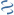MultiGeneBlast hits

Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP019797 : Rhodothermus marinus AA3-38 DNA Total score: 1.0 Cumulative Blast bit score: 599
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
RmaAA338_05190
dihydrodipicolinate synthase family protein
Accession: BBM71653
Location: 579420-580337
NCBI BlastP on this gene
Accession: BBM71653
Location: 579420-580337
NCBI BlastP on this gene
RmaAA338_05180
RmaAA338_05170
galactose-1-phosphate uridylyltransferase
Accession: BBM71651
Location: 577667-578722
NCBI BlastP on this gene
Accession: BBM71651
Location: 577667-578722
NCBI BlastP on this gene
RmaAA338_05160
yidK
galK_1
hypothetical protein
Accession: BBM71648
Location: 572338-574851
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBM71648
Location: 572338-574851
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RmaAA338_05130
RmaAA338_05120
RmaAA338_05110
RmaAA338_05100
RmaAA338_05090
RmaAA338_05080
RmaAA338_05070
copA_1
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP019796 : Rhodothermus marinus AA2-13 DNA Total score: 1.0 Cumulative Blast bit score: 599
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
RmaAA213_05210
dihydrodipicolinate synthase family protein
Accession: BBM68674
Location: 577195-578112
NCBI BlastP on this gene
Accession: BBM68674
Location: 577195-578112
NCBI BlastP on this gene
RmaAA213_05200
RmaAA213_05190
galactose-1-phosphate uridylyltransferase
Accession: BBM68672
Location: 575442-576497
NCBI BlastP on this gene
Accession: BBM68672
Location: 575442-576497
NCBI BlastP on this gene
RmaAA213_05180
yidK
galK_1
hypothetical protein
Accession: BBM68669
Location: 570113-572626
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBM68669
Location: 570113-572626
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RmaAA213_05150
RmaAA213_05140
RmaAA213_05130
RmaAA213_05120
CRISPR-associated endonuclease Cas1
Accession: BBM68665
Location: 566446-567339
NCBI BlastP on this gene
Accession: BBM68665
Location: 566446-567339
NCBI BlastP on this gene
cas1
RmaAA213_05100
type I-E CRISPR-associated protein Cas5/CasD
Accession: BBM68663
Location: 564880-565641
NCBI BlastP on this gene
Accession: BBM68663
Location: 564880-565641
NCBI BlastP on this gene
RmaAA213_05090
type I-E CRISPR-associated protein
Accession: BBM68662
Location: 563612-564883
NCBI BlastP on this gene
Accession: BBM68662
Location: 563612-564883
NCBI BlastP on this gene
RmaAA213_05080
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP043306 : Cellvibrio japonicus strain ADPT1-KOJIBIOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 588
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FY115_02380
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI18344
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: QEI18344
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
FY115_02375
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI18343
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QEI18343
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
FY115_02370
sulfite exporter TauE/SafE family protein
Accession: QEI18342
Location: 595510-596292
NCBI BlastP on this gene
Accession: QEI18342
Location: 595510-596292
NCBI BlastP on this gene
FY115_02365
FY115_02360
twin-arginine translocase subunit TatC
Accession: QEI18340
Location: 594225-595040
NCBI BlastP on this gene
Accession: QEI18340
Location: 594225-595040
NCBI BlastP on this gene
tatC
DUF839 domain-containing protein
Accession: QEI18339
Location: 592703-594100
NCBI BlastP on this gene
Accession: QEI18339
Location: 592703-594100
NCBI BlastP on this gene
FY115_02350
PEP-CTERM sorting domain-containing protein
Accession: QEI18338
Location: 592119-592676
NCBI BlastP on this gene
Accession: QEI18338
Location: 592119-592676
NCBI BlastP on this gene
FY115_02345
twin-arginine translocase TatA/TatE family subunit
Accession: QEI18337
Location: 591697-591900
NCBI BlastP on this gene
Accession: QEI18337
Location: 591697-591900
NCBI BlastP on this gene
tatA
glutathione S-transferase family protein
Accession: QEI18336
Location: 590945-591628
NCBI BlastP on this gene
Accession: QEI18336
Location: 590945-591628
NCBI BlastP on this gene
FY115_02335
type II/IV secretion system protein
Accession: QEI18335
Location: 589082-590857
NCBI BlastP on this gene
Accession: QEI18335
Location: 589082-590857
NCBI BlastP on this gene
FY115_02330
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP043305 : Cellvibrio japonicus strain ADPT2-NIGEROSE chromosome. Total score: 1.0 Cumulative Blast bit score: 588
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FY116_02380
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI14764
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: QEI14764
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
FY116_02375
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI14763
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QEI14763
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
FY116_02370
sulfite exporter TauE/SafE family protein
Accession: QEI14762
Location: 595510-596292
NCBI BlastP on this gene
Accession: QEI14762
Location: 595510-596292
NCBI BlastP on this gene
FY116_02365
FY116_02360
twin-arginine translocase subunit TatC
Accession: QEI14760
Location: 594225-595040
NCBI BlastP on this gene
Accession: QEI14760
Location: 594225-595040
NCBI BlastP on this gene
tatC
DUF839 domain-containing protein
Accession: QEI14759
Location: 592703-594100
NCBI BlastP on this gene
Accession: QEI14759
Location: 592703-594100
NCBI BlastP on this gene
FY116_02350
PEP-CTERM sorting domain-containing protein
Accession: QEI14758
Location: 592119-592676
NCBI BlastP on this gene
Accession: QEI14758
Location: 592119-592676
NCBI BlastP on this gene
FY116_02345
twin-arginine translocase TatA/TatE family subunit
Accession: QEI14757
Location: 591697-591900
NCBI BlastP on this gene
Accession: QEI14757
Location: 591697-591900
NCBI BlastP on this gene
tatA
glutathione S-transferase family protein
Accession: QEI14756
Location: 590945-591628
NCBI BlastP on this gene
Accession: QEI14756
Location: 590945-591628
NCBI BlastP on this gene
FY116_02335
type II/IV secretion system protein
Accession: QEI14755
Location: 589082-590857
NCBI BlastP on this gene
Accession: QEI14755
Location: 589082-590857
NCBI BlastP on this gene
FY116_02330
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP043304 : Cellvibrio japonicus strain ADPT3-ISOMALTOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 588
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FY117_02380
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI11190
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: QEI11190
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
FY117_02375
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI11189
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QEI11189
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
FY117_02370
sulfite exporter TauE/SafE family protein
Accession: QEI11188
Location: 595510-596292
NCBI BlastP on this gene
Accession: QEI11188
Location: 595510-596292
NCBI BlastP on this gene
FY117_02365
FY117_02360
twin-arginine translocase subunit TatC
Accession: QEI11186
Location: 594225-595040
NCBI BlastP on this gene
Accession: QEI11186
Location: 594225-595040
NCBI BlastP on this gene
tatC
DUF839 domain-containing protein
Accession: QEI11185
Location: 592703-594100
NCBI BlastP on this gene
Accession: QEI11185
Location: 592703-594100
NCBI BlastP on this gene
FY117_02350
PEP-CTERM sorting domain-containing protein
Accession: QEI11184
Location: 592119-592676
NCBI BlastP on this gene
Accession: QEI11184
Location: 592119-592676
NCBI BlastP on this gene
FY117_02345
twin-arginine translocase TatA/TatE family subunit
Accession: QEI11183
Location: 591697-591900
NCBI BlastP on this gene
Accession: QEI11183
Location: 591697-591900
NCBI BlastP on this gene
tatA
glutathione S-transferase family protein
Accession: QEI11182
Location: 590945-591628
NCBI BlastP on this gene
Accession: QEI11182
Location: 590945-591628
NCBI BlastP on this gene
FY117_02335
type II/IV secretion system protein
Accession: QEI11181
Location: 589082-590857
NCBI BlastP on this gene
Accession: QEI11181
Location: 589082-590857
NCBI BlastP on this gene
FY117_02330
