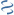MultiGeneBlast hits

Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP003557 : Melioribacter roseus P3M Total score: 1.0 Cumulative Blast bit score: 149
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
6-pyruvoyl tetrahydrobiopterin synthase
Accession: AFN73796
Location: 625444-625860
NCBI BlastP on this gene
Accession: AFN73796
Location: 625444-625860
NCBI BlastP on this gene
MROS_0553
MROS_0554
Tetratricopeptide TPR 2 repeat protein
Accession: AFN73798
Location: 626605-627378
NCBI BlastP on this gene
Accession: AFN73798
Location: 626605-627378
NCBI BlastP on this gene
MROS_0555
metal dependent phosphohydrolase
Accession: AFN73799
Location: 627395-628399
NCBI BlastP on this gene
Accession: AFN73799
Location: 627395-628399
NCBI BlastP on this gene
MROS_0556
MROS_0557
PAS/PAC sensor hybrid histidine kinase
Accession: AFN73801
Location: 628924-630408
NCBI BlastP on this gene
Accession: AFN73801
Location: 628924-630408
NCBI BlastP on this gene
MROS_0558
MROS_0559
pectate lyase, putative, pel1G
Accession: AFN73803
Location: 632851-634728
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 15 %
E-value: 1e-33
NCBI BlastP on this gene
Accession: AFN73803
Location: 632851-634728
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 15 %
E-value: 1e-33
NCBI BlastP on this gene
MROS_0560
MROS_0561
MROS_0562
MROS_0563
MROS_0564
MROS_0565
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP022660 : Bacteroides thetaiotaomicron F9-2 DNA, nearly complete genome. Total score: 1.0 Cumulative Blast bit score: 149
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
BatF92_20560
BatF92_20570
hybrid sensor histidine kinase/response regulator
Accession: BCA50116
Location: 2730071-2734417
NCBI BlastP on this gene
Accession: BCA50116
Location: 2730071-2734417
NCBI BlastP on this gene
BatF92_20580
polysaccharide lyase
Accession: BCA50117
Location: 2734481-2736163
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 149
Sequence coverage: 90 %
E-value: 4e-36
NCBI BlastP on this gene
Accession: BCA50117
Location: 2734481-2736163
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 149
Sequence coverage: 90 %
E-value: 4e-36
NCBI BlastP on this gene
BatF92_20590
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP045915 : Gracilibacillus sp. SCU50 chromosome Total score: 1.0 Cumulative Blast bit score: 147
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
tRNA pseudouridine(38-40) synthase TruA
Accession: QGH32749
Location: 203993-204742
NCBI BlastP on this gene
Accession: QGH32749
Location: 203993-204742
NCBI BlastP on this gene
truA
rplM
rpsI
phosphatase PAP2 family protein
Accession: QGH32752
Location: 206293-209283
NCBI BlastP on this gene
Accession: QGH32752
Location: 206293-209283
NCBI BlastP on this gene
GI584_01100
DJ-1/PfpI/YhbO family deglycase/protease
Accession: QGH32753
Location: 209745-210257
NCBI BlastP on this gene
Accession: QGH32753
Location: 209745-210257
NCBI BlastP on this gene
GI584_01105
pectinesterase
Accession: QGH32754
Location: 210614-213865
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 147
Sequence coverage: 65 %
E-value: 3e-34
NCBI BlastP on this gene
Accession: QGH32754
Location: 210614-213865
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 147
Sequence coverage: 65 %
E-value: 3e-34
NCBI BlastP on this gene
GI584_01110
GI584_01115
LLM class flavin-dependent oxidoreductase
Accession: QGH32756
Location: 214720-215775
NCBI BlastP on this gene
Accession: QGH32756
Location: 214720-215775
NCBI BlastP on this gene
GI584_01120
GI584_01125
TAXI family TRAP transporter solute-binding subunit
Accession: QGH32758
Location: 216523-217578
NCBI BlastP on this gene
Accession: QGH32758
Location: 216523-217578
NCBI BlastP on this gene
GI584_01130
DUF1850 domain-containing protein
Accession: QGH32759
Location: 217568-218140
NCBI BlastP on this gene
Accession: QGH32759
Location: 217568-218140
NCBI BlastP on this gene
GI584_01135
TRAP transporter fused permease subunit
Accession: QGH32760
Location: 218166-220388
NCBI BlastP on this gene
Accession: QGH32760
Location: 218166-220388
NCBI BlastP on this gene
GI584_01140
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP012937 : Bacteroides thetaiotaomicron strain 7330 Total score: 1.0 Cumulative Blast bit score: 147
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
lacZ_7
Btheta7330_02595
tmoS_6
hypothetical protein
Accession: ALJ42135
Location: 3159908-3161590
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
Accession: ALJ42135
Location: 3159908-3161590
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
Btheta7330_02593
Btheta7330_02592
RNA polymerase sigma factor SigM
Accession: ALJ42133
Location: 3156017-3156568
NCBI BlastP on this gene
Accession: ALJ42133
Location: 3156017-3156568
NCBI BlastP on this gene
sigM
Btheta7330_02590
ybjJ
yjjP
Btheta7330_02587
Btheta7330_02586
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AE015928 : Bacteroides thetaiotaomicron VPI-5482 Total score: 1.0 Cumulative Blast bit score: 147
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
BT_0983
BT_0982
two-component system sensor histidine
Accession: AAO76088
Location: 1200747-1205165
NCBI BlastP on this gene
Accession: AAO76088
Location: 1200747-1205165
NCBI BlastP on this gene
BT_0981
putative pectate lyase
Accession: AAO76087
Location: 1199001-1200683
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
Accession: AAO76087
Location: 1199001-1200683
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
BT_0980
Pectin lyase fold/virulence factor
Accession: AAO76086
Location: 1195838-1198933
NCBI BlastP on this gene
Accession: AAO76086
Location: 1195838-1198933
NCBI BlastP on this gene
BT_0979
BT_0978
BT_0977
BT_0976
conserved hypothetical protein, putative integral membrane protein
Accession: AAO76082
Location: 1192543-1193310
NCBI BlastP on this gene
Accession: AAO76082
Location: 1192543-1193310
NCBI BlastP on this gene
BT_0975
conserved hypothetical protein, putative integral membrane protein
Accession: AAO76081
Location: 1192047-1192541
NCBI BlastP on this gene
Accession: AAO76081
Location: 1192047-1192541
NCBI BlastP on this gene
BT_0974
conserved hypothetical protein
Accession: AAO76080
Location: 1191305-1192033
NCBI BlastP on this gene
Accession: AAO76080
Location: 1191305-1192033
NCBI BlastP on this gene
BT_0973
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP017774 : Flavobacterium commune strain PK15 chromosome Total score: 1.0 Cumulative Blast bit score: 146
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
pectate lyase
Accession: APA01008
Location: 3067202-3069991
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 146
Sequence coverage: 92 %
E-value: 3e-34
NCBI BlastP on this gene
Accession: APA01008
Location: 3067202-3069991
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 146
Sequence coverage: 92 %
E-value: 3e-34
NCBI BlastP on this gene
BIW12_12820
BIW12_12815
BIW12_12810
glycosyl transferase family 2
Accession: APA00237
Location: 3063936-3065366
NCBI BlastP on this gene
Accession: APA00237
Location: 3063936-3065366
NCBI BlastP on this gene
BIW12_12805
BIW12_12800
BIW12_12795
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP031728 : Cellvibrio sp. KY-GH-1 chromosome Total score: 1.0 Cumulative Blast bit score: 144
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
D0C16_17605
D0C16_17610
PLP-dependent aminotransferase family protein
Accession: QEY17647
Location: 4105230-4106639
NCBI BlastP on this gene
Accession: QEY17647
Location: 4105230-4106639
NCBI BlastP on this gene
D0C16_17615
cytochrome c oxidase accessory protein CcoG
Accession: QEY17648
Location: 4106678-4108093
NCBI BlastP on this gene
Accession: QEY17648
Location: 4106678-4108093
NCBI BlastP on this gene
ccoG
D0C16_17625
LysR family transcriptional regulator
Accession: QEY17649
Location: 4108775-4109704
NCBI BlastP on this gene
Accession: QEY17649
Location: 4108775-4109704
NCBI BlastP on this gene
D0C16_17630
D0C16_17635
pectate lyase
Accession: QEY17651
Location: 4111657-4113348
BlastP hit with VTO45599.1
Percentage identity: 33 %
BlastP bit score: 144
Sequence coverage: 13 %
E-value: 5e-32
NCBI BlastP on this gene
Accession: QEY17651
Location: 4111657-4113348
BlastP hit with VTO45599.1
Percentage identity: 33 %
BlastP bit score: 144
Sequence coverage: 13 %
E-value: 5e-32
NCBI BlastP on this gene
D0C16_17640
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP043306 : Cellvibrio japonicus strain ADPT1-KOJIBIOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 143
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
FY115_06270
spore coat protein U domain-containing protein
Accession: QEI19025
Location: 1493198-1493728
NCBI BlastP on this gene
Accession: QEI19025
Location: 1493198-1493728
NCBI BlastP on this gene
FY115_06265
spore coat protein U domain-containing protein
Accession: QEI19024
Location: 1492632-1493174
NCBI BlastP on this gene
Accession: QEI19024
Location: 1492632-1493174
NCBI BlastP on this gene
FY115_06260
FY115_06255
FY115_06250
FY115_06245
FY115_06240
FY115_06235
FY115_06230
