 Cumulative Blast bit score: 159
pectate lyase, putative, pel1G
Accession: ACE83184
Location: 3704298-3706496
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: ACE83184
Location: 3704298-3706496
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 159
Sequence coverage: 18 %
E-value: 2e-36
NCBI BlastP on this gene
pel1G
rhdA
CJA_3118
orn
pectin methylesterase putative, pme8B
Accession: ACE83435
Location: 3701180-3701401
NCBI BlastP on this gene
Accession: ACE83435
Location: 3701180-3701401
NCBI BlastP on this gene
pme8B
CJA_3115
CJA_3114
CJA_3113
putative dipeptidyl peptidase IV
Accession: ACE86065
Location: 3698344-3699336
NCBI BlastP on this gene
Accession: ACE86065
Location: 3698344-3699336
NCBI BlastP on this gene
CJA_3112
CJA_3111
CJA_3110
288. : AP019371 Actinoplanes sp. OR16 DNA Total score: 1.0 Cumulative Blast bit score: 158
glyceraldehyde 3-phosphate reductase
Accession: BBH65236
Location: 2069915-2070910
NCBI BlastP on this gene
Accession: BBH65236
Location: 2069915-2070910
NCBI BlastP on this gene
ACTI_19210
sulfotransferase family protein
Accession: BBH65237
Location: 2070896-2071813
NCBI BlastP on this gene
Accession: BBH65237
Location: 2070896-2071813
NCBI BlastP on this gene
ACTI_19220
ACTI_19230
ACTI_19240
ACTI_19250
ACTI_19260
ACTI_19270
ACTI_19280
hypothetical protein
Accession: BBH65244
Location: 2077591-2079135
BlastP hit with VTO45599.1
Percentage identity: 37 %
BlastP bit score: 158
Sequence coverage: 12 %
E-value: 1e-36
NCBI BlastP on this gene
Accession: BBH65244
Location: 2077591-2079135
BlastP hit with VTO45599.1
Percentage identity: 37 %
BlastP bit score: 158
Sequence coverage: 12 %
E-value: 1e-36
NCBI BlastP on this gene
ACTI_19290
ACTI_19300
ACTI_19310
ACTI_19320
ACTI_19330
289. : CP020991 Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 157
3-oxoacyl-(acyl-carrier-protein) reductase
Accession: AUO20008
Location: 2212386-2213138
NCBI BlastP on this gene
Accession: AUO20008
Location: 2212386-2213138
NCBI BlastP on this gene
B9O19_01859
B9O19_01860
3-oxoacyl-(acyl-carrier-protein) synthase 2
Accession: AUO20010
Location: 2213664-2214902
NCBI BlastP on this gene
Accession: AUO20010
Location: 2213664-2214902
NCBI BlastP on this gene
B9O19_01861
GCN5-related N-acetyltransferase
Accession: AUO20011
Location: 2214976-2215989
NCBI BlastP on this gene
Accession: AUO20011
Location: 2214976-2215989
NCBI BlastP on this gene
B9O19_01862
B9O19_01863
B9O19_01864
B9O19_01865
copper amine oxidase-like domain-containing protein
Accession: AUO20015
Location: 2218246-2219430
NCBI BlastP on this gene
Accession: AUO20015
Location: 2218246-2219430
NCBI BlastP on this gene
B9O19_01866
pectate lyase family 1
Accession: AUO20016
Location: 2219740-2222430
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 157
Sequence coverage: 90 %
E-value: 5e-38
NCBI BlastP on this gene
Accession: AUO20016
Location: 2219740-2222430
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 157
Sequence coverage: 90 %
E-value: 5e-38
NCBI BlastP on this gene
B9O19_01867
B9O19_01868
B9O19_01869
S-layer domain-containing protein
Accession: AUO20019
Location: 2224870-2230200
NCBI BlastP on this gene
Accession: AUO20019
Location: 2224870-2230200
NCBI BlastP on this gene
B9O19_01870
290. : CP048113 Chitinophaga sp. H33E-04 chromosome Total score: 1.0 Cumulative Blast bit score: 154
T9SS type A sorting domain-containing protein
Accession: QHS62610