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP000934 : Cellvibrio japonicus Ueda107 Total score: 1.0 Cumulative Blast bit score: 587
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
carbohydrate binding protein, cbp35C
Accession: ACE84402
Location: 599129-606433
NCBI BlastP on this gene
Accession: ACE84402
Location: 599129-606433
NCBI BlastP on this gene
cbp35C
CJA_0493
arabinogalactan endo-1,4-beta-galactosidase, putative, gal53B
Accession: ACE86078
Location: 597653-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 299
Sequence coverage: 87 %
E-value: 3e-94
NCBI BlastP on this gene
Accession: ACE86078
Location: 597653-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 299
Sequence coverage: 87 %
E-value: 3e-94
NCBI BlastP on this gene
gal53B
arabinogalactan endo-1,4-beta-galactosidase, putative, gal53A
Accession: ACE85511
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: ACE85511
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
gal53A-1
CJA_0490
CJA_0489
Sec-independent protein translocase TatC
Accession: ACE85846
Location: 594225-595040
NCBI BlastP on this gene
Accession: ACE85846
Location: 594225-595040
NCBI BlastP on this gene
CJA_0488
Tat (twin-arginine translocation) pathway signal sequence domain protein
Accession: ACE83245
Location: 592703-594100
NCBI BlastP on this gene
Accession: ACE83245
Location: 592703-594100
NCBI BlastP on this gene
CJA_0487
PEP-CTERM putative exosortase interaction domain protein
Accession: ACE84061
Location: 592119-592676
NCBI BlastP on this gene
Accession: ACE84061
Location: 592119-592676
NCBI BlastP on this gene
CJA_0486
twin arginine-targeting protein translocase,
Accession: ACE85402
Location: 591691-591900
NCBI BlastP on this gene
Accession: ACE85402
Location: 591691-591900
NCBI BlastP on this gene
CJA_0485
putative glutathione S-transferase
Accession: ACE85636
Location: 590945-591628
NCBI BlastP on this gene
Accession: ACE85636
Location: 590945-591628
NCBI BlastP on this gene
CJA_0484
type IV pilus biogenesis protein
Accession: ACE86015
Location: 589058-590857
NCBI BlastP on this gene
Accession: ACE86015
Location: 589058-590857
NCBI BlastP on this gene
CJA_0483
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP039703 : Clostridium butyricum strain 29-1 plasmid p-butyl_plas_29-1 Total score: 1.0 Cumulative Blast bit score: 572
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FBD77_19750
response regulator transcription factor
Accession: QCJ04649
Location: 594955-595665
NCBI BlastP on this gene
Accession: QCJ04649
Location: 594955-595665
NCBI BlastP on this gene
FBD77_19745
FtsX-like permease family protein
Accession: FBD77_19740
Location: 592190-594783
NCBI BlastP on this gene
Accession: FBD77_19740
Location: 592190-594783
NCBI BlastP on this gene
FBD77_19740
ABC transporter ATP-binding protein
Accession: QCJ04648
Location: 591504-592190
NCBI BlastP on this gene
Accession: QCJ04648
Location: 591504-592190
NCBI BlastP on this gene
FBD77_19735
HAMP domain-containing histidine kinase
Accession: QCJ04647
Location: 590431-591420
NCBI BlastP on this gene
Accession: QCJ04647
Location: 590431-591420
NCBI BlastP on this gene
FBD77_19730
response regulator transcription factor
Accession: QCJ04646
Location: 589758-590444
NCBI BlastP on this gene
Accession: QCJ04646
Location: 589758-590444
NCBI BlastP on this gene
FBD77_19725
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ04645
Location: 588228-589253
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: QCJ04645
Location: 588228-589253
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
FBD77_19720
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ04644
Location: 586817-587815
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: QCJ04644
Location: 586817-587815
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
FBD77_19715
FBD77_19710
FBD77_19705
DUF3502 domain-containing protein
Accession: QCJ04641
Location: 582723-584267
NCBI BlastP on this gene
Accession: QCJ04641
Location: 582723-584267
NCBI BlastP on this gene
FBD77_19700
DUF3502 domain-containing protein
Accession: QCJ04640
Location: 580948-582486
NCBI BlastP on this gene
Accession: QCJ04640
Location: 580948-582486
NCBI BlastP on this gene
FBD77_19695
carbohydrate ABC transporter permease
Accession: QCJ04639
Location: 579860-580750
NCBI BlastP on this gene
Accession: QCJ04639
Location: 579860-580750
NCBI BlastP on this gene
FBD77_19690
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP016333 : Clostridium butyricum strain TK520 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
BBB49_18970
two-component system response regulator
Accession: AOR96152
Location: 601479-602189
NCBI BlastP on this gene
Accession: AOR96152
Location: 601479-602189
NCBI BlastP on this gene
BBB49_18975
BBB49_18980
peptide ABC transporter ATP-binding protein
Accession: AOR96154
Location: 604955-605641
NCBI BlastP on this gene
Accession: AOR96154
Location: 604955-605641
NCBI BlastP on this gene
BBB49_18985
two-component sensor histidine kinase
Accession: AOR96155
Location: 605725-606714
NCBI BlastP on this gene
Accession: AOR96155
Location: 605725-606714
NCBI BlastP on this gene
BBB49_18990
BBB49_18995
arabinogalactan endo-1,4-beta-galactosidase
Accession: AOR96157
Location: 607894-608919
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: AOR96157
Location: 607894-608919
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
BBB49_19000
arabinogalactan endo-1,4-beta-galactosidase
Accession: AOR96340
Location: 609332-610330
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: AOR96340
Location: 609332-610330
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
BBB49_19005
BBB49_19010
BBB49_19015
BBB49_19020
BBB49_19025
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP014705 : Clostridium butyricum strain TOA chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
AZ909_19500
two-component system response regulator
Accession: ANF16241
Location: 642248-642958
NCBI BlastP on this gene
Accession: ANF16241
Location: 642248-642958
NCBI BlastP on this gene
AZ909_19505
AZ909_19510
peptide ABC transporter ATP-binding protein
Accession: ANF16243
Location: 645724-646410
NCBI BlastP on this gene
Accession: ANF16243
Location: 645724-646410
NCBI BlastP on this gene
AZ909_19515
two-component sensor histidine kinase
Accession: ANF16244
Location: 646494-647483
NCBI BlastP on this gene
Accession: ANF16244
Location: 646494-647483
NCBI BlastP on this gene
AZ909_19520
AZ909_19525
arabinogalactan endo-1,4-beta-galactosidase
Accession: ANF16246
Location: 648663-649688
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: ANF16246
Location: 648663-649688
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
AZ909_19530
arabinogalactan endo-1,4-beta-galactosidase
Accession: ANF16398
Location: 650101-651099
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: ANF16398
Location: 650101-651099
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
AZ909_19535
AZ909_19540
AZ909_19545
AZ909_19550
AZ909_19555
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP013489 : Clostridium butyricum strain KNU-L09 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
ATN24_17010
two-component system response regulator
Accession: ALR90169
Location: 6115-6825
NCBI BlastP on this gene
Accession: ALR90169
Location: 6115-6825
NCBI BlastP on this gene
ATN24_17015
ATN24_17020
peptide ABC transporter ATP-binding protein
Accession: ALR90171
Location: 9591-10277
NCBI BlastP on this gene
Accession: ALR90171
Location: 9591-10277
NCBI BlastP on this gene
ATN24_17025
ATN24_17030
two-component system response regulator
Accession: ALR90173
Location: 11337-12023
NCBI BlastP on this gene
Accession: ALR90173
Location: 11337-12023
NCBI BlastP on this gene
ATN24_17035
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALR90174
Location: 12530-13555
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: ALR90174
Location: 12530-13555
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
ATN24_17040
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALR90819
Location: 13968-14966
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: ALR90819
Location: 13968-14966
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
ATN24_17045
ATN24_17050
ATN24_17055
ATN24_17060
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP013353 : Clostridium butyricum strain JKY6D1 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
ATD26_19465