pectate lyase
Accession: QEI19017
Location: 1485472-1486998
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
Accession: QEI19017
Location: 1485472-1486998
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
FY115_06225
FY115_06220
FY115_06215
DUF4442 domain-containing protein
Accession: QEI19014
Location: 1483283-1483762
NCBI BlastP on this gene
Accession: QEI19014
Location: 1483283-1483762
NCBI BlastP on this gene
FY115_06210
FY115_06205
PilZ domain-containing protein
Accession: QEI19012
Location: 1481689-1482066
NCBI BlastP on this gene
Accession: QEI19012
Location: 1481689-1482066
NCBI BlastP on this gene
FY115_06200
PilZ domain-containing protein
Accession: QEI19011
Location: 1481290-1481673
NCBI BlastP on this gene
Accession: QEI19011
Location: 1481290-1481673
NCBI BlastP on this gene
FY115_06195
amino acid ABC transporter substrate-binding protein
Accession: QEI19010
Location: 1480419-1481276
NCBI BlastP on this gene
Accession: QEI19010
Location: 1480419-1481276
NCBI BlastP on this gene
FY115_06190
radA
FY115_06180
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP023344 : Nibricoccus aquaticus strain HZ-65 chromosome Total score: 1.0 Cumulative Blast bit score: 143
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
C4-dicarboxylate ABC transporter permease
Accession: ATC63056
Location: 743952-744704
NCBI BlastP on this gene
Accession: ATC63056
Location: 743952-744704
NCBI BlastP on this gene
CMV30_03255
CMV30_03260
CMV30_03265
CMV30_03270
CMV30_03275
CMV30_03280
xylB
pectate lyase
Accession: ATC63061
Location: 751163-753994
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 143
Sequence coverage: 92 %
E-value: 5e-33
NCBI BlastP on this gene
Accession: ATC63061
Location: 751163-753994
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 143
Sequence coverage: 92 %
E-value: 5e-33
NCBI BlastP on this gene
CMV30_03290
CMV30_03295
CMV30_03300
CMV30_03305
CMV30_03310
CMV30_03315
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP019799 : Cellvibrio sp. PSBB023 chromosome Total score: 1.0 Cumulative Blast bit score: 143
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
B0D95_14040
B0D95_14045
B0D95_14050
B0D95_14055
Twin-arginine translocation pathway signal
Accession: AQT61091
Location: 3227229-3227783
NCBI BlastP on this gene
Accession: AQT61091
Location: 3227229-3227783
NCBI BlastP on this gene
B0D95_14060
B0D95_14065
pectate lyase
Accession: AQT61093
Location: 3228144-3229679
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 7e-32
NCBI BlastP on this gene
Accession: AQT61093
Location: 3228144-3229679
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 7e-32
NCBI BlastP on this gene
B0D95_14070
B0D95_14075
B0D95_14080
B0D95_14085
B0D95_14090
B0D95_14095
B0D95_14100
B0D95_14105
peptidoglycan-binding protein
Accession: AQT61101
Location: 3236331-3237008
NCBI BlastP on this gene
Accession: AQT61101
Location: 3236331-3237008
NCBI BlastP on this gene
B0D95_14110
type VI secretion protein VgrG
Accession: AQT61102
Location: 3237012-3238802
NCBI BlastP on this gene
Accession: AQT61102
Location: 3237012-3238802
NCBI BlastP on this gene
B0D95_14115
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP000934 : Cellvibrio japonicus Ueda107 Total score: 1.0 Cumulative Blast bit score: 143
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
fimC
putative type 1 pili pilus subunit
Accession: ACE82822
Location: 1493194-1493724
NCBI BlastP on this gene
Accession: ACE82822
Location: 1493194-1493724
NCBI BlastP on this gene
CJA_1285
Spore Coat Protein U domain protein
Accession: ACE84644
Location: 1492628-1493170
NCBI BlastP on this gene
Accession: ACE84644
Location: 1492628-1493170
NCBI BlastP on this gene
CJA_1284
CJA_1283
CJA_1282
CJA_1281
CJA_1280
CJA_1279
CJA_1278
pectate lyase III
Accession: ACE86187
Location: 1485468-1486994
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
Accession: ACE86187
Location: 1485468-1486994
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
pelC
CJA_1275
CJA_1276
CJA_1274
conserved hypothetical protein
Accession: ACE84224
Location: 1483279-1483758
NCBI BlastP on this gene
Accession: ACE84224
Location: 1483279-1483758
NCBI BlastP on this gene
CJA_1273
probable NAGC-like transcription regulator yajF
Accession: ACE85345
Location: 1482291-1483211
NCBI BlastP on this gene
Accession: ACE85345
Location: 1482291-1483211
NCBI BlastP on this gene
CJA_1272
conserved hypothetical protein
Accession: ACE86214
Location: 1481685-1482062
NCBI BlastP on this gene
Accession: ACE86214
Location: 1481685-1482062
NCBI BlastP on this gene
CJA_1271
CJA_1270
conserved hypothetical protein
Accession: ACE85956
Location: 1480415-1481272
NCBI BlastP on this gene
Accession: ACE85956
Location: 1480415-1481272
NCBI BlastP on this gene
CJA_1268
radA
CJA_1267
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AM746676 : Sorangium cellulosum 'So ce 56' complete genome. Total score: 1.0 Cumulative Blast bit score: 143
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Probable pectate lyase precursor
Accession: CAN95995
Location: 8188518-8190302
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 143
Sequence coverage: 63 %
E-value: 6e-34
NCBI BlastP on this gene
Accession: CAN95995
Location: 8188518-8190302
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 143
Sequence coverage: 63 %
E-value: 6e-34
NCBI BlastP on this gene
sce5832
sugE
sce5830
sce5829
sce5828
sce5827
sce5826
sce5825
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP042913 : Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 142
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Pr1d_37640
Pr1d_37630
Pr1d_37620
Pr1d_37610
tetD
Pr1d_37590
Pr1d_37580
hypothetical protein
Accession: QEG36443
Location: 4508967-4510544
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 108 %
E-value: 1e-33
NCBI BlastP on this gene
Accession: QEG36443
Location: 4508967-4510544
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 108 %
E-value: 1e-33
NCBI BlastP on this gene
Pr1d_37570
pelL_2
Pr1d_37550
Pr1d_37540
D-allose-binding periplasmic protein precursor
Accession: QEG36439
Location: 4501478-4502509
NCBI BlastP on this gene
Accession: QEG36439
Location: 4501478-4502509
NCBI BlastP on this gene
alsB_2
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP002993 : Streptomyces sp. SirexAA-E Total score: 1.0 Cumulative Blast bit score: 142
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
SACTE_1310
conserved hypothetical protein
Accession: AEN09229
Location: 1469659-1470360
NCBI BlastP on this gene
Accession: AEN09229
Location: 1469659-1470360
NCBI BlastP on this gene
SACTE_1309
RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: AEN09228
Location: 1469087-1469659
NCBI BlastP on this gene
Accession: AEN09228
Location: 1469087-1469659
NCBI BlastP on this gene
SACTE_1308
penicillin-binding protein transpeptidase
Accession: AEN09227
Location: 1467366-1468817
NCBI BlastP on this gene
Accession: AEN09227
Location: 1467366-1468817
NCBI BlastP on this gene
SACTE_1307
GCN5-related N-acetyltransferase
Accession: AEN09226
Location: 1466780-1467259
NCBI BlastP on this gene
Accession: AEN09226
Location: 1466780-1467259
NCBI BlastP on this gene
SACTE_1306
transcriptional regulator, IclR family
Accession: AEN09225
Location: 1465981-1466775
NCBI BlastP on this gene
Accession: AEN09225
Location: 1465981-1466775
NCBI BlastP on this gene
SACTE_1305
SACTE_1304
protein of unknown function DUF1349
Accession: AEN09223
Location: 1463683-1464285
NCBI BlastP on this gene
Accession: AEN09223
Location: 1463683-1464285
NCBI BlastP on this gene
SACTE_1303
Pectate lyase/Amb allergen
Accession: AEN09222
Location: 1462500-1463474
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 142
Sequence coverage: 14 %
E-value: 1e-32
NCBI BlastP on this gene
Accession: AEN09222
Location: 1462500-1463474
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 142
Sequence coverage: 14 %
E-value: 1e-32
NCBI BlastP on this gene
SACTE_1302
serine/threonine protein kinase
Accession: AEN09221
Location: 1461363-1462157
NCBI BlastP on this gene
Accession: AEN09221
Location: 1461363-1462157
NCBI BlastP on this gene
SACTE_1301
SACTE_1299
SACTE_1298
transcriptional regulator, MarR family
Accession: AEN09218
Location: 1458601-1459188
NCBI BlastP on this gene
Accession: AEN09218
Location: 1458601-1459188
NCBI BlastP on this gene
SACTE_1297
12-oxophytodienoate reductase
Accession: AEN09217
Location: 1457350-1458417
NCBI BlastP on this gene
Accession: AEN09217
Location: 1457350-1458417
NCBI BlastP on this gene
SACTE_1296
transcriptional regulator, LacI family
Accession: AEN09216
Location: 1456302-1457336
NCBI BlastP on this gene
Accession: AEN09216
Location: 1456302-1457336
NCBI BlastP on this gene
SACTE_1295
binding-protein-dependent transport systems inner membrane component
Accession: AEN09215
Location: 1455399-1456256
NCBI BlastP on this gene
Accession: AEN09215
Location: 1455399-1456256
NCBI BlastP on this gene
SACTE_1294
binding-protein-dependent transport systems inner membrane component
Accession: AEN09214
Location: 1454476-1455402
NCBI BlastP on this gene