Location: 6079946-6081601
NCBI BlastP on this gene
Accession: QHS62610
Location: 6079946-6081601
NCBI BlastP on this gene
GWR21_24420
GWR21_24425
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHS62612
Location: 6085364-6087127
NCBI BlastP on this gene
Accession: QHS62612
Location: 6085364-6087127
NCBI BlastP on this gene
GWR21_24430
DUF3826 domain-containing protein
Accession: QHS62613
Location: 6087189-6089294
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 154
Sequence coverage: 94 %
E-value: 1e-37
NCBI BlastP on this gene
Accession: QHS62613
Location: 6087189-6089294
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 154
Sequence coverage: 94 %
E-value: 1e-37
NCBI BlastP on this gene
GWR21_24435
GWR21_24440
DUF4038 domain-containing protein
Accession: QHS62615
Location: 6092389-6093792
NCBI BlastP on this gene
Accession: QHS62615
Location: 6092389-6093792
NCBI BlastP on this gene
GWR21_24445
glycoside hydrolase family 43 protein
Accession: QHS62616
Location: 6093807-6094709
NCBI BlastP on this gene
Accession: QHS62616
Location: 6093807-6094709
NCBI BlastP on this gene
GWR21_24450
GWR21_24455
291. : CP021904 Alkalitalea saponilacus strain SC/BZ-SP2 chromosome Total score: 1.0 Cumulative Blast bit score: 154
tsaB
CDL62_08555
efflux RND transporter periplasmic adaptor subunit
Accession: ASB49187
Location: 2188471-2189553
NCBI BlastP on this gene
Accession: ASB49187
Location: 2188471-2189553
NCBI BlastP on this gene
CDL62_08560
ABC transporter ATP-binding protein
Accession: ASB49188
Location: 2189622-2190902
NCBI BlastP on this gene
Accession: ASB49188
Location: 2189622-2190902
NCBI BlastP on this gene
CDL62_08565
CDL62_08570
macrolide ABC transporter ATP-binding protein
Accession: ASB51107
Location: 2192321-2193043
NCBI BlastP on this gene
Accession: ASB51107
Location: 2192321-2193043
NCBI BlastP on this gene
CDL62_08575
hypothetical protein
Accession: ASB49190
Location: 2193569-2196439
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 154
Sequence coverage: 14 %
E-value: 3e-34
NCBI BlastP on this gene
Accession: ASB49190
Location: 2193569-2196439
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 154
Sequence coverage: 14 %
E-value: 3e-34
NCBI BlastP on this gene
CDL62_08580
pflB
pyruvate formate-lyase 1-activating enzyme
Accession: ASB49192
Location: 2198990-2199805
NCBI BlastP on this gene
Accession: ASB49192
Location: 2198990-2199805
NCBI BlastP on this gene
pflA
CDL62_08595
CDL62_08600
CDL62_08605
292. : LT622246 Bacteroides ovatus V975 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 153
BACOV975_03474
BACOV975_03475
two-component system sensor histidine
Accession: SCV09682
Location: 4621311-4625741
NCBI BlastP on this gene
Accession: SCV09682
Location: 4621311-4625741
NCBI BlastP on this gene
BACOV975_03476
hypothetical protein
Accession: SCV09683
Location: 4625800-4627482
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: SCV09683
Location: 4625800-4627482
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
BACOV975_03477
293. : FP929033 Bacteroides xylanisolvens XB1A draft genome. Total score: 1.0 Cumulative Blast bit score: 153
Beta-galactosidase/beta-glucuronidase
Accession: CBK65668
Location: 490785-493439
NCBI BlastP on this gene
Accession: CBK65668
Location: 490785-493439
NCBI BlastP on this gene
BXY_04110
His Kinase A (phosphoacceptor) domain./Histidine
Accession: CBK65669
Location: 493613-498010
NCBI BlastP on this gene