two-component system response regulator
Accession: ALS19054
Location: 573180-573890
NCBI BlastP on this gene
Accession: ALS19054
Location: 573180-573890
NCBI BlastP on this gene
ATD26_19470
ATD26_19475
peptide ABC transporter ATP-binding protein
Accession: ALS19056
Location: 576656-577342
NCBI BlastP on this gene
Accession: ALS19056
Location: 576656-577342
NCBI BlastP on this gene
ATD26_19480
ATD26_19485
two-component system response regulator
Accession: ALS19058
Location: 578402-579088
NCBI BlastP on this gene
Accession: ALS19058
Location: 578402-579088
NCBI BlastP on this gene
ATD26_19490
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALS19059
Location: 579595-580620
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: ALS19059
Location: 579595-580620
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
ATD26_19495
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALS19254
Location: 581033-582031
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: ALS19254
Location: 581033-582031
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
ATD26_19500
ATD26_19505
ATD26_19510
ATD26_19515
ATD26_19520
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP039706 : Clostridium butyricum strain 4-1 plasmid p-butyl_plas_4-1 Total score: 1.0 Cumulative Blast bit score: 571
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FBD76_18270
response regulator transcription factor
Accession: QCJ08290
Location: 130688-131398
NCBI BlastP on this gene
Accession: QCJ08290
Location: 130688-131398
NCBI BlastP on this gene
FBD76_18275
FBD76_18280
ABC transporter ATP-binding protein
Accession: QCJ08292
Location: 134164-134850
NCBI BlastP on this gene
Accession: QCJ08292
Location: 134164-134850
NCBI BlastP on this gene
FBD76_18285
HAMP domain-containing histidine kinase
Accession: QCJ08293
Location: 134934-135923
NCBI BlastP on this gene
Accession: QCJ08293
Location: 134934-135923
NCBI BlastP on this gene
FBD76_18290
response regulator transcription factor
Accession: QCJ08294
Location: 135910-136596
NCBI BlastP on this gene
Accession: QCJ08294
Location: 135910-136596
NCBI BlastP on this gene
FBD76_18295
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ08295
Location: 137102-138127
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
Accession: QCJ08295
Location: 137102-138127
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
FBD76_18300
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ08778
Location: 138540-139538
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: QCJ08778
Location: 138540-139538
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
FBD76_18305
FBD76_18310
extracellular solute-binding protein
Accession: QCJ08297
Location: 142088-143632
NCBI BlastP on this gene
Accession: QCJ08297
Location: 142088-143632
NCBI BlastP on this gene
FBD76_18315
DUF3502 domain-containing protein
Accession: QCJ08298
Location: 143869-145407
NCBI BlastP on this gene
Accession: QCJ08298
Location: 143869-145407
NCBI BlastP on this gene
FBD76_18320
carbohydrate ABC transporter permease
Accession: QCJ08299
Location: 145605-146495
NCBI BlastP on this gene
Accession: QCJ08299
Location: 145605-146495
NCBI BlastP on this gene
FBD76_18325
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP030776 : Clostridium butyricum strain S-45-5 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 571
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
DRB99_19095
DRB99_19100
DRB99_19105
ABC transporter ATP-binding protein
Accession: AXB87024
Location: 402998-403684
NCBI BlastP on this gene
Accession: AXB87024
Location: 402998-403684
NCBI BlastP on this gene
DRB99_19110
DRB99_19115
DRB99_19120
arabinogalactan endo-1,4-beta-galactosidase
Accession: AXB87027
Location: 405936-406961
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
Accession: AXB87027
Location: 405936-406961
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
DRB99_19125
arabinogalactan endo-1,4-beta-galactosidase
Accession: AXB87306
Location: 407374-408372
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: AXB87306
Location: 407374-408372
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
DRB99_19130
DRB99_19135
extracellular solute-binding protein
Accession: AXB87029
Location: 410922-412466
NCBI BlastP on this gene
Accession: AXB87029
Location: 410922-412466
NCBI BlastP on this gene
DRB99_19140
DUF3502 domain-containing protein
Accession: AXB87030
Location: 412703-414241
NCBI BlastP on this gene
Accession: AXB87030
Location: 412703-414241
NCBI BlastP on this gene
DRB99_19145
carbohydrate ABC transporter permease
Accession: AXB87031
Location: 414439-415329
NCBI BlastP on this gene
Accession: AXB87031
Location: 414439-415329
NCBI BlastP on this gene
DRB99_19150
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP012643 : Rufibacter tibetensis strain 1351 Total score: 1.0 Cumulative Blast bit score: 521
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
DC20_01105
DC20_01110
DC20_01115
DC20_01120
SusC/RagA family TonB-linked outer membrane protein
Accession: ALI97834
Location: 297461-300445
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 521
Sequence coverage: 105 %
E-value: 2e-164
NCBI BlastP on this gene
Accession: ALI97834
Location: 297461-300445
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 521
Sequence coverage: 105 %
E-value: 2e-164
NCBI BlastP on this gene
DC20_01125
DC20_01130
DC20_01135
DC20_01140
DC20_01145
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP017422 : Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 520
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FLA_3189
FLA_3190
FLA_3191
rhamnogalacturonan acetylesterase
Accession: BAV07169
Location: 3897284-3899215
NCBI BlastP on this gene
Accession: BAV07169
Location: 3897284-3899215
NCBI BlastP on this gene
FLA_3192
FLA_3193
outer membrane protein SusC, starch binding
Accession: BAV07171
Location: 3900777-3904079
BlastP hit with EDO09628.1
Percentage identity: 36 %
BlastP bit score: 520
Sequence coverage: 102 %
E-value: 5e-163
NCBI BlastP on this gene
Accession: BAV07171
Location: 3900777-3904079
BlastP hit with EDO09628.1
Percentage identity: 36 %
BlastP bit score: 520
Sequence coverage: 102 %
E-value: 5e-163
NCBI BlastP on this gene
FLA_3194
FLA_3195
RNA polymerase ECF-type sigma factor
Accession: BAV07173
Location: 3905487-3906071
NCBI BlastP on this gene
Accession: BAV07173
Location: 3905487-3906071
NCBI BlastP on this gene
FLA_3196
FLA_3197
FLA_3198
transcriptional regulator, AraC family
Accession: BAV07176
Location: 3908412-3909401
NCBI BlastP on this gene
Accession: BAV07176
Location: 3908412-3909401
NCBI BlastP on this gene
FLA_3199
FLA_3200
FLA_3201
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP017774 : Flavobacterium commune strain PK15 chromosome Total score: 1.0 Cumulative Blast bit score: 506
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
SusC/RagA family TonB-linked outer membrane protein
Accession: AOZ99793
Location: 2446153-2449416
BlastP hit with EDO09628.1
Percentage identity: 35 %
BlastP bit score: 506
Sequence coverage: 103 %
E-value: 6e-158
NCBI BlastP on this gene
Accession: AOZ99793
Location: 2446153-2449416
BlastP hit with EDO09628.1
Percentage identity: 35 %
BlastP bit score: 506
Sequence coverage: 103 %
E-value: 6e-158
NCBI BlastP on this gene
BIW12_10290
BIW12_10285
BIW12_10280
BIW12_10275
BIW12_10270
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP018694 : Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 496
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
SusC, outer membrane protein
Accession: BBE18918
Location: 3446242-3449412
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 497
Sequence coverage: 101 %
E-value: 1e-154
NCBI BlastP on this gene
Accession: BBE18918
Location: 3446242-3449412
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 497
Sequence coverage: 101 %
E-value: 1e-154
NCBI BlastP on this gene
AQPE_3088
AQPE_3087
RNA polymerase ECF-type sigma factor
Accession: BBE18916
Location: 3444182-3444805
NCBI BlastP on this gene
Accession: BBE18916
Location: 3444182-3444805
NCBI BlastP on this gene
AQPE_3086
AQPE_3085
AQPE_3084
two-component system sensor histidine kinase
Accession: BBE18913
Location: 3440824-3442479
NCBI BlastP on this gene
Accession: BBE18913
Location: 3440824-3442479
NCBI BlastP on this gene
AQPE_3083
AQPE_3082