Accession: AEN09214
Location: 1454476-1455402
NCBI BlastP on this gene
SACTE_1293
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
LT629740 : Mucilaginibacter mallensis strain MP1X4 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 140
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
SAMN05216490_2657
SAMN05216490_2658
SAMN05216490_2659
2-desacetyl-2-hydroxyethyl bacteriochlorophyllide A dehydrogenase
Accession: SDT17721
Location: 3216537-3217553
NCBI BlastP on this gene
Accession: SDT17721
Location: 3216537-3217553
NCBI BlastP on this gene
SAMN05216490_2660
hypothetical protein
Accession: SDT17775
Location: 3217813-3219468
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 140
Sequence coverage: 90 %
E-value: 6e-33
NCBI BlastP on this gene
Accession: SDT17775
Location: 3217813-3219468
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 140
Sequence coverage: 90 %
E-value: 6e-33
NCBI BlastP on this gene
SAMN05216490_2661
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP036525 : Planctomycetes bacterium K22_7 chromosome Total score: 1.0 Cumulative Blast bit score: 140
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
K227x_60210
Serine/threonine-protein kinase PrkC
Accession: QDT07594
Location: 7964682-7967261
NCBI BlastP on this gene
Accession: QDT07594
Location: 7964682-7967261
NCBI BlastP on this gene
prkC_24
ECF RNA polymerase sigma-E factor
Accession: QDT07595
Location: 7967261-7967938
NCBI BlastP on this gene
Accession: QDT07595
Location: 7967261-7967938
NCBI BlastP on this gene
rpoE_6
K227x_60240
K227x_60250
hypothetical protein
Accession: QDT07598
Location: 7971214-7972689
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 140
Sequence coverage: 68 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: QDT07598
Location: 7971214-7972689
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 140
Sequence coverage: 68 %
E-value: 2e-33
NCBI BlastP on this gene
K227x_60260
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP020991 : Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 140
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
B9O19_01124
B9O19_01125
ribose 5-phosphate isomerase B
Accession: AUO19289
Location: 1277606-1278046
NCBI BlastP on this gene
Accession: AUO19289
Location: 1277606-1278046
NCBI BlastP on this gene
B9O19_01126
B9O19_01127
B9O19_01128
pectate lyase family 1
Accession: AUO19292
Location: 1283067-1286006
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 63 %
E-value: 5e-32
NCBI BlastP on this gene
Accession: AUO19292
Location: 1283067-1286006
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 63 %
E-value: 5e-32
NCBI BlastP on this gene
B9O19_01129
metallo-beta-lactamase family protein
Accession: AUO19293
Location: 1287427-1288278
NCBI BlastP on this gene
Accession: AUO19293
Location: 1287427-1288278
NCBI BlastP on this gene
B9O19_01130
B9O19_01131
cell wall repeat-containing protein
Accession: AUO19295
Location: 1289360-1292212
NCBI BlastP on this gene
Accession: AUO19295
Location: 1289360-1292212
NCBI BlastP on this gene
pmpB
transcriptional regulator,TetR family
Accession: AUO19296
Location: 1292343-1292999
NCBI BlastP on this gene
Accession: AUO19296
Location: 1292343-1292999
NCBI BlastP on this gene
B9O19_01133
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP012672 : Sorangium cellulosum strain So ce836 chromosome Total score: 1.0 Cumulative Blast bit score: 140
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
pectate lyase
Accession: AUX35189
Location: 9964704-9966125
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 64 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: AUX35189
Location: 9964704-9966125
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 64 %
E-value: 2e-33
NCBI BlastP on this gene
pel
SOCE836_073770
SOCE836_073760
SAM-dependent methyltransferase
Accession: AUX35186
Location: 9962227-9962991
NCBI BlastP on this gene
Accession: AUX35186
Location: 9962227-9962991
NCBI BlastP on this gene
smtA
SOCE836_073740
SOCE836_073730
SOCE836_073720
SOCE836_073710
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP049143 : Roseimicrobium sp. ORNL1 chromosome Total score: 1.0 Cumulative Blast bit score: 139
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
G5S37_30630
G5S37_30635
G5S37_30640
tRNA uridine-5-carboxymethylaminomethyl(34) synthesis enzyme MnmG
Accession: QIF05675
Location: 7571327-7573225
NCBI BlastP on this gene
Accession: QIF05675
Location: 7571327-7573225
NCBI BlastP on this gene
mnmG
DUF1801 domain-containing protein
Accession: QIF05676
Location: 7573466-7573858
NCBI BlastP on this gene
Accession: QIF05676
Location: 7573466-7573858
NCBI BlastP on this gene
G5S37_30650
metallopeptidase family protein
Accession: QIF05677
Location: 7573869-7574240
NCBI BlastP on this gene
Accession: QIF05677
Location: 7573869-7574240
NCBI BlastP on this gene
G5S37_30655
hypothetical protein
Accession: QIF05678
Location: 7574328-7575779
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 139
Sequence coverage: 76 %
E-value: 9e-33
NCBI BlastP on this gene
Accession: QIF05678
Location: 7574328-7575779
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 139
Sequence coverage: 76 %
E-value: 9e-33
NCBI BlastP on this gene
G5S37_30660
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP036349 : Planctomycetes bacterium Spa11 chromosome. Total score: 1.0 Cumulative Blast bit score: 138
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Spa11_01540
Spa11_01530
Spa11_01520
pelL_1
Pectate lyase
Accession: QDV71981
Location: 173272-175455
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 68 %
E-value: 9e-32
NCBI BlastP on this gene
Accession: QDV71981
Location: 173272-175455
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 68 %
E-value: 9e-32
NCBI BlastP on this gene
Spa11_01500
Spa11_01490
Spa11_01480
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV71978
Location: 170372-170866
NCBI BlastP on this gene
Accession: QDV71978
Location: 170372-170866
NCBI BlastP on this gene
fabZ_2
acpP_1
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV71976
Location: 169265-169822
NCBI BlastP on this gene
Accession: QDV71976
Location: 169265-169822
NCBI BlastP on this gene
fabZ_1
3-oxoacyl-[acyl-carrier-protein] synthase 2
Accession: QDV71975
Location: 167738-169009
NCBI BlastP on this gene
Accession: QDV71975
Location: 167738-169009
NCBI BlastP on this gene
fabF_1
pgl_1
Spa11_01420
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP029255 : Flavobacterium crocinum strain HYN0056 chromosome Total score: 1.0 Cumulative Blast bit score: 138
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
HYN56_13410
HYN56_13405
HYN56_13400
3-phosphoserine/phosphohydroxythreonine transaminase
Accession: AWK05172
Location: 2989808-2990878
NCBI BlastP on this gene
Accession: AWK05172
Location: 2989808-2990878
NCBI BlastP on this gene
HYN56_13395
3-phosphoglycerate dehydrogenase
Accession: AWK07434
Location: 2988761-2989711
NCBI BlastP on this gene
Accession: AWK07434
Location: 2988761-2989711
NCBI BlastP on this gene
HYN56_13390
HYN56_13385
HYN56_13380
HYN56_13375
pectate lyase
Accession: AWK07433
Location: 2983853-2986600
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 138
Sequence coverage: 87 %
E-value: 2e-31
NCBI BlastP on this gene
Accession: AWK07433
Location: 2983853-2986600
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 138
Sequence coverage: 87 %
E-value: 2e-31
NCBI BlastP on this gene
HYN56_13370
HYN56_13365
ABC transporter ATP-binding protein
Accession: AWK05167
Location: 2982224-2983102
NCBI BlastP on this gene
Accession: AWK05167
Location: 2982224-2983102
NCBI BlastP on this gene
HYN56_13360
HYN56_13355
HYN56_13350
HYN56_13345
HYN56_13340
HYN56_13335
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP028159 : Plantactinospora sp. BB1 chromosome Total score: 1.0 Cumulative Blast bit score: 138
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
C6W10_01930
C6W10_01925
C6W10_01920
TetR family transcriptional regulator
Accession: AVT35422
Location: 474725-475405
NCBI BlastP on this gene
Accession: AVT35422
Location: 474725-475405
NCBI BlastP on this gene
C6W10_01915
C6W10_01910
C6W10_01905
C6W10_01900
C6W10_01895
aminoglycoside phosphotransferase
Accession: AVT35418
Location: 470481-471299
NCBI BlastP on this gene
Accession: AVT35418
Location: 470481-471299
NCBI BlastP on this gene
C6W10_01890
pectate lyase
Accession: AVT41103
Location: 468731-469912
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
Accession: AVT41103
Location: 468731-469912
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
C6W10_01885
C6W10_01880
TetR family transcriptional regulator
Accession: AVT35416
Location: 467386-467976
NCBI BlastP on this gene
Accession: AVT35416
Location: 467386-467976
NCBI BlastP on this gene
C6W10_01875
C6W10_01870
C6W10_01865
C6W10_01860
FAD-binding molybdopterin dehydrogenase
Accession: AVT35412
Location: 461701-462678
NCBI BlastP on this gene
Accession: AVT35412
Location: 461701-462678
NCBI BlastP on this gene
C6W10_01855
C6W10_01850