Accession: CBK65669
Location: 493613-498010
NCBI BlastP on this gene
BXY_04120
hypothetical protein
Accession: CBK65670
Location: 498069-499751
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: CBK65670
Location: 498069-499751
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
BXY_04130
BXY_04140
RNA polymerase sigma factor, sigma-70 family
Accession: CBK65672
Location: 503078-503629
NCBI BlastP on this gene
Accession: CBK65672
Location: 503078-503629
NCBI BlastP on this gene
BXY_04150
BXY_04160
BXY_04170
Uncharacterized conserved protein
Accession: CBK65675
Location: 505413-506180
NCBI BlastP on this gene
Accession: CBK65675
Location: 505413-506180
NCBI BlastP on this gene
BXY_04180
BXY_04190
BXY_04200
294. : CP046397 Bacteroides ovatus strain FDAARGOS_733 chromosome Total score: 1.0 Cumulative Blast bit score: 153
FOC41_03120
FOC41_03125
polysaccharide lyase
Accession: QGT70025
Location: 848992-850674
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: QGT70025
Location: 848992-850674
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
FOC41_03130
FOC41_03135
sigma-70 family RNA polymerase sigma factor
Accession: QGT74087
Location: 854064-854555
NCBI BlastP on this gene
Accession: QGT74087
Location: 854064-854555
NCBI BlastP on this gene
FOC41_03140
FOC41_03145
FOC41_03150
FOC41_03155
FOC41_03160
FOC41_03165
295. : CP041230 Bacteroides xylanisolvens strain H207 chromosome Total score: 1.0 Cumulative Blast bit score: 153
FKZ68_16475
FKZ68_16480
polysaccharide lyase
Accession: QDH55717
Location: 4324837-4326519
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: QDH55717
Location: 4324837-4326519
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
FKZ68_16485
296. : CP012938 Bacteroides ovatus strain ATCC 8483 Total score: 1.0 Cumulative Blast bit score: 153
lacZ_6
tmoS_2
hypothetical protein
Accession: ALJ45347
Location: 858860-860542
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Accession: ALJ45347
Location: 858860-860542
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 153
Sequence coverage: 89 %
E-value: 2e-37
NCBI BlastP on this gene
Bovatus_00681
Bovatus_00682
RNA polymerase sigma factor SigM
Accession: ALJ45349
Location: 863872-864420
NCBI BlastP on this gene
Accession: ALJ45349
Location: 863872-864420
NCBI BlastP on this gene
sigM_1
Bovatus_00684
ybjJ
yjjP_1
Bovatus_00687
Bovatus_00688
297. : CP050831 Bacteroides sp. CBA7301 chromosome Total score: 1.0 Cumulative Blast bit score: 150
BacF7301_17805
BacF7301_17810
BacF7301_17815
polysaccharide lyase
Accession: QIU95892
Location: 4832610-4834292
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 89 %
E-value: 2e-36
NCBI BlastP on this gene
Accession: QIU95892
Location: 4832610-4834292
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 150
Sequence coverage: 89 %
E-value: 2e-36
NCBI BlastP on this gene
BacF7301_17820
298. : CP039396 Muribaculum sp. H5 chromosome. Total score: 1.0 Cumulative Blast bit score: 150
E7747_06865
E7747_06870
DUF5119 domain-containing protein
Accession: QCD42021
Location: 1584612-1585550
NCBI BlastP on this gene
Accession: QCD42021
Location: 1584612-1585550
NCBI BlastP on this gene
E7747_06875
DUF3575 domain-containing protein
Accession: QCD42022
Location: 1585547-1586746
NCBI BlastP on this gene
Accession: QCD42022
Location: 1585547-1586746
NCBI BlastP on this gene
E7747_06880
methylmalonyl Co-A mutase-associated GTPase MeaB