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP049857 : Dysgonomonas sp. HDW5A chromosome Total score: 1.0 Cumulative Blast bit score: 491
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
helix-turn-helix transcriptional regulator
Accession: QIK59404
Location: 1427691-1428116
NCBI BlastP on this gene
Accession: QIK59404
Location: 1427691-1428116
NCBI BlastP on this gene
G7050_05960
glycoside hydrolase family 97 protein
Accession: QIK59403
Location: 1425506-1427500
NCBI BlastP on this gene
Accession: QIK59403
Location: 1425506-1427500
NCBI BlastP on this gene
G7050_05955
G7050_05950
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIK59401
Location: 1422093-1423607
NCBI BlastP on this gene
Accession: QIK59401
Location: 1422093-1423607
NCBI BlastP on this gene
G7050_05945
TonB-dependent receptor
Accession: QIK59400
Location: 1419121-1422057
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 491
Sequence coverage: 102 %
E-value: 2e-153
NCBI BlastP on this gene
Accession: QIK59400
Location: 1419121-1422057
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 491
Sequence coverage: 102 %
E-value: 2e-153
NCBI BlastP on this gene
G7050_05940
G7050_05935
DUF5004 domain-containing protein
Accession: QIK59398
Location: 1417190-1417720
NCBI BlastP on this gene
Accession: QIK59398
Location: 1417190-1417720
NCBI BlastP on this gene
G7050_05930
G7050_05925
glycoside hydrolase family 97 protein
Accession: QIK59396
Location: 1412878-1414860
NCBI BlastP on this gene
Accession: QIK59396
Location: 1412878-1414860
NCBI BlastP on this gene
G7050_05920
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP049858 : Dysgonomonas sp. HDW5B chromosome Total score: 1.0 Cumulative Blast bit score: 488
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
helix-turn-helix transcriptional regulator
Accession: QIK53955
Location: 1519538-1519963
NCBI BlastP on this gene
Accession: QIK53955
Location: 1519538-1519963
NCBI BlastP on this gene
G7051_06225
glycoside hydrolase family 97 protein
Accession: QIK53954
Location: 1517444-1519438
NCBI BlastP on this gene
Accession: QIK53954
Location: 1517444-1519438
NCBI BlastP on this gene
G7051_06220
G7051_06215
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIK53952
Location: 1514152-1515666
NCBI BlastP on this gene
Accession: QIK53952
Location: 1514152-1515666
NCBI BlastP on this gene
G7051_06210
TonB-dependent receptor
Accession: QIK53951
Location: 1511180-1514116
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 488
Sequence coverage: 102 %
E-value: 5e-152
NCBI BlastP on this gene
Accession: QIK53951
Location: 1511180-1514116
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 488
Sequence coverage: 102 %
E-value: 5e-152
NCBI BlastP on this gene
G7051_06205
G7051_06200
DUF5004 domain-containing protein
Accession: QIK53949
Location: 1509278-1509808
NCBI BlastP on this gene
Accession: QIK53949
Location: 1509278-1509808
NCBI BlastP on this gene
G7051_06195
G7051_06190
glycoside hydrolase family 97 protein
Accession: QIK53947
Location: 1504966-1506948
NCBI BlastP on this gene
Accession: QIK53947
Location: 1504966-1506948
NCBI BlastP on this gene
G7051_06185
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP018694 : Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 488
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
AQPE_0220
AQPE_0219
AQPE_0218
AQPE_0217
SusC, outer membrane protein
Accession: BBE16079
Location: 257808-261026
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 488
Sequence coverage: 103 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: BBE16079
Location: 257808-261026
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 488
Sequence coverage: 103 %
E-value: 3e-151
NCBI BlastP on this gene
AQPE_0216
AQPE_0215
RNA polymerase ECF-type sigma factor
Accession: BBE16077
Location: 255774-256430
NCBI BlastP on this gene
Accession: BBE16077
Location: 255774-256430
NCBI BlastP on this gene
AQPE_0214
AQPE_0213
AQPE_0212
RNA polymerase ECF-type sigma factor
Accession: BBE16074
Location: 253457-254053
NCBI BlastP on this gene
Accession: BBE16074
Location: 253457-254053
NCBI BlastP on this gene
AQPE_0211
AQPE_0210
AQPE_0209
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP003557 : Melioribacter roseus P3M Total score: 1.0 Cumulative Blast bit score: 485
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
nicotinate-nucleotide pyrophosphorylase
Accession: AFN74194
Location: 1088098-1088952
NCBI BlastP on this gene
Accession: AFN74194
Location: 1088098-1088952
NCBI BlastP on this gene
MROS_0953
MROS_0954
MROS_0955
MROS_0956
phosphoribulokinase/uridine kinase
Accession: AFN74198
Location: 1091578-1092921
NCBI BlastP on this gene
Accession: AFN74198
Location: 1091578-1092921
NCBI BlastP on this gene
MROS_0957
MROS_0958
MROS_0959
Glycosyl hydrolase family 5
Accession: AFN74201
Location: 1094686-1097229
BlastP hit with EDO09630.1
Percentage identity: 34 %
BlastP bit score: 485
Sequence coverage: 100 %
E-value: 7e-154
NCBI BlastP on this gene
Accession: AFN74201
Location: 1094686-1097229
BlastP hit with EDO09630.1
Percentage identity: 34 %
BlastP bit score: 485
Sequence coverage: 100 %
E-value: 7e-154
NCBI BlastP on this gene
MROS_0960
arabinogalactan endo-1,4-beta-galactosidase
Accession: AFN74202
Location: 1097226-1099451
NCBI BlastP on this gene
Accession: AFN74202
Location: 1097226-1099451
NCBI BlastP on this gene
MROS_0961
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP050956 : Parabacteroides distasonis strain FDAARGOS_615 chromosome. Total score: 1.0 Cumulative Blast bit score: 483
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
FOB23_06895
FOB23_06890
galactosamine-6-phosphate isomerase
Accession: QIX64863
Location: 1618136-1618858
NCBI BlastP on this gene
Accession: QIX64863
Location: 1618136-1618858
NCBI BlastP on this gene
FOB23_06885
DUF1080 domain-containing protein
Accession: QIX64862
Location: 1617323-1618129
NCBI BlastP on this gene
Accession: QIX64862
Location: 1617323-1618129
NCBI BlastP on this gene
FOB23_06880
Gfo/Idh/MocA family oxidoreductase
Accession: QIX64861
Location: 1615680-1617224
NCBI BlastP on this gene
Accession: QIX64861
Location: 1615680-1617224
NCBI BlastP on this gene
FOB23_06875
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIX64860
Location: 1614039-1615607
NCBI BlastP on this gene
Accession: QIX64860
Location: 1614039-1615607
NCBI BlastP on this gene
FOB23_06870
TonB-dependent receptor
Accession: QIX64859
Location: 1610876-1614019
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 7e-150
NCBI BlastP on this gene
Accession: QIX64859
Location: 1610876-1614019
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 7e-150
NCBI BlastP on this gene
FOB23_06865
DUF4974 domain-containing protein
Accession: QIX64858
Location: 1609639-1610613
NCBI BlastP on this gene
Accession: QIX64858
Location: 1609639-1610613
NCBI BlastP on this gene
FOB23_06860
N-acetylmuramoyl-L-alanine amidase
Accession: QIX64857
Location: 1609098-1609568
NCBI BlastP on this gene
Accession: QIX64857
Location: 1609098-1609568
NCBI BlastP on this gene
FOB23_06855
FOB23_06850
FOB23_06845
FOB23_06840
FOB23_06835
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP034563 : Flammeovirga pectinis strain L12M1 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 483
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
TonB-dependent receptor
Accession: AZQ65029
Location: 604756-607773
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 484
Sequence coverage: 107 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: AZQ65029
Location: 604756-607773
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 484
Sequence coverage: 107 %
E-value: 2e-150
NCBI BlastP on this gene
EI427_22680
EI427_22675
hybrid sensor histidine kinase/response regulator
Accession: AZQ65027
Location: 598510-602772
NCBI BlastP on this gene
Accession: AZQ65027
Location: 598510-602772
NCBI BlastP on this gene
EI427_22670
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP019729 : Parabacteroides distasonis NBRC 113806 DNA Total score: 1.0 Cumulative Blast bit score: 483
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
DN0286_31590
DN0286_31580
glucosamine-6-phosphate deaminase
Accession: BBK92871
Location: 3824100-3824822
NCBI BlastP on this gene
Accession: BBK92871
Location: 3824100-3824822
NCBI BlastP on this gene
agaI
DN0286_31560
DN0286_31550
DN0286_31540
SusC/RagA family TonB-linked outer membrane protein