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP028158 : Plantactinospora sp. BC1 chromosome Total score: 1.0 Cumulative Blast bit score: 138
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
C6361_05390
C6361_05385
C6361_05380
TetR family transcriptional regulator
Accession: AVT29017
Location: 1306163-1306843
NCBI BlastP on this gene
Accession: AVT29017
Location: 1306163-1306843
NCBI BlastP on this gene
C6361_05375
C6361_05370
C6361_05365
C6361_05360
C6361_05355
aminoglycoside phosphotransferase
Accession: AVT29013
Location: 1301895-1302713
NCBI BlastP on this gene
Accession: AVT29013
Location: 1301895-1302713
NCBI BlastP on this gene
C6361_05350
pectate lyase
Accession: AVT34190
Location: 1300144-1301325
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
Accession: AVT34190
Location: 1300144-1301325
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
C6361_05345
C6361_05340
C6361_05335
TetR family transcriptional regulator
Accession: AVT29011
Location: 1298799-1299389
NCBI BlastP on this gene
Accession: AVT29011
Location: 1298799-1299389
NCBI BlastP on this gene
C6361_05330
C6361_05325
C6361_05320
carbon monoxide dehydrogenase
Accession: AVT29008
Location: 1294145-1296361
NCBI BlastP on this gene
Accession: AVT29008
Location: 1294145-1296361
NCBI BlastP on this gene
C6361_05315
FAD-binding molybdopterin dehydrogenase
Accession: AVT29007
Location: 1293171-1294148
NCBI BlastP on this gene
Accession: AVT29007
Location: 1293171-1294148
NCBI BlastP on this gene
C6361_05310
C6361_05305
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP020991 : Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 138
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
B9O19_01438
hybrid sensor histidine kinase/response regulator
Accession: AUO19600
Location: 1669649-1672846
NCBI BlastP on this gene
Accession: AUO19600
Location: 1669649-1672846
NCBI BlastP on this gene
B9O19_01439
B9O19_01440
B9O19_01441
pectate lyase family 1
Accession: AUO19603
Location: 1674376-1677705
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 53 %
E-value: 2e-31
NCBI BlastP on this gene
Accession: AUO19603
Location: 1674376-1677705
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 53 %
E-value: 2e-31
NCBI BlastP on this gene
B9O19_01442
B9O19_01443
GCN5-related N-acetyltransferase
Accession: AUO19605
Location: 1682734-1683315
NCBI BlastP on this gene
Accession: AUO19605
Location: 1682734-1683315
NCBI BlastP on this gene
B9O19_01444
DNA-directed RNA polymerase, beta subunit
Accession: AUO19606
Location: 1683684-1687511
NCBI BlastP on this gene
Accession: AUO19606
Location: 1683684-1687511
NCBI BlastP on this gene
B9O19_01445
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP036350 : Planctomycetes bacterium K2D chromosome Total score: 1.0 Cumulative Blast bit score: 137
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
K2D_01040
K2D_01030
K2D_01020
pelL_1
Pectate lyase
Accession: QDV76522
Location: 117208-119391
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 137
Sequence coverage: 68 %
E-value: 1e-31
NCBI BlastP on this gene
Accession: QDV76522
Location: 117208-119391
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 137
Sequence coverage: 68 %
E-value: 1e-31
NCBI BlastP on this gene
K2D_01000
K2D_00990
K2D_00980
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV76519
Location: 114315-114809
NCBI BlastP on this gene
Accession: QDV76519
Location: 114315-114809
NCBI BlastP on this gene
fabZ_2
acpP_1
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV76517
Location: 113208-113765
NCBI BlastP on this gene
Accession: QDV76517
Location: 113208-113765
NCBI BlastP on this gene
fabZ_1
3-oxoacyl-[acyl-carrier-protein] synthase 2
Accession: QDV76516
Location: 111681-112952
NCBI BlastP on this gene
Accession: QDV76516
Location: 111681-112952
NCBI BlastP on this gene
fabF_1
pgl_1
K2D_00920
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP041253 : Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 133
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
DUF5108 domain-containing protein
Accession: QDH80551
Location: 4224053-4225654
NCBI BlastP on this gene
Accession: QDH80551
Location: 4224053-4225654
NCBI BlastP on this gene
FKX85_16485
fasciclin domain-containing protein
Accession: QDH80550
Location: 4222452-4224041
NCBI BlastP on this gene
Accession: QDH80550
Location: 4222452-4224041
NCBI BlastP on this gene
FKX85_16480
SusC/RagA family TonB-linked outer membrane protein
Accession: QDH80549
Location: 4219345-4222446
NCBI BlastP on this gene
Accession: QDH80549
Location: 4219345-4222446
NCBI BlastP on this gene
FKX85_16475
RagB/SusD family nutrient uptake outer membrane protein
Accession: QDH81607
Location: 4217699-4219333
NCBI BlastP on this gene
Accession: QDH81607
Location: 4217699-4219333
NCBI BlastP on this gene
FKX85_16470
pectate lyase
Accession: QDH80548
Location: 4215703-4216980
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 133
Sequence coverage: 63 %
E-value: 5e-31
NCBI BlastP on this gene
Accession: QDH80548
Location: 4215703-4216980
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 133
Sequence coverage: 63 %
E-value: 5e-31
NCBI BlastP on this gene
FKX85_16465
FKX85_16460
glycoside hydrolase family 88 protein
Accession: QDH80546
Location: 4212092-4213276
NCBI BlastP on this gene
Accession: QDH80546
Location: 4212092-4213276
NCBI BlastP on this gene
FKX85_16455
glycoside hydrolase family 28 protein
Accession: QDH80545
Location: 4210390-4211814
NCBI BlastP on this gene
Accession: QDH80545
Location: 4210390-4211814
NCBI BlastP on this gene
FKX85_16450
FKX85_16445
T9SS type A sorting domain-containing protein
Accession: QDH80543
Location: 4204275-4209011
NCBI BlastP on this gene
Accession: QDH80543
Location: 4204275-4209011
NCBI BlastP on this gene
FKX85_16440
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP030041 : Echinicola strongylocentroti strain MEBiC08714 chromosome Total score: 1.0 Cumulative Blast bit score: 132
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
DN752_23955
fasciclin domain-containing protein
Accession: AWW33336
Location: 6149045-6150634
NCBI BlastP on this gene
Accession: AWW33336
Location: 6149045-6150634
NCBI BlastP on this gene
DN752_23960
SusC/RagA family TonB-linked outer membrane protein
Accession: AWW33337
Location: 6150640-6153741
NCBI BlastP on this gene
Accession: AWW33337
Location: 6150640-6153741
NCBI BlastP on this gene
DN752_23965
DN752_23970
pectate lyase
Accession: AWW32936
Location: 6155953-6157230
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 132
Sequence coverage: 62 %
E-value: 9e-31
NCBI BlastP on this gene
Accession: AWW32936
Location: 6155953-6157230
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 132
Sequence coverage: 62 %
E-value: 9e-31
NCBI BlastP on this gene
DN752_23975
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP031010 : Alteromonas sp. RKMC-009 chromosome Total score: 1.0 Cumulative Blast bit score: 101
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
Tat pathway signal sequence domain protein
Accession: AYA65921
Location: 4295818-4298523
NCBI BlastP on this gene
Accession: AYA65921
Location: 4295818-4298523
NCBI BlastP on this gene
DS731_18945
glycoside hydrolase family 105 protein
Accession: AYA66676
Location: 4298793-4299899
NCBI BlastP on this gene
Accession: AYA66676
Location: 4298793-4299899
NCBI BlastP on this gene
DS731_18950
DS731_18955
DS731_18960
DS731_18965
IclR family transcriptional regulator
Accession: AYA65924
Location: 4303653-4304426
BlastP hit with VTO45596.1
Percentage identity: 33 %
BlastP bit score: 101
Sequence coverage: 88 %
E-value: 4e-22
NCBI BlastP on this gene
Accession: AYA65924
Location: 4303653-4304426
BlastP hit with VTO45596.1
Percentage identity: 33 %
BlastP bit score: 101
Sequence coverage: 88 %
E-value: 4e-22
NCBI BlastP on this gene
DS731_18970
DS731_18975
DeoR/GlpR transcriptional regulator
Accession: AYA65926
Location: 4305496-4306284
NCBI BlastP on this gene
Accession: AYA65926
Location: 4305496-4306284
NCBI BlastP on this gene
DS731_18980
DS731_18985
DS731_18990
formimidoylglutamate deiminase
Accession: AYA65929
Location: 4311710-4313086
NCBI BlastP on this gene
Accession: AYA65929
Location: 4311710-4313086
NCBI BlastP on this gene
DS731_18995
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
AP021859 : Alteromonas sp. I4 DNA Total score: 1.0 Cumulative Blast bit score: 77
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
IclR family transcriptional regulator
Accession: BBO29965
Location: 4997441-4998136
BlastP hit with VTO45596.1
Percentage identity: 32 %
BlastP bit score: 77
Sequence coverage: 47 %
E-value: 1e-13
NCBI BlastP on this gene
Accession: BBO29965
Location: 4997441-4998136
BlastP hit with VTO45596.1
Percentage identity: 32 %
BlastP bit score: 77
Sequence coverage: 47 %
E-value: 1e-13
NCBI BlastP on this gene
AltI4_43530
2-dehydro-3-deoxygluconokinase
Accession: BBO29964
Location: 4996474-4997427
NCBI BlastP on this gene
Accession: BBO29964
Location: 4996474-4997427
NCBI BlastP on this gene
kdgK_2
AltI4_43510
glycerol-3-phosphate acyltransferase
Accession: BBO29962
Location: 4993502-4995985