Accession: QCD42023
Location: 1586985-1588070
NCBI BlastP on this gene
Accession: QCD42023
Location: 1586985-1588070
NCBI BlastP on this gene
meaB
DUF1573 domain-containing protein
Accession: QCD42024
Location: 1588096-1589178
NCBI BlastP on this gene
Accession: QCD42024
Location: 1588096-1589178
NCBI BlastP on this gene
E7747_06890
DUF1573 domain-containing protein
Accession: QCD42025
Location: 1589222-1589602
NCBI BlastP on this gene
Accession: QCD42025
Location: 1589222-1589602
NCBI BlastP on this gene
E7747_06895
E7747_06900
hypothetical protein
Accession: E7747_06905
Location: 1590130-1592649
BlastP hit with VTO45597.1
Percentage identity: 39 %
BlastP bit score: 150
Sequence coverage: 61 %
E-value: 1e-35
NCBI BlastP on this gene
Accession: E7747_06905
Location: 1590130-1592649
BlastP hit with VTO45597.1
Percentage identity: 39 %
BlastP bit score: 150
Sequence coverage: 61 %
E-value: 1e-35
NCBI BlastP on this gene
E7747_06905
E7747_06910
leucine-rich repeat domain-containing protein
Accession: QCD42028
Location: 1595441-1597144
NCBI BlastP on this gene
Accession: QCD42028
Location: 1595441-1597144
NCBI BlastP on this gene
E7747_06915
E7747_06920
SPOR domain-containing protein
Accession: QCD42030
Location: 1598420-1598953
NCBI BlastP on this gene
Accession: QCD42030
Location: 1598420-1598953
NCBI BlastP on this gene
E7747_06925
E7747_06930
299. : CP036298 Planctomycetes bacterium Q31a chromosome Total score: 1.0 Cumulative Blast bit score: 149
hypothetical protein
Accession: QDV26505
Location: 6324637-6326205
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 69 %
E-value: 4e-36
NCBI BlastP on this gene
Accession: QDV26505
Location: 6324637-6326205
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 69 %
E-value: 4e-36
NCBI BlastP on this gene
Q31a_48790
Q31a_48780
Q31a_48770
Autoinducer 2 sensor kinase/phosphatase LuxQ
Accession: QDV26502
Location: 6317932-6322041
NCBI BlastP on this gene
Accession: QDV26502
Location: 6317932-6322041
NCBI BlastP on this gene
luxQ_2
300. : CP023344 Nibricoccus aquaticus strain HZ-65 chromosome Total score: 1.0 Cumulative Blast bit score: 149
NADH-quinone oxidoreductase subunit C
Accession: ATC64901
Location: 3384889-3385515
NCBI BlastP on this gene
Accession: ATC64901
Location: 3384889-3385515
NCBI BlastP on this gene
CMV30_13510
NADH-quinone oxidoreductase subunit B
Accession: ATC64900
Location: 3384331-3384837
NCBI BlastP on this gene
Accession: ATC64900
Location: 3384331-3384837
NCBI BlastP on this gene
CMV30_13505
CMV30_13500
hybrid sensor histidine kinase/response regulator
Accession: ATC64898
Location: 3380688-3382604
NCBI BlastP on this gene
Accession: ATC64898
Location: 3380688-3382604
NCBI BlastP on this gene
CMV30_13490
DNA-binding response regulator
Accession: ATC64897
Location: 3379793-3380668
NCBI BlastP on this gene
Accession: ATC64897
Location: 3379793-3380668
NCBI BlastP on this gene
CMV30_13485
CMV30_13480
CMV30_13475
Por secretion system protein
Accession: ATC64894
Location: 3375115-3378627
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 83 %
E-value: 5e-35
NCBI BlastP on this gene
Accession: ATC64894
Location: 3375115-3378627
BlastP hit with VTO45597.1
Percentage identity: 32 %
BlastP bit score: 149
Sequence coverage: 83 %
E-value: 5e-35
NCBI BlastP on this gene
CMV30_13470
CMV30_13465
CMV30_13460
CMV30_13455
CMV30_13450


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.