Accession: BBK92867
Location: 3816822-3819983
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 6e-150
NCBI BlastP on this gene
Accession: BBK92867
Location: 3816822-3819983
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 6e-150
NCBI BlastP on this gene
DN0286_31530
iron dicitrate transporter FecR
Accession: BBK92866
Location: 3815603-3816577
NCBI BlastP on this gene
Accession: BBK92866
Location: 3815603-3816577
NCBI BlastP on this gene
DN0286_31520
N-acetylmuramoyl-L-alanine amidase
Accession: BBK92865
Location: 3815062-3815532
NCBI BlastP on this gene
Accession: BBK92865
Location: 3815062-3815532
NCBI BlastP on this gene
DN0286_31510
DN0286_31500
DN0286_31490
DN0286_31480
DN0286_31470
DN0286_31460
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP040468 : Parabacteroides distasonis strain CavFT-hAR46 chromosome Total score: 1.0 Cumulative Blast bit score: 481
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
TonB-dependent receptor
Accession: QCY57978
Location: 4269079-4272222
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 481
Sequence coverage: 100 %
E-value: 1e-148
NCBI BlastP on this gene
Accession: QCY57978
Location: 4269079-4272222
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 481
Sequence coverage: 100 %
E-value: 1e-148
NCBI BlastP on this gene
FE931_18250
DUF4974 domain-containing protein
Accession: QCY57977
Location: 4267843-4268817
NCBI BlastP on this gene
Accession: QCY57977
Location: 4267843-4268817
NCBI BlastP on this gene
FE931_18245
N-acetylmuramoyl-L-alanine amidase
Accession: QCY57976
Location: 4267302-4267772
NCBI BlastP on this gene
Accession: QCY57976
Location: 4267302-4267772
NCBI BlastP on this gene
FE931_18240
FE931_18235
FE931_18230
FE931_18225
FE931_18220
FE931_18215
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP022754 : Parabacteroides sp. CT06 chromosome Total score: 1.0 Cumulative Blast bit score: 479
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
SusC/RagA family TonB-linked outer membrane protein
Accession: AST55569
Location: 4699591-4702734
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 2e-148
NCBI BlastP on this gene
Accession: AST55569
Location: 4699591-4702734
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 2e-148
NCBI BlastP on this gene
CI960_20600
CI960_20595
N-acetylmuramoyl-L-alanine amidase
Accession: AST55567
Location: 4697814-4698284
NCBI BlastP on this gene
Accession: AST55567
Location: 4697814-4698284
NCBI BlastP on this gene
CI960_20590
CI960_20585
CI960_20580
CI960_20575
CI960_20570
CI960_20565
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP000140 : Parabacteroides distasonis ATCC 8503 Total score: 1.0 Cumulative Blast bit score: 479
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
putative outer membrane protein, probably involved in nutrient binding
Accession: ABR44581
Location: 3420940-3424152
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 4e-148
NCBI BlastP on this gene
Accession: ABR44581
Location: 3420940-3424152
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 4e-148
NCBI BlastP on this gene
BDI_2873
BDI_2872
N-acetylmuramoyl-L-alanine amidase
Accession: ABR44579
Location: 3419232-3419702
NCBI BlastP on this gene
Accession: ABR44579
Location: 3419232-3419702
NCBI BlastP on this gene
BDI_2871
conserved hypothetical protein
Accession: ABR44578
Location: 3418801-3419235
NCBI BlastP on this gene
Accession: ABR44578
Location: 3418801-3419235
NCBI BlastP on this gene
BDI_2870
conserved hypothetical protein
Accession: ABR44577
Location: 3417769-3418398
NCBI BlastP on this gene
Accession: ABR44577
Location: 3417769-3418398
NCBI BlastP on this gene
BDI_2869
conserved hypothetical protein
Accession: ABR44576
Location: 3416466-3417704
NCBI BlastP on this gene
Accession: ABR44576
Location: 3416466-3417704
NCBI BlastP on this gene
BDI_2868
conserved hypothetical protein
Accession: ABR44575
Location: 3416064-3416285
NCBI BlastP on this gene
Accession: ABR44575
Location: 3416064-3416285
NCBI BlastP on this gene
BDI_2867
BDI_2866
conserved hypothetical protein
Accession: ABR44573
Location: 3415130-3415357
NCBI BlastP on this gene
Accession: ABR44573
Location: 3415130-3415357
NCBI BlastP on this gene
BDI_2865
BDI_2864
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP012801 : Bacteroides cellulosilyticus strain WH2 Total score: 1.0 Cumulative Blast bit score: 476
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
BcellWH2_03128
BcellWH2_03129
BcellWH2_03130
TonB dependent receptor
Accession: ALJ60368
Location: 4010059-4013079
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 476
Sequence coverage: 104 %
E-value: 2e-147
NCBI BlastP on this gene
Accession: ALJ60368
Location: 4010059-4013079
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 476
Sequence coverage: 104 %
E-value: 2e-147
NCBI BlastP on this gene
BcellWH2_03131
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP003560 : Flammeovirga sp. MY04 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 474
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
Ribonucleotide reductase of class Ia, beta subunit
Accession: ANQ48086
Location: 898308-899285
NCBI BlastP on this gene
Accession: ANQ48086
Location: 898308-899285
NCBI BlastP on this gene
MY04_0704
Ribonucleotide reductase of class Ia, alpha subunit
Accession: ANQ48087
Location: 899328-901016
NCBI BlastP on this gene
Accession: ANQ48087
Location: 899328-901016
NCBI BlastP on this gene
MY04_0705
Endonuclease/exonuclease/phosphatase family protein
Accession: ANQ48088
Location: 901858-902895
NCBI BlastP on this gene
Accession: ANQ48088
Location: 901858-902895
NCBI BlastP on this gene
MY04_0706
MY04_0707
SusC-like TonB-dependent receptor
Accession: ANQ48090
Location: 904675-907602
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 474
Sequence coverage: 104 %
E-value: 1e-146
NCBI BlastP on this gene
Accession: ANQ48090
Location: 904675-907602
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 474
Sequence coverage: 104 %
E-value: 1e-146
NCBI BlastP on this gene
MY04_0708
MY04_0709
Phosphoserine phosphatase/homoserine phosphotransferase bifunctional protein
Accession: ANQ48092
Location: 912287-913288
NCBI BlastP on this gene
Accession: ANQ48092
Location: 912287-913288
NCBI BlastP on this gene
MY04_0710
Aldose epimerase family protein
Accession: ANQ48093
Location: 913433-914329
NCBI BlastP on this gene
Accession: ANQ48093
Location: 913433-914329
NCBI BlastP on this gene
MY04_0711
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP031965 : Aquimarina sp. AD10 chromosome Total score: 1.0 Cumulative Blast bit score: 472
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
D1816_20035
D1816_20030
RagB/SusD family nutrient uptake outer membrane protein
Accession: AXT62551
Location: 4829941-4831428
NCBI BlastP on this gene
Accession: AXT62551
Location: 4829941-4831428
NCBI BlastP on this gene
D1816_20025
TonB-dependent receptor
Accession: AXT62550
Location: 4826939-4829917
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 472
Sequence coverage: 102 %
E-value: 6e-146
NCBI BlastP on this gene
Accession: AXT62550
Location: 4826939-4829917
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 472
Sequence coverage: 102 %
E-value: 6e-146
NCBI BlastP on this gene
D1816_20020
D1816_20015
LacI family transcriptional regulator
Accession: AXT62548
Location: 4823257-4824288
NCBI BlastP on this gene
Accession: AXT62548
Location: 4823257-4824288
NCBI BlastP on this gene
D1816_20010
LacI family transcriptional regulator
Accession: AXT62547
Location: 4822205-4823218
NCBI BlastP on this gene
Accession: AXT62547
Location: 4822205-4823218
NCBI BlastP on this gene
D1816_20005
D1816_20000
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP011531 : Bacteroides dorei CL03T12C01 Total score: 1.0 Cumulative Blast bit score: 472
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
ABI39_03940
ABI39_03935
glycerophosphodiester phosphodiesterase
Accession: AND18685
Location: 1098725-1099537
NCBI BlastP on this gene
Accession: AND18685
Location: 1098725-1099537
NCBI BlastP on this gene
ABI39_03930
glycan metabolism protein RagB
Accession: AND18684
Location: 1097011-1098651
NCBI BlastP on this gene
Accession: AND18684
Location: 1097011-1098651
NCBI BlastP on this gene
ABI39_03925
TonB-dependent receptor
Accession: AND18683
Location: 1093967-1096990
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 472