NCBI BlastP on this gene
Accession: BBO29962
Location: 4993502-4995985
NCBI BlastP on this gene
plsB
AltI4_43490
AltI4_43480
AltI4_43470
AltI4_43460
GGDEF domain-containing protein
Accession: BBO29957
Location: 4988299-4989600
NCBI BlastP on this gene
Accession: BBO29957
Location: 4988299-4989600
NCBI BlastP on this gene
AltI4_43450
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
CP022434 : Nocardiopsis dassonvillei strain HZNU_N_1 chromosome Total score: 1.0 Cumulative Blast bit score: 61
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
CGQ36_18510
formimidoylglutamate deiminase
Accession: ASU61355
Location: 4447874-4449385
NCBI BlastP on this gene
Accession: ASU61355
Location: 4447874-4449385
NCBI BlastP on this gene
CGQ36_18515
CGQ36_18520
CGQ36_18525
hutH
IclR family transcriptional regulator
Accession: ASU59440
Location: 4454155-4454916
BlastP hit with VTO45596.1
Percentage identity: 31 %
BlastP bit score: 61
Sequence coverage: 61 %
E-value: 7e-08
NCBI BlastP on this gene
Accession: ASU59440
Location: 4454155-4454916
BlastP hit with VTO45596.1
Percentage identity: 31 %
BlastP bit score: 61
Sequence coverage: 61 %
E-value: 7e-08
NCBI BlastP on this gene
CGQ36_18535
CGQ36_18540
Query: Alteromonas macleodii isolate Alteromonas macleodii 83-1 genome
301. : CP003557 Melioribacter roseus P3M Total score: 1.0 Cumulative Blast bit score: 149
ALT831_03876
ALT831_03877
ALT831_03878
ALT831_03879
6-pyruvoyl tetrahydrobiopterin synthase
Accession: AFN73796
Location: 625444-625860
NCBI BlastP on this gene
Accession: AFN73796
Location: 625444-625860
NCBI BlastP on this gene
MROS_0553
MROS_0554
Tetratricopeptide TPR 2 repeat protein
Accession: AFN73798
Location: 626605-627378
NCBI BlastP on this gene
Accession: AFN73798
Location: 626605-627378
NCBI BlastP on this gene
MROS_0555
metal dependent phosphohydrolase
Accession: AFN73799
Location: 627395-628399
NCBI BlastP on this gene
Accession: AFN73799
Location: 627395-628399
NCBI BlastP on this gene
MROS_0556
MROS_0557
PAS/PAC sensor hybrid histidine kinase
Accession: AFN73801
Location: 628924-630408
NCBI BlastP on this gene
Accession: AFN73801
Location: 628924-630408
NCBI BlastP on this gene
MROS_0558
MROS_0559
pectate lyase, putative, pel1G
Accession: AFN73803
Location: 632851-634728
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 15 %
E-value: 1e-33
NCBI BlastP on this gene
Accession: AFN73803
Location: 632851-634728
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 149
Sequence coverage: 15 %
E-value: 1e-33
NCBI BlastP on this gene
MROS_0560
MROS_0561
MROS_0562
MROS_0563
MROS_0564
MROS_0565
302. : AP022660 Bacteroides thetaiotaomicron F9-2 DNA, nearly complete genome. Total score: 1.0 Cumulative Blast bit score: 149
BatF92_20560
BatF92_20570
hybrid sensor histidine kinase/response regulator
Accession: BCA50116
Location: 2730071-2734417
NCBI BlastP on this gene
Accession: BCA50116
Location: 2730071-2734417
NCBI BlastP on this gene
BatF92_20580
polysaccharide lyase
Accession: BCA50117
Location: 2734481-2736163
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 149
Sequence coverage: 90 %
E-value: 4e-36
NCBI BlastP on this gene
Accession: BCA50117
Location: 2734481-2736163
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 149
Sequence coverage: 90 %
E-value: 4e-36
NCBI BlastP on this gene
BatF92_20590
303. : CP045915 Gracilibacillus sp. SCU50 chromosome Total score: 1.0 Cumulative Blast bit score: 147
tRNA pseudouridine(38-40) synthase TruA
Accession: QGH32749
Location: 203993-204742
NCBI BlastP on this gene
Accession: QGH32749
Location: 203993-204742
NCBI BlastP on this gene
truA
rplM
rpsI
phosphatase PAP2 family protein
Accession: QGH32752
Location: 206293-209283
NCBI BlastP on this gene
Accession: QGH32752
Location: 206293-209283
NCBI BlastP on this gene
GI584_01100
DJ-1/PfpI/YhbO family deglycase/protease
Accession: QGH32753
Location: 209745-210257
NCBI BlastP on this gene
Accession: QGH32753
Location: 209745-210257
NCBI BlastP on this gene
GI584_01105
pectinesterase
Accession: QGH32754
Location: 210614-213865
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 147
Sequence coverage: 65 %
E-value: 3e-34
NCBI BlastP on this gene
Accession: QGH32754
Location: 210614-213865
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 147
Sequence coverage: 65 %
E-value: 3e-34
NCBI BlastP on this gene
GI584_01110
GI584_01115
LLM class flavin-dependent oxidoreductase
Accession: QGH32756
Location: 214720-215775
NCBI BlastP on this gene
Accession: QGH32756
Location: 214720-215775
NCBI BlastP on this gene
GI584_01120
GI584_01125
TAXI family TRAP transporter solute-binding subunit
Accession: QGH32758
Location: 216523-217578
NCBI BlastP on this gene
Accession: QGH32758
Location: 216523-217578
NCBI BlastP on this gene
GI584_01130
DUF1850 domain-containing protein
Accession: QGH32759
Location: 217568-218140
NCBI BlastP on this gene
Accession: QGH32759
Location: 217568-218140
NCBI BlastP on this gene
GI584_01135
TRAP transporter fused permease subunit
Accession: QGH32760
Location: 218166-220388
NCBI BlastP on this gene
Accession: QGH32760
Location: 218166-220388
NCBI BlastP on this gene
GI584_01140
304. : CP012937 Bacteroides thetaiotaomicron strain 7330 Total score: 1.0 Cumulative Blast bit score: 147
lacZ_7
Btheta7330_02595
tmoS_6
hypothetical protein
Accession: ALJ42135
Location: 3159908-3161590
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
Accession: ALJ42135
Location: 3159908-3161590
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
Btheta7330_02593
Btheta7330_02592
RNA polymerase sigma factor SigM
Accession: ALJ42133
Location: 3156017-3156568
NCBI BlastP on this gene
Accession: ALJ42133
Location: 3156017-3156568
NCBI BlastP on this gene
sigM
Btheta7330_02590
ybjJ
yjjP
Btheta7330_02587
Btheta7330_02586
305. : AE015928 Bacteroides thetaiotaomicron VPI-5482 Total score: 1.0 Cumulative Blast bit score: 147
BT_0983
BT_0982
two-component system sensor histidine
Accession: AAO76088
Location: 1200747-1205165
NCBI BlastP on this gene
Accession: AAO76088
Location: 1200747-1205165
NCBI BlastP on this gene
BT_0981
putative pectate lyase
Accession: AAO76087
Location: 1199001-1200683
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
Accession: AAO76087
Location: 1199001-1200683
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 147
Sequence coverage: 90 %
E-value: 2e-35
NCBI BlastP on this gene
BT_0980
Pectin lyase fold/virulence factor
Accession: AAO76086
Location: 1195838-1198933
NCBI BlastP on this gene
Accession: AAO76086
Location: 1195838-1198933
NCBI BlastP on this gene
BT_0979
BT_0978
BT_0977
BT_0976
conserved hypothetical protein, putative integral membrane protein
Accession: AAO76082
Location: 1192543-1193310
NCBI BlastP on this gene
Accession: AAO76082
Location: 1192543-1193310
NCBI BlastP on this gene
BT_0975
conserved hypothetical protein, putative integral membrane protein
Accession: AAO76081
Location: 1192047-1192541
NCBI BlastP on this gene
Accession: AAO76081
Location: 1192047-1192541
NCBI BlastP on this gene
BT_0974
conserved hypothetical protein
Accession: AAO76080
Location: 1191305-1192033
NCBI BlastP on this gene
Accession: AAO76080
Location: 1191305-1192033
NCBI BlastP on this gene
BT_0973
306. : CP017774 Flavobacterium commune strain PK15 chromosome Total score: 1.0 Cumulative Blast bit score: 146
pectate lyase
Accession: APA01008
Location: 3067202-3069991
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 146
Sequence coverage: 92 %
E-value: 3e-34
NCBI BlastP on this gene
Accession: APA01008
Location: 3067202-3069991
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 146
Sequence coverage: 92 %
E-value: 3e-34
NCBI BlastP on this gene
BIW12_12820
BIW12_12815
BIW12_12810
glycosyl transferase family 2
Accession: APA00237
Location: 3063936-3065366
NCBI BlastP on this gene
Accession: APA00237
Location: 3063936-3065366
NCBI BlastP on this gene
BIW12_12805
BIW12_12800
BIW12_12795
307. : CP031728 Cellvibrio sp. KY-GH-1 chromosome Total score: 1.0 Cumulative Blast bit score: 144
D0C16_17605
D0C16_17610
PLP-dependent aminotransferase family protein
Accession: QEY17647
Location: 4105230-4106639
NCBI BlastP on this gene
Accession: QEY17647
Location: 4105230-4106639
NCBI BlastP on this gene
D0C16_17615
cytochrome c oxidase accessory protein CcoG
Accession: QEY17648
Location: 4106678-4108093
NCBI BlastP on this gene
Accession: QEY17648
Location: 4106678-4108093
NCBI BlastP on this gene
ccoG
D0C16_17625
LysR family transcriptional regulator
Accession: QEY17649
Location: 4108775-4109704
NCBI BlastP on this gene
Accession: QEY17649
Location: 4108775-4109704
NCBI BlastP on this gene
D0C16_17630
D0C16_17635
pectate lyase
Accession: QEY17651
Location: 4111657-4113348
BlastP hit with VTO45599.1
Percentage identity: 33 %
BlastP bit score: 144
Sequence coverage: 13 %
E-value: 5e-32
NCBI BlastP on this gene
Accession: QEY17651
Location: 4111657-4113348
BlastP hit with VTO45599.1
Percentage identity: 33 %
BlastP bit score: 144
Sequence coverage: 13 %
E-value: 5e-32
NCBI BlastP on this gene
D0C16_17640