Sequence coverage: 105 %
E-value: 1e-145
NCBI BlastP on this gene
Accession: AND18683
Location: 1093967-1096990
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 472
Sequence coverage: 105 %
E-value: 1e-145
NCBI BlastP on this gene
ABI39_03920
ABI39_03915
alpha-N-acetylglucosaminidase
Accession: AND18681
Location: 1090501-1092765
NCBI BlastP on this gene
Accession: AND18681
Location: 1090501-1092765
NCBI BlastP on this gene
ABI39_03910
ABI39_03905
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP043529 : Bacteroides vulgatus strain VIC01 chromosome Total score: 1.0 Cumulative Blast bit score: 471
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
Exo-poly-alpha-D-galacturonosidase
Accession: QEW34965
Location: 521778-523190
NCBI BlastP on this gene
Accession: QEW34965
Location: 521778-523190
NCBI BlastP on this gene
pehX_1
VIC01_00415
Glycerophosphodiester phosphodiesterase
Accession: QEW34967
Location: 525569-526381
NCBI BlastP on this gene
Accession: QEW34967
Location: 525569-526381
NCBI BlastP on this gene
glpQ
susD_2
TonB-dependent receptor SusC
Accession: QEW34969
Location: 528116-531139
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 3e-145
NCBI BlastP on this gene
Accession: QEW34969
Location: 528116-531139
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 3e-145
NCBI BlastP on this gene
susC_12
VIC01_00419
VIC01_00420
todS_4
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP013020 : Bacteroides vulgatus strain mpk genome. Total score: 1.0 Cumulative Blast bit score: 471
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
BvMPK_0613
BvMPK_0612
BvMPK_0611
Glycerophosphoryl diester phosphodiesterase
Accession: ALK83231
Location: 717256-718065
NCBI BlastP on this gene
Accession: ALK83231
Location: 717256-718065
NCBI BlastP on this gene
BvMPK_0610
BvMPK_0609
SusC, outer membrane protein involved in starch binding
Accession: ALK83229
Location: 712498-715521
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 2e-145
NCBI BlastP on this gene
Accession: ALK83229
Location: 712498-715521
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 2e-145
NCBI BlastP on this gene
BvMPK_0608
N-acetylglucosamine related transporter, NagX
Accession: ALK83228
Location: 711304-712422
NCBI BlastP on this gene
Accession: ALK83228
Location: 711304-712422
NCBI BlastP on this gene
BvMPK_0607
BvMPK_0606
BvMPK_0605
BvMPK_0604
two-component system sensor histidine
Accession: ALK83224
Location: 703690-707910
NCBI BlastP on this gene
Accession: ALK83224
Location: 703690-707910
NCBI BlastP on this gene
BvMPK_0603
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP048115 : Mucilaginibacter sp. 14171R-50 chromosome Total score: 1.0 Cumulative Blast bit score: 469
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
VCBS repeat-containing protein
Accession: QHS57762
Location: 4475985-4479320
NCBI BlastP on this gene
Accession: QHS57762
Location: 4475985-4479320
NCBI BlastP on this gene
GWR56_20245
VCBS repeat-containing protein
Accession: QHS57763
Location: 4479327-4482623
NCBI BlastP on this gene
Accession: QHS57763
Location: 4479327-4482623
NCBI BlastP on this gene
GWR56_20250
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHS57764
Location: 4482710-4484248
NCBI BlastP on this gene
Accession: QHS57764
Location: 4482710-4484248
NCBI BlastP on this gene
GWR56_20255
SusC/RagA family TonB-linked outer membrane protein
Accession: QHS57765
Location: 4484262-4487297
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 469
Sequence coverage: 107 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: QHS57765
Location: 4484262-4487297
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 469
Sequence coverage: 107 %
E-value: 2e-144
NCBI BlastP on this gene
GWR56_20260
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP032869 : Mucilaginibacter sp. HYN0043 chromosome Total score: 1.0 Cumulative Blast bit score: 468
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
HYN43_010400
HYN43_010405
RagB/SusD family nutrient uptake outer membrane protein
Accession: AYL95674
Location: 2639607-2641169
NCBI BlastP on this gene
Accession: AYL95674
Location: 2639607-2641169
NCBI BlastP on this gene
HYN43_010410
SusC/RagA family TonB-linked outer membrane protein
Accession: AYL95675
Location: 2641156-2644197
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 468
Sequence coverage: 107 %
E-value: 4e-144
NCBI BlastP on this gene
Accession: AYL95675
Location: 2641156-2644197
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 468
Sequence coverage: 107 %
E-value: 4e-144
NCBI BlastP on this gene
HYN43_010415
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP029255 : Flavobacterium crocinum strain HYN0056 chromosome Total score: 1.0 Cumulative Blast bit score: 464
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
HYN56_21310
HYN56_21315
HYN56_21320
RagB/SusD family nutrient uptake outer membrane protein
Accession: AWK06633
Location: 5005723-5007234
NCBI BlastP on this gene
Accession: AWK06633
Location: 5005723-5007234
NCBI BlastP on this gene
HYN56_21325
SusC/RagA family TonB-linked outer membrane protein
Accession: AWK07537
Location: 5007262-5010243
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 465
Sequence coverage: 105 %
E-value: 4e-143
NCBI BlastP on this gene
Accession: AWK07537
Location: 5007262-5010243
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 465
Sequence coverage: 105 %
E-value: 4e-143
NCBI BlastP on this gene
HYN56_21330
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP030261 : Flavobacterium sp. HYN0086 chromosome Total score: 1.0 Cumulative Blast bit score: 464
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
HYN86_17735
HYN86_17740
glycoside hydrolase family 97 protein
Accession: AXB58339
Location: 4112970-4114988
NCBI BlastP on this gene
Accession: AXB58339
Location: 4112970-4114988
NCBI BlastP on this gene
HYN86_17745
HYN86_17750
RagB/SusD family nutrient uptake outer membrane protein
Accession: AXB58341
Location: 4116458-4117969
NCBI BlastP on this gene
Accession: AXB58341
Location: 4116458-4117969
NCBI BlastP on this gene
HYN86_17755
SusC/RagA family TonB-linked outer membrane protein
Accession: AXB58342
Location: 4117998-4120979
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 105 %
E-value: 6e-143
NCBI BlastP on this gene
Accession: AXB58342
Location: 4117998-4120979
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 105 %
E-value: 6e-143
NCBI BlastP on this gene
HYN86_17760
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP028136 : Gramella fulva strain SH35 Total score: 1.0 Cumulative Blast bit score: 464
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
DUF4861 domain-containing protein
Accession: AVR47419
Location: 2298992-2300293
NCBI BlastP on this gene
Accession: AVR47419
Location: 2298992-2300293
NCBI BlastP on this gene
C7S20_10250
C7S20_10245
C7S20_10240
RagB/SusD family nutrient uptake outer membrane protein
Accession: AVR45611
Location: 2293252-2294820
NCBI BlastP on this gene
Accession: AVR45611
Location: 2293252-2294820
NCBI BlastP on this gene
C7S20_10235
TonB-dependent receptor
Accession: AVR45610
Location: 2290258-2293239
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 104 %
E-value: 9e-143
NCBI BlastP on this gene
Accession: AVR45610
Location: 2290258-2293239
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 104 %
E-value: 9e-143
NCBI BlastP on this gene
C7S20_10230
C7S20_10225
SusC/RagA family TonB-linked outer membrane protein
Accession: AVR45608
Location: 2284759-2287923
NCBI BlastP on this gene
Accession: AVR45608
Location: 2284759-2287923
NCBI BlastP on this gene
C7S20_10220
C7S20_10215
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP031965 : Aquimarina sp. AD10 chromosome Total score: 1.0 Cumulative Blast bit score: 462
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
SusC/RagA family TonB-linked outer membrane protein
Accession: AXT62743
Location: 5099255-5102542
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 463
Sequence coverage: 103 %
E-value: 2e-141
NCBI BlastP on this gene
Accession: AXT62743
Location: 5099255-5102542
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 463
Sequence coverage: 103 %
E-value: 2e-141
NCBI BlastP on this gene
D1816_21150
DUF4974 domain-containing protein
Accession: AXT62742
Location: 5097952-5099118
NCBI BlastP on this gene
Accession: AXT62742
Location: 5097952-5099118
NCBI BlastP on this gene
D1816_21145
RNA polymerase sigma-70 factor
Accession: AXT62741
Location: 5097256-5097852
NCBI BlastP on this gene
Accession: AXT62741
Location: 5097256-5097852
NCBI BlastP on this gene
D1816_21140
D1816_21135
DUF2452 domain-containing protein
Accession: AXT62739
Location: 5095059-5095457
NCBI BlastP on this gene
Accession: AXT62739
Location: 5095059-5095457
NCBI BlastP on this gene
D1816_21130
D1816_21125
deoxyribodipyrimidine photolyase
Accession: AXT62737
Location: 5093292-5094782
NCBI BlastP on this gene
Accession: AXT62737
Location: 5093292-5094782
NCBI BlastP on this gene
D1816_21120
cryptochrome/photolyase family protein
Accession: AXT62736
Location: 5091770-5093302
NCBI BlastP on this gene
Accession: AXT62736
Location: 5091770-5093302
NCBI BlastP on this gene
D1816_21115
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP003346 : Echinicola vietnamensis DSM 17526 Total score: 1.0 Cumulative Blast bit score: 462
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
Protein of unknown function (DUF2726)
Accession: AGA76749
Location: 508718-509575
NCBI BlastP on this gene
Accession: AGA76749
Location: 508718-509575
NCBI BlastP on this gene
Echvi_0464
Echvi_0465
Echvi_0466
Echvi_0467
Echvi_0468
TonB-linked outer membrane protein, SusC/RagA family
Accession: AGA76754
Location: 514799-517885
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 463
Sequence coverage: 102 %
E-value: 5e-142
NCBI BlastP on this gene
Accession: AGA76754
Location: 514799-517885
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 463
Sequence coverage: 102 %
E-value: 5e-142
NCBI BlastP on this gene
Echvi_0469
beta-ketoacyl-acyl-carrier-protein synthase II
Accession: AGA76755
Location: 518337-519581
NCBI BlastP on this gene
Accession: AGA76755
Location: 518337-519581
NCBI BlastP on this gene
Echvi_0470
transcriptional regulator, tetR family
Accession: AGA76756
Location: 519578-520138
NCBI BlastP on this gene
Accession: AGA76756
Location: 519578-520138
NCBI BlastP on this gene
Echvi_0471
Echvi_0472
putative oxidoreductase, aryl-alcohol dehydrogenase like protein
Accession: AGA76758
Location: 520723-521754
NCBI BlastP on this gene
Accession: AGA76758
Location: 520723-521754
NCBI BlastP on this gene
Echvi_0473
Echvi_0474
Echvi_0475
Calcineurin-like phosphoesterase
Accession: AGA76761
Location: 522675-523334
NCBI BlastP on this gene
Accession: AGA76761
Location: 522675-523334
NCBI BlastP on this gene
Echvi_0476
Echvi_0477
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP003349 : Solitalea canadensis DSM 3403 Total score: 1.0 Cumulative Blast bit score: 459
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
TonB-linked outer membrane protein, SusC/RagA family
Accession: AFD07962
Location: 3474885-3477860
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 459
Sequence coverage: 100 %
E-value: 8e-141
NCBI BlastP on this gene
Accession: AFD07962
Location: 3474885-3477860
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 459
Sequence coverage: 100 %
E-value: 8e-141
NCBI BlastP on this gene
Solca_2943
Solca_2942
Solca_2941
Solca_2940
Solca_2939
dehydrogenase of unknown specificity, short-chain alcohol dehydrogenase like protein
Accession: AFD07957
Location: 3469445-3470197
NCBI BlastP on this gene
Accession: AFD07957
Location: 3469445-3470197
NCBI BlastP on this gene
Solca_2938
dehydrogenase of unknown specificity, short-chain alcohol dehydrogenase like protein
Accession: AFD07956
Location: 3468652-3469407
NCBI BlastP on this gene
Accession: AFD07956
Location: 3468652-3469407
NCBI BlastP on this gene
Solca_2937
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP014773 : Mucilaginibacter sp. PAMC 26640 chromosome Total score: 1.0 Cumulative Blast bit score: 457
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
A0256_11575
A0256_11570
A0256_11565
hypothetical protein
Accession: AMR32010
Location: 2645392-2648421
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 4e-140
NCBI BlastP on this gene
Accession: AMR32010
Location: 2645392-2648421
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 4e-140
NCBI BlastP on this gene
A0256_11560
A0256_11555
adenine phosphoribosyltransferase
Accession: AMR32008
Location: 2642424-2642951
NCBI BlastP on this gene
Accession: AMR32008
Location: 2642424-2642951
NCBI BlastP on this gene
A0256_11550
A0256_11545
A0256_11540
A0256_11535
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP001807 : Rhodothermus marinus DSM 4252 Total score: 1.0 Cumulative Blast bit score: 457
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
Rmar_1759
Rmar_1760
Rmar_1761
TonB-dependent receptor plug
Accession: ACY48646
Location: 2061694-2064765
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 103 %
E-value: 4e-140
NCBI BlastP on this gene
Accession: ACY48646
Location: 2061694-2064765
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 103 %
E-value: 4e-140
NCBI BlastP on this gene
Rmar_1762
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP019797 : Rhodothermus marinus AA3-38 DNA Total score: 1.0 Cumulative Blast bit score: 457
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
RmaAA338_19320
RmaAA338_19330
RmaAA338_19340
SusC/RagA family TonB-linked outer membrane protein
Accession: BBM73070
Location: 2255167-2258229
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: BBM73070
Location: 2255167-2258229
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
RmaAA338_19350
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
AP019796 : Rhodothermus marinus AA2-13 DNA Total score: 1.0 Cumulative Blast bit score: 457
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
RmaAA213_19270
RmaAA213_19280
RmaAA213_19290
SusC/RagA family TonB-linked outer membrane protein
Accession: BBM70084
Location: 2257950-2261012
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: BBM70084
Location: 2257950-2261012
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
RmaAA213_19300
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP048409 : Draconibacterium sp. M1 chromosome Total score: 1.0 Cumulative Blast bit score: 456
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
TonB-dependent receptor
Accession: QIA07694
Location: 2023131-2026043
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 104 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: QIA07694
Location: 2023131-2026043
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 104 %
E-value: 5e-140
NCBI BlastP on this gene
G0Q07_08125
substrate-binding domain-containing protein
Accession: QIA07693
Location: 2020007-2022661
NCBI BlastP on this gene
Accession: QIA07693
Location: 2020007-2022661
NCBI BlastP on this gene
G0Q07_08120
G0Q07_08115
G0Q07_08110
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP034562 : Flammeovirga pectinis strain L12M1 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 456
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
TonB-dependent receptor
Accession: AZQ62316
Location: 2184855-2187782
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 105 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: AZQ62316
Location: 2184855-2187782
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 105 %
E-value: 5e-140
NCBI BlastP on this gene
EI427_08710
EI427_08705
DUF1349 domain-containing protein
Accession: AZQ62314
Location: 2179522-2180223
NCBI BlastP on this gene
Accession: AZQ62314
Location: 2179522-2180223
NCBI BlastP on this gene
EI427_08700
aldose 1-epimerase family protein
Accession: AZQ62313
Location: 2178480-2179400
NCBI BlastP on this gene
Accession: AZQ62313
Location: 2178480-2179400
NCBI BlastP on this gene
EI427_08695
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP025791 : Flavivirga eckloniae strain ECD14 chromosome Total score: 1.0 Cumulative Blast bit score: 456
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
C1H87_07285
C1H87_07290
C1H87_07295
C1H87_07300
C1H87_07305
C1H87_07310
TonB-dependent receptor
Accession: AUP78528
Location: 1753729-1756725
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 456
Sequence coverage: 101 %
E-value: 1e-139
NCBI BlastP on this gene
Accession: AUP78528
Location: 1753729-1756725
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 456
Sequence coverage: 101 %
E-value: 1e-139
NCBI BlastP on this gene
C1H87_07315
C1H87_07320
C1H87_07325
C1H87_07330