308. : CP043306 Cellvibrio japonicus strain ADPT1-KOJIBIOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 143
FY115_06270
spore coat protein U domain-containing protein
Accession: QEI19025
Location: 1493198-1493728
NCBI BlastP on this gene
Accession: QEI19025
Location: 1493198-1493728
NCBI BlastP on this gene
FY115_06265
spore coat protein U domain-containing protein
Accession: QEI19024
Location: 1492632-1493174
NCBI BlastP on this gene
Accession: QEI19024
Location: 1492632-1493174
NCBI BlastP on this gene
FY115_06260
FY115_06255
FY115_06250
FY115_06245
FY115_06240
FY115_06235
FY115_06230
pectate lyase
Accession: QEI19017
Location: 1485472-1486998
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
Accession: QEI19017
Location: 1485472-1486998
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
FY115_06225
FY115_06220
FY115_06215
DUF4442 domain-containing protein
Accession: QEI19014
Location: 1483283-1483762
NCBI BlastP on this gene
Accession: QEI19014
Location: 1483283-1483762
NCBI BlastP on this gene
FY115_06210
FY115_06205
PilZ domain-containing protein
Accession: QEI19012
Location: 1481689-1482066
NCBI BlastP on this gene
Accession: QEI19012
Location: 1481689-1482066
NCBI BlastP on this gene
FY115_06200
PilZ domain-containing protein
Accession: QEI19011
Location: 1481290-1481673
NCBI BlastP on this gene
Accession: QEI19011
Location: 1481290-1481673
NCBI BlastP on this gene
FY115_06195
amino acid ABC transporter substrate-binding protein
Accession: QEI19010
Location: 1480419-1481276
NCBI BlastP on this gene
Accession: QEI19010
Location: 1480419-1481276
NCBI BlastP on this gene
FY115_06190
radA
FY115_06180
309. : CP023344 Nibricoccus aquaticus strain HZ-65 chromosome Total score: 1.0 Cumulative Blast bit score: 143
C4-dicarboxylate ABC transporter permease
Accession: ATC63056
Location: 743952-744704
NCBI BlastP on this gene
Accession: ATC63056
Location: 743952-744704
NCBI BlastP on this gene
CMV30_03255
CMV30_03260
CMV30_03265
CMV30_03270
CMV30_03275
CMV30_03280
xylB
pectate lyase
Accession: ATC63061
Location: 751163-753994
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 143
Sequence coverage: 92 %
E-value: 5e-33
NCBI BlastP on this gene
Accession: ATC63061
Location: 751163-753994
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 143
Sequence coverage: 92 %
E-value: 5e-33
NCBI BlastP on this gene
CMV30_03290
CMV30_03295
CMV30_03300
CMV30_03305
CMV30_03310
CMV30_03315
310. : CP019799 Cellvibrio sp. PSBB023 chromosome Total score: 1.0 Cumulative Blast bit score: 143
B0D95_14040
B0D95_14045
B0D95_14050
B0D95_14055
Twin-arginine translocation pathway signal
Accession: AQT61091
Location: 3227229-3227783
NCBI BlastP on this gene
Accession: AQT61091
Location: 3227229-3227783
NCBI BlastP on this gene
B0D95_14060
B0D95_14065
pectate lyase
Accession: AQT61093
Location: 3228144-3229679
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 7e-32
NCBI BlastP on this gene
Accession: AQT61093
Location: 3228144-3229679
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 7e-32
NCBI BlastP on this gene
B0D95_14070
B0D95_14075
B0D95_14080
B0D95_14085
B0D95_14090
B0D95_14095
B0D95_14100
B0D95_14105
peptidoglycan-binding protein
Accession: AQT61101
Location: 3236331-3237008
NCBI BlastP on this gene
Accession: AQT61101
Location: 3236331-3237008
NCBI BlastP on this gene
B0D95_14110
type VI secretion protein VgrG
Accession: AQT61102
Location: 3237012-3238802
NCBI BlastP on this gene
Accession: AQT61102
Location: 3237012-3238802
NCBI BlastP on this gene
B0D95_14115
311. : CP000934 Cellvibrio japonicus Ueda107 Total score: 1.0 Cumulative Blast bit score: 143
fimC
putative type 1 pili pilus subunit
Accession: ACE82822
Location: 1493194-1493724
NCBI BlastP on this gene
Accession: ACE82822
Location: 1493194-1493724
NCBI BlastP on this gene
CJA_1285
Spore Coat Protein U domain protein
Accession: ACE84644
Location: 1492628-1493170
NCBI BlastP on this gene
Accession: ACE84644
Location: 1492628-1493170
NCBI BlastP on this gene
CJA_1284
CJA_1283
CJA_1282
CJA_1281
CJA_1280
CJA_1279
CJA_1278
pectate lyase III
Accession: ACE86187
Location: 1485468-1486994
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
Accession: ACE86187
Location: 1485468-1486994
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 143
Sequence coverage: 13 %
E-value: 8e-32
NCBI BlastP on this gene
pelC
CJA_1275
CJA_1276
CJA_1274
conserved hypothetical protein
Accession: ACE84224
Location: 1483279-1483758
NCBI BlastP on this gene
Accession: ACE84224
Location: 1483279-1483758
NCBI BlastP on this gene
CJA_1273
probable NAGC-like transcription regulator yajF
Accession: ACE85345
Location: 1482291-1483211
NCBI BlastP on this gene
Accession: ACE85345
Location: 1482291-1483211
NCBI BlastP on this gene
CJA_1272
conserved hypothetical protein
Accession: ACE86214
Location: 1481685-1482062
NCBI BlastP on this gene
Accession: ACE86214
Location: 1481685-1482062
NCBI BlastP on this gene
CJA_1271
CJA_1270
conserved hypothetical protein
Accession: ACE85956
Location: 1480415-1481272
NCBI BlastP on this gene
Accession: ACE85956
Location: 1480415-1481272
NCBI BlastP on this gene
CJA_1268
radA
CJA_1267
312. : AM746676 Sorangium cellulosum 'So ce 56' complete genome. Total score: 1.0 Cumulative Blast bit score: 143
Probable pectate lyase precursor
Accession: CAN95995
Location: 8188518-8190302
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 143
Sequence coverage: 63 %
E-value: 6e-34
NCBI BlastP on this gene
Accession: CAN95995
Location: 8188518-8190302
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 143
Sequence coverage: 63 %
E-value: 6e-34
NCBI BlastP on this gene
sce5832
sugE
sce5830
sce5829
sce5828
sce5827
sce5826
sce5825
313. : CP042913 Bythopirellula goksoyri strain Pr1d chromosome. Total score: 1.0 Cumulative Blast bit score: 142
Pr1d_37640
Pr1d_37630
Pr1d_37620
Pr1d_37610
tetD
Pr1d_37590
Pr1d_37580
hypothetical protein
Accession: QEG36443
Location: 4508967-4510544
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 108 %
E-value: 1e-33
NCBI BlastP on this gene
Accession: QEG36443
Location: 4508967-4510544
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 142
Sequence coverage: 108 %
E-value: 1e-33
NCBI BlastP on this gene
Pr1d_37570
pelL_2
Pr1d_37550
Pr1d_37540
D-allose-binding periplasmic protein precursor
Accession: QEG36439
Location: 4501478-4502509
NCBI BlastP on this gene
Accession: QEG36439
Location: 4501478-4502509
NCBI BlastP on this gene
alsB_2
314. : CP002993 Streptomyces sp. SirexAA-E Total score: 1.0 Cumulative Blast bit score: 142
SACTE_1310
conserved hypothetical protein
Accession: AEN09229
Location: 1469659-1470360
NCBI BlastP on this gene
Accession: AEN09229
Location: 1469659-1470360
NCBI BlastP on this gene
SACTE_1309
RNA polymerase, sigma-24 subunit, ECF subfamily
Accession: AEN09228
Location: 1469087-1469659
NCBI BlastP on this gene
Accession: AEN09228
Location: 1469087-1469659
NCBI BlastP on this gene
SACTE_1308
penicillin-binding protein transpeptidase
Accession: AEN09227
Location: 1467366-1468817
NCBI BlastP on this gene
Accession: AEN09227
Location: 1467366-1468817
NCBI BlastP on this gene
SACTE_1307
GCN5-related N-acetyltransferase
Accession: AEN09226
Location: 1466780-1467259
NCBI BlastP on this gene
Accession: AEN09226
Location: 1466780-1467259
NCBI BlastP on this gene
SACTE_1306
transcriptional regulator, IclR family
Accession: AEN09225
Location: 1465981-1466775
NCBI BlastP on this gene
Accession: AEN09225
Location: 1465981-1466775
NCBI BlastP on this gene
SACTE_1305
SACTE_1304
protein of unknown function DUF1349
Accession: AEN09223
Location: 1463683-1464285
NCBI BlastP on this gene
Accession: AEN09223
Location: 1463683-1464285
NCBI BlastP on this gene
SACTE_1303
Pectate lyase/Amb allergen
Accession: AEN09222
Location: 1462500-1463474
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 142
Sequence coverage: 14 %
E-value: 1e-32
NCBI BlastP on this gene
Accession: AEN09222
Location: 1462500-1463474
BlastP hit with VTO45599.1
Percentage identity: 34 %
BlastP bit score: 142
Sequence coverage: 14 %
E-value: 1e-32
NCBI BlastP on this gene
SACTE_1302
serine/threonine protein kinase
Accession: AEN09221
Location: 1461363-1462157
NCBI BlastP on this gene
Accession: AEN09221
Location: 1461363-1462157
NCBI BlastP on this gene
SACTE_1301
SACTE_1299
SACTE_1298
transcriptional regulator, MarR family
Accession: AEN09218
Location: 1458601-1459188
NCBI BlastP on this gene
Accession: AEN09218
Location: 1458601-1459188
NCBI BlastP on this gene
SACTE_1297
12-oxophytodienoate reductase
Accession: AEN09217
Location: 1457350-1458417
NCBI BlastP on this gene
Accession: AEN09217
Location: 1457350-1458417
NCBI BlastP on this gene
SACTE_1296
transcriptional regulator, LacI family
Accession: AEN09216
Location: 1456302-1457336
NCBI BlastP on this gene
Accession: AEN09216
Location: 1456302-1457336
NCBI BlastP on this gene
SACTE_1295
binding-protein-dependent transport systems inner membrane component
Accession: AEN09215
Location: 1455399-1456256
NCBI BlastP on this gene
Accession: AEN09215
Location: 1455399-1456256
NCBI BlastP on this gene
SACTE_1294
binding-protein-dependent transport systems inner membrane component
Accession: AEN09214
Location: 1454476-1455402
NCBI BlastP on this gene
Accession: AEN09214
Location: 1454476-1455402
NCBI BlastP on this gene
SACTE_1293
315. : LT629740 Mucilaginibacter mallensis strain MP1X4 genome assembly, chromosome: I. Total score: 1.0 Cumulative Blast bit score: 140
SAMN05216490_2657
SAMN05216490_2658
SAMN05216490_2659
2-desacetyl-2-hydroxyethyl bacteriochlorophyllide A dehydrogenase
Accession: SDT17721
Location: 3216537-3217553
NCBI BlastP on this gene
Accession: SDT17721
Location: 3216537-3217553
NCBI BlastP on this gene
SAMN05216490_2660
hypothetical protein
Accession: SDT17775
Location: 3217813-3219468
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 140
Sequence coverage: 90 %
E-value: 6e-33
NCBI BlastP on this gene
Accession: SDT17775
Location: 3217813-3219468
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 140
Sequence coverage: 90 %
E-value: 6e-33
NCBI BlastP on this gene
SAMN05216490_2661
316. : CP036525 Planctomycetes bacterium K22_7 chromosome Total score: 1.0 Cumulative Blast bit score: 140
K227x_60210
Serine/threonine-protein kinase PrkC
Accession: QDT07594
Location: 7964682-7967261
NCBI BlastP on this gene
Accession: QDT07594
Location: 7964682-7967261
NCBI BlastP on this gene
prkC_24
ECF RNA polymerase sigma-E factor
Accession: QDT07595
Location: 7967261-7967938
NCBI BlastP on this gene
Accession: QDT07595
Location: 7967261-7967938
NCBI BlastP on this gene
rpoE_6
K227x_60240
K227x_60250
hypothetical protein
Accession: QDT07598
Location: 7971214-7972689
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 140
Sequence coverage: 68 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: QDT07598
Location: 7971214-7972689
BlastP hit with VTO45597.1
Percentage identity: 33 %
BlastP bit score: 140
Sequence coverage: 68 %
E-value: 2e-33
NCBI BlastP on this gene
K227x_60260
317. : CP020991 Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 140
B9O19_01124
B9O19_01125
ribose 5-phosphate isomerase B
Accession: AUO19289
Location: 1277606-1278046
NCBI BlastP on this gene
Accession: AUO19289
Location: 1277606-1278046
NCBI BlastP on this gene
B9O19_01126
B9O19_01127
B9O19_01128
pectate lyase family 1
Accession: AUO19292
Location: 1283067-1286006
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 63 %
E-value: 5e-32
NCBI BlastP on this gene
Accession: AUO19292
Location: 1283067-1286006
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 63 %
E-value: 5e-32
NCBI BlastP on this gene
B9O19_01129
metallo-beta-lactamase family protein
Accession: AUO19293
Location: 1287427-1288278
NCBI BlastP on this gene
Accession: AUO19293
Location: 1287427-1288278
NCBI BlastP on this gene
B9O19_01130
B9O19_01131
cell wall repeat-containing protein
Accession: AUO19295
Location: 1289360-1292212
NCBI BlastP on this gene
Accession: AUO19295
Location: 1289360-1292212
NCBI BlastP on this gene
pmpB
transcriptional regulator,TetR family
Accession: AUO19296
Location: 1292343-1292999
NCBI BlastP on this gene
Accession: AUO19296
Location: 1292343-1292999
NCBI BlastP on this gene
B9O19_01133
318. : CP012672 Sorangium cellulosum strain So ce836 chromosome Total score: 1.0 Cumulative Blast bit score: 140
pectate lyase
Accession: AUX35189
Location: 9964704-9966125
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 64 %
E-value: 2e-33
NCBI BlastP on this gene
Accession: AUX35189
Location: 9964704-9966125
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 140
Sequence coverage: 64 %
E-value: 2e-33
NCBI BlastP on this gene
pel
SOCE836_073770
SOCE836_073760
SAM-dependent methyltransferase
Accession: AUX35186
Location: 9962227-9962991
NCBI BlastP on this gene
Accession: AUX35186
Location: 9962227-9962991
NCBI BlastP on this gene
smtA
SOCE836_073740
SOCE836_073730
SOCE836_073720
SOCE836_073710
319. : CP049143 Roseimicrobium sp. ORNL1 chromosome Total score: 1.0 Cumulative Blast bit score: 139
G5S37_30630
G5S37_30635
G5S37_30640
tRNA uridine-5-carboxymethylaminomethyl(34) synthesis enzyme MnmG
Accession: QIF05675
Location: 7571327-7573225
NCBI BlastP on this gene
Accession: QIF05675
Location: 7571327-7573225
NCBI BlastP on this gene
mnmG
DUF1801 domain-containing protein
Accession: QIF05676
Location: 7573466-7573858
NCBI BlastP on this gene
Accession: QIF05676
Location: 7573466-7573858
NCBI BlastP on this gene
G5S37_30650
metallopeptidase family protein
Accession: QIF05677
Location: 7573869-7574240
NCBI BlastP on this gene
Accession: QIF05677
Location: 7573869-7574240
NCBI BlastP on this gene
G5S37_30655
hypothetical protein
Accession: QIF05678
Location: 7574328-7575779
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 139
Sequence coverage: 76 %
E-value: 9e-33
NCBI BlastP on this gene
Accession: QIF05678
Location: 7574328-7575779
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 139
Sequence coverage: 76 %
E-value: 9e-33
NCBI BlastP on this gene
G5S37_30660
320. : CP036349 Planctomycetes bacterium Spa11 chromosome. Total score: 1.0 Cumulative Blast bit score: 138
Spa11_01540
Spa11_01530
Spa11_01520
pelL_1
Pectate lyase
Accession: QDV71981
Location: 173272-175455
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 68 %
E-value: 9e-32
NCBI BlastP on this gene
Accession: QDV71981
Location: 173272-175455
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 68 %
E-value: 9e-32
NCBI BlastP on this gene
Spa11_01500
Spa11_01490
Spa11_01480
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV71978
Location: 170372-170866
NCBI BlastP on this gene
Accession: QDV71978
Location: 170372-170866
NCBI BlastP on this gene
fabZ_2
acpP_1
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV71976
Location: 169265-169822
NCBI BlastP on this gene
Accession: QDV71976
Location: 169265-169822
NCBI BlastP on this gene
fabZ_1
3-oxoacyl-[acyl-carrier-protein] synthase 2
Accession: QDV71975
Location: 167738-169009
NCBI BlastP on this gene
Accession: QDV71975
Location: 167738-169009
NCBI BlastP on this gene
fabF_1
pgl_1
Spa11_01420
321. : CP029255 Flavobacterium crocinum strain HYN0056 chromosome Total score: 1.0 Cumulative Blast bit score: 138
HYN56_13410
HYN56_13405
HYN56_13400
3-phosphoserine/phosphohydroxythreonine transaminase
Accession: AWK05172
Location: 2989808-2990878
NCBI BlastP on this gene
Accession: AWK05172
Location: 2989808-2990878
NCBI BlastP on this gene
HYN56_13395
3-phosphoglycerate dehydrogenase
Accession: AWK07434
Location: 2988761-2989711
NCBI BlastP on this gene
Accession: AWK07434
Location: 2988761-2989711
NCBI BlastP on this gene
HYN56_13390
HYN56_13385
HYN56_13380
HYN56_13375
pectate lyase
Accession: AWK07433
Location: 2983853-2986600
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 138
Sequence coverage: 87 %
E-value: 2e-31
NCBI BlastP on this gene
Accession: AWK07433
Location: 2983853-2986600
BlastP hit with VTO45597.1
Percentage identity: 31 %
BlastP bit score: 138
Sequence coverage: 87 %
E-value: 2e-31
NCBI BlastP on this gene
HYN56_13370
HYN56_13365
ABC transporter ATP-binding protein
Accession: AWK05167
Location: 2982224-2983102
NCBI BlastP on this gene
Accession: AWK05167
Location: 2982224-2983102
NCBI BlastP on this gene
HYN56_13360
HYN56_13355
HYN56_13350
HYN56_13345
HYN56_13340
HYN56_13335
322. : CP028159 Plantactinospora sp. BB1 chromosome Total score: 1.0 Cumulative Blast bit score: 138
C6W10_01930
C6W10_01925
C6W10_01920
TetR family transcriptional regulator
Accession: AVT35422
Location: 474725-475405
NCBI BlastP on this gene
Accession: AVT35422
Location: 474725-475405
NCBI BlastP on this gene
C6W10_01915
C6W10_01910
C6W10_01905
C6W10_01900
C6W10_01895
aminoglycoside phosphotransferase
Accession: AVT35418
Location: 470481-471299
NCBI BlastP on this gene
Accession: AVT35418
Location: 470481-471299
NCBI BlastP on this gene
C6W10_01890
pectate lyase
Accession: AVT41103
Location: 468731-469912
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
Accession: AVT41103
Location: 468731-469912
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
C6W10_01885
C6W10_01880
TetR family transcriptional regulator
Accession: AVT35416
Location: 467386-467976
NCBI BlastP on this gene
Accession: AVT35416
Location: 467386-467976
NCBI BlastP on this gene
C6W10_01875
C6W10_01870
C6W10_01865
C6W10_01860
FAD-binding molybdopterin dehydrogenase
Accession: AVT35412
Location: 461701-462678
NCBI BlastP on this gene
Accession: AVT35412
Location: 461701-462678
NCBI BlastP on this gene
C6W10_01855
C6W10_01850
323. : CP028158 Plantactinospora sp. BC1 chromosome Total score: 1.0 Cumulative Blast bit score: 138
C6361_05390
C6361_05385
C6361_05380
TetR family transcriptional regulator
Accession: AVT29017
Location: 1306163-1306843
NCBI BlastP on this gene
Accession: AVT29017
Location: 1306163-1306843
NCBI BlastP on this gene
C6361_05375
C6361_05370
C6361_05365
C6361_05360
C6361_05355
aminoglycoside phosphotransferase
Accession: AVT29013
Location: 1301895-1302713
NCBI BlastP on this gene
Accession: AVT29013
Location: 1301895-1302713
NCBI BlastP on this gene
C6361_05350
pectate lyase
Accession: AVT34190
Location: 1300144-1301325
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
Accession: AVT34190
Location: 1300144-1301325
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 58 %
E-value: 7e-33
NCBI BlastP on this gene
C6361_05345
C6361_05340
C6361_05335
TetR family transcriptional regulator
Accession: AVT29011
Location: 1298799-1299389
NCBI BlastP on this gene
Accession: AVT29011
Location: 1298799-1299389
NCBI BlastP on this gene
C6361_05330
C6361_05325
C6361_05320
carbon monoxide dehydrogenase
Accession: AVT29008
Location: 1294145-1296361
NCBI BlastP on this gene
Accession: AVT29008
Location: 1294145-1296361
NCBI BlastP on this gene
C6361_05315
FAD-binding molybdopterin dehydrogenase
Accession: AVT29007
Location: 1293171-1294148
NCBI BlastP on this gene
Accession: AVT29007
Location: 1293171-1294148
NCBI BlastP on this gene
C6361_05310
C6361_05305
324. : CP020991 Monoglobus pectinilyticus strain 14 chromosome. Total score: 1.0 Cumulative Blast bit score: 138
B9O19_01438
hybrid sensor histidine kinase/response regulator
Accession: AUO19600
Location: 1669649-1672846
NCBI BlastP on this gene
Accession: AUO19600
Location: 1669649-1672846
NCBI BlastP on this gene
B9O19_01439
B9O19_01440
B9O19_01441
pectate lyase family 1
Accession: AUO19603
Location: 1674376-1677705
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 53 %
E-value: 2e-31
NCBI BlastP on this gene
Accession: AUO19603
Location: 1674376-1677705
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 138
Sequence coverage: 53 %
E-value: 2e-31
NCBI BlastP on this gene
B9O19_01442
B9O19_01443
GCN5-related N-acetyltransferase
Accession: AUO19605
Location: 1682734-1683315
NCBI BlastP on this gene
Accession: AUO19605
Location: 1682734-1683315
NCBI BlastP on this gene
B9O19_01444
DNA-directed RNA polymerase, beta subunit
Accession: AUO19606
Location: 1683684-1687511
NCBI BlastP on this gene
Accession: AUO19606
Location: 1683684-1687511
NCBI BlastP on this gene
B9O19_01445
325. : CP036350 Planctomycetes bacterium K2D chromosome Total score: 1.0 Cumulative Blast bit score: 137
K2D_01040
K2D_01030
K2D_01020
pelL_1
Pectate lyase
Accession: QDV76522
Location: 117208-119391
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 137
Sequence coverage: 68 %
E-value: 1e-31
NCBI BlastP on this gene
Accession: QDV76522
Location: 117208-119391
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 137
Sequence coverage: 68 %
E-value: 1e-31
NCBI BlastP on this gene
K2D_01000
K2D_00990
K2D_00980
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV76519
Location: 114315-114809
NCBI BlastP on this gene
Accession: QDV76519
Location: 114315-114809
NCBI BlastP on this gene
fabZ_2
acpP_1
3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ
Accession: QDV76517
Location: 113208-113765
NCBI BlastP on this gene
Accession: QDV76517
Location: 113208-113765
NCBI BlastP on this gene
fabZ_1
3-oxoacyl-[acyl-carrier-protein] synthase 2
Accession: QDV76516
Location: 111681-112952
NCBI BlastP on this gene
Accession: QDV76516
Location: 111681-112952
NCBI BlastP on this gene
fabF_1
pgl_1
K2D_00920
326. : CP041253 Echinicola sp. LN3S3 chromosome Total score: 1.0 Cumulative Blast bit score: 133
DUF5108 domain-containing protein
Accession: QDH80551
Location: 4224053-4225654
NCBI BlastP on this gene
Accession: QDH80551
Location: 4224053-4225654
NCBI BlastP on this gene
FKX85_16485
fasciclin domain-containing protein
Accession: QDH80550
Location: 4222452-4224041
NCBI BlastP on this gene
Accession: QDH80550
Location: 4222452-4224041
NCBI BlastP on this gene
FKX85_16480
SusC/RagA family TonB-linked outer membrane protein
Accession: QDH80549
Location: 4219345-4222446
NCBI BlastP on this gene
Accession: QDH80549
Location: 4219345-4222446
NCBI BlastP on this gene
FKX85_16475
RagB/SusD family nutrient uptake outer membrane protein
Accession: QDH81607
Location: 4217699-4219333
NCBI BlastP on this gene
Accession: QDH81607
Location: 4217699-4219333
NCBI BlastP on this gene
FKX85_16470
pectate lyase
Accession: QDH80548
Location: 4215703-4216980
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 133
Sequence coverage: 63 %
E-value: 5e-31
NCBI BlastP on this gene
Accession: QDH80548
Location: 4215703-4216980
BlastP hit with VTO45597.1
Percentage identity: 35 %
BlastP bit score: 133
Sequence coverage: 63 %
E-value: 5e-31
NCBI BlastP on this gene
FKX85_16465
FKX85_16460
glycoside hydrolase family 88 protein
Accession: QDH80546
Location: 4212092-4213276
NCBI BlastP on this gene
Accession: QDH80546
Location: 4212092-4213276
NCBI BlastP on this gene
FKX85_16455
glycoside hydrolase family 28 protein
Accession: QDH80545
Location: 4210390-4211814
NCBI BlastP on this gene
Accession: QDH80545
Location: 4210390-4211814
NCBI BlastP on this gene
FKX85_16450
FKX85_16445
T9SS type A sorting domain-containing protein
Accession: QDH80543
Location: 4204275-4209011
NCBI BlastP on this gene
Accession: QDH80543
Location: 4204275-4209011
NCBI BlastP on this gene
FKX85_16440
327. : CP030041 Echinicola strongylocentroti strain MEBiC08714 chromosome Total score: 1.0 Cumulative Blast bit score: 132
DN752_23955
fasciclin domain-containing protein
Accession: AWW33336
Location: 6149045-6150634
NCBI BlastP on this gene
Accession: AWW33336
Location: 6149045-6150634
NCBI BlastP on this gene
DN752_23960
SusC/RagA family TonB-linked outer membrane protein
Accession: AWW33337
Location: 6150640-6153741
NCBI BlastP on this gene
Accession: AWW33337
Location: 6150640-6153741
NCBI BlastP on this gene
DN752_23965
DN752_23970
pectate lyase
Accession: AWW32936
Location: 6155953-6157230
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 132
Sequence coverage: 62 %
E-value: 9e-31
NCBI BlastP on this gene
Accession: AWW32936
Location: 6155953-6157230
BlastP hit with VTO45597.1
Percentage identity: 36 %
BlastP bit score: 132
Sequence coverage: 62 %
E-value: 9e-31
NCBI BlastP on this gene
DN752_23975
328. : CP031010 Alteromonas sp. RKMC-009 chromosome Total score: 1.0 Cumulative Blast bit score: 101
Tat pathway signal sequence domain protein
Accession: AYA65921
Location: 4295818-4298523
NCBI BlastP on this gene
Accession: AYA65921
Location: 4295818-4298523
NCBI BlastP on this gene
DS731_18945
glycoside hydrolase family 105 protein
Accession: AYA66676
Location: 4298793-4299899
NCBI BlastP on this gene
Accession: AYA66676
Location: 4298793-4299899
NCBI BlastP on this gene
DS731_18950
DS731_18955
DS731_18960
DS731_18965
IclR family transcriptional regulator
Accession: AYA65924
Location: 4303653-4304426
BlastP hit with VTO45596.1
Percentage identity: 33 %
BlastP bit score: 101
Sequence coverage: 88 %
E-value: 4e-22
NCBI BlastP on this gene
Accession: AYA65924
Location: 4303653-4304426
BlastP hit with VTO45596.1
Percentage identity: 33 %
BlastP bit score: 101
Sequence coverage: 88 %
E-value: 4e-22
NCBI BlastP on this gene
DS731_18970
DS731_18975
DeoR/GlpR transcriptional regulator
Accession: AYA65926
Location: 4305496-4306284
NCBI BlastP on this gene
Accession: AYA65926
Location: 4305496-4306284
NCBI BlastP on this gene
DS731_18980
DS731_18985
DS731_18990
formimidoylglutamate deiminase
Accession: AYA65929
Location: 4311710-4313086
NCBI BlastP on this gene
Accession: AYA65929
Location: 4311710-4313086
NCBI BlastP on this gene
DS731_18995
329. : AP021859 Alteromonas sp. I4 DNA Total score: 1.0 Cumulative Blast bit score: 77
IclR family transcriptional regulator
Accession: BBO29965
Location: 4997441-4998136
BlastP hit with VTO45596.1
Percentage identity: 32 %
BlastP bit score: 77
Sequence coverage: 47 %
E-value: 1e-13
NCBI BlastP on this gene
Accession: BBO29965
Location: 4997441-4998136
BlastP hit with VTO45596.1
Percentage identity: 32 %
BlastP bit score: 77
Sequence coverage: 47 %
E-value: 1e-13
NCBI BlastP on this gene
AltI4_43530
2-dehydro-3-deoxygluconokinase
Accession: BBO29964
Location: 4996474-4997427
NCBI BlastP on this gene
Accession: BBO29964
Location: 4996474-4997427
NCBI BlastP on this gene
kdgK_2
AltI4_43510
glycerol-3-phosphate acyltransferase
Accession: BBO29962
Location: 4993502-4995985
NCBI BlastP on this gene
Accession: BBO29962
Location: 4993502-4995985
NCBI BlastP on this gene
plsB
AltI4_43490
AltI4_43480
AltI4_43470
AltI4_43460
GGDEF domain-containing protein
Accession: BBO29957
Location: 4988299-4989600
NCBI BlastP on this gene
Accession: BBO29957
Location: 4988299-4989600
NCBI BlastP on this gene
AltI4_43450
330. : CP022434 Nocardiopsis dassonvillei strain HZNU_N_1 chromosome Total score: 1.0 Cumulative Blast bit score: 61
CGQ36_18510
formimidoylglutamate deiminase
Accession: ASU61355
Location: 4447874-4449385
NCBI BlastP on this gene
Accession: ASU61355
Location: 4447874-4449385
NCBI BlastP on this gene
CGQ36_18515
CGQ36_18520
CGQ36_18525
hutH
IclR family transcriptional regulator
Accession: ASU59440
Location: 4454155-4454916
BlastP hit with VTO45596.1
Percentage identity: 31 %
BlastP bit score: 61
Sequence coverage: 61 %
E-value: 7e-08
NCBI BlastP on this gene
Accession: ASU59440
Location: 4454155-4454916
BlastP hit with VTO45596.1
Percentage identity: 31 %
BlastP bit score: 61
Sequence coverage: 61 %
E-value: 7e-08
NCBI BlastP on this gene
CGQ36_18535
CGQ36_18540


Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.