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP023254 : Chitinophaga sp. MD30 chromosome. Total score: 1.0 Cumulative Blast bit score: 456
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
metal-independent alpha-mannosidase
Accession: ASZ10680
Location: 1676942-1678372
NCBI BlastP on this gene
Accession: ASZ10680
Location: 1676942-1678372
NCBI BlastP on this gene
CK934_06640
CK934_06635
RagB/SusD family nutrient uptake outer membrane protein
Accession: ASZ10678
Location: 1671695-1673245
NCBI BlastP on this gene
Accession: ASZ10678
Location: 1671695-1673245
NCBI BlastP on this gene
CK934_06630
hypothetical protein
Accession: ASZ10677
Location: 1668704-1671673
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 102 %
E-value: 1e-139
NCBI BlastP on this gene
Accession: ASZ10677
Location: 1668704-1671673
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 102 %
E-value: 1e-139
NCBI BlastP on this gene
CK934_06625
CK934_06620
CK934_06615
CK934_06610
tRNA-specific adenosine deaminase
Accession: ASZ10673
Location: 1662805-1663245
NCBI BlastP on this gene
Accession: ASZ10673
Location: 1662805-1663245
NCBI BlastP on this gene
CK934_06605
TIGR00730 family Rossman fold protein
Accession: ASZ10672
Location: 1662196-1662780
NCBI BlastP on this gene
Accession: ASZ10672
Location: 1662196-1662780
NCBI BlastP on this gene
CK934_06600
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
CP050956 : Parabacteroides distasonis strain FDAARGOS_615 chromosome. Total score: 1.0 Cumulative Blast bit score: 454
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
multidrug DMT transporter permease
Accession: QIX66393
Location: 3696407-3697408
NCBI BlastP on this gene
Accession: QIX66393
Location: 3696407-3697408
NCBI BlastP on this gene
FOB23_15395
FOB23_15390
ADP-ribosylglycohydrolase family protein
Accession: QIX66391
Location: 3693815-3695371
NCBI BlastP on this gene
Accession: QIX66391
Location: 3693815-3695371
NCBI BlastP on this gene
FOB23_15385
rbsK
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIX66389
Location: 3691335-3692852
NCBI BlastP on this gene
Accession: QIX66389
Location: 3691335-3692852
NCBI BlastP on this gene
FOB23_15375
TonB-dependent receptor
Accession: QIX66388
Location: 3688350-3691322
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 454
Sequence coverage: 105 %
E-value: 3e-139
NCBI BlastP on this gene
Accession: QIX66388
Location: 3688350-3691322
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 454
Sequence coverage: 105 %
E-value: 3e-139
NCBI BlastP on this gene
FOB23_15370
FOB23_15365
helix-turn-helix transcriptional regulator
Accession: QIX66386
Location: 3686168-3686521
NCBI BlastP on this gene
Accession: QIX66386
Location: 3686168-3686521
NCBI BlastP on this gene
FOB23_15360
FOB23_15355
FOB23_15350
FOB23_15345
FOB23_15340
Query: Bacteroides ovatus ATCC 8483 B_ovatus-MSIQ_Cont457, whole genome
451. : AP019797 Rhodothermus marinus AA3-38 DNA Total score: 1.0 Cumulative Blast bit score: 599
BACOVA_05488
BACOVA_05489
BACOVA_05490
BACOVA_05491
BACOVA_05492
BACOVA_05493
RmaAA338_05190
dihydrodipicolinate synthase family protein
Accession: BBM71653
Location: 579420-580337
NCBI BlastP on this gene
Accession: BBM71653
Location: 579420-580337
NCBI BlastP on this gene
RmaAA338_05180
RmaAA338_05170
galactose-1-phosphate uridylyltransferase
Accession: BBM71651
Location: 577667-578722
NCBI BlastP on this gene
Accession: BBM71651
Location: 577667-578722
NCBI BlastP on this gene
RmaAA338_05160
yidK
galK_1
hypothetical protein
Accession: BBM71648
Location: 572338-574851
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBM71648
Location: 572338-574851
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RmaAA338_05130
RmaAA338_05120
RmaAA338_05110
RmaAA338_05100
RmaAA338_05090
RmaAA338_05080
RmaAA338_05070
copA_1
452. : AP019796 Rhodothermus marinus AA2-13 DNA Total score: 1.0 Cumulative Blast bit score: 599
RmaAA213_05210
dihydrodipicolinate synthase family protein
Accession: BBM68674
Location: 577195-578112
NCBI BlastP on this gene
Accession: BBM68674
Location: 577195-578112
NCBI BlastP on this gene
RmaAA213_05200
RmaAA213_05190
galactose-1-phosphate uridylyltransferase
Accession: BBM68672
Location: 575442-576497
NCBI BlastP on this gene
Accession: BBM68672
Location: 575442-576497
NCBI BlastP on this gene
RmaAA213_05180
yidK
galK_1
hypothetical protein
Accession: BBM68669
Location: 570113-572626
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
Accession: BBM68669
Location: 570113-572626
BlastP hit with EDO09630.1
Percentage identity: 39 %
BlastP bit score: 599
Sequence coverage: 99 %
E-value: 0.0
NCBI BlastP on this gene
RmaAA213_05150
RmaAA213_05140
RmaAA213_05130
RmaAA213_05120
CRISPR-associated endonuclease Cas1
Accession: BBM68665
Location: 566446-567339
NCBI BlastP on this gene
Accession: BBM68665
Location: 566446-567339
NCBI BlastP on this gene
cas1
RmaAA213_05100
type I-E CRISPR-associated protein Cas5/CasD
Accession: BBM68663
Location: 564880-565641
NCBI BlastP on this gene
Accession: BBM68663
Location: 564880-565641
NCBI BlastP on this gene
RmaAA213_05090
type I-E CRISPR-associated protein
Accession: BBM68662
Location: 563612-564883
NCBI BlastP on this gene
Accession: BBM68662
Location: 563612-564883
NCBI BlastP on this gene
RmaAA213_05080
453. : CP043306 Cellvibrio japonicus strain ADPT1-KOJIBIOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 588
FY115_02380
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI18344
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: QEI18344
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
FY115_02375
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI18343
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QEI18343
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
FY115_02370
sulfite exporter TauE/SafE family protein
Accession: QEI18342
Location: 595510-596292
NCBI BlastP on this gene
Accession: QEI18342
Location: 595510-596292
NCBI BlastP on this gene
FY115_02365
FY115_02360
twin-arginine translocase subunit TatC
Accession: QEI18340
Location: 594225-595040
NCBI BlastP on this gene
Accession: QEI18340
Location: 594225-595040
NCBI BlastP on this gene
tatC
DUF839 domain-containing protein
Accession: QEI18339
Location: 592703-594100
NCBI BlastP on this gene
Accession: QEI18339
Location: 592703-594100
NCBI BlastP on this gene
FY115_02350
PEP-CTERM sorting domain-containing protein
Accession: QEI18338
Location: 592119-592676
NCBI BlastP on this gene
Accession: QEI18338
Location: 592119-592676
NCBI BlastP on this gene
FY115_02345
twin-arginine translocase TatA/TatE family subunit
Accession: QEI18337
Location: 591697-591900
NCBI BlastP on this gene
Accession: QEI18337
Location: 591697-591900
NCBI BlastP on this gene
tatA
glutathione S-transferase family protein
Accession: QEI18336
Location: 590945-591628
NCBI BlastP on this gene
Accession: QEI18336
Location: 590945-591628
NCBI BlastP on this gene
FY115_02335
type II/IV secretion system protein
Accession: QEI18335
Location: 589082-590857
NCBI BlastP on this gene
Accession: QEI18335
Location: 589082-590857
NCBI BlastP on this gene
FY115_02330
454. : CP043305 Cellvibrio japonicus strain ADPT2-NIGEROSE chromosome. Total score: 1.0 Cumulative Blast bit score: 588
FY116_02380
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI14764
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: QEI14764
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
FY116_02375
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI14763
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QEI14763
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
FY116_02370
sulfite exporter TauE/SafE family protein
Accession: QEI14762
Location: 595510-596292
NCBI BlastP on this gene
Accession: QEI14762
Location: 595510-596292
NCBI BlastP on this gene
FY116_02365
FY116_02360
twin-arginine translocase subunit TatC
Accession: QEI14760
Location: 594225-595040
NCBI BlastP on this gene
Accession: QEI14760
Location: 594225-595040
NCBI BlastP on this gene
tatC
DUF839 domain-containing protein
Accession: QEI14759
Location: 592703-594100
NCBI BlastP on this gene
Accession: QEI14759
Location: 592703-594100
NCBI BlastP on this gene
FY116_02350
PEP-CTERM sorting domain-containing protein
Accession: QEI14758
Location: 592119-592676
NCBI BlastP on this gene
Accession: QEI14758
Location: 592119-592676
NCBI BlastP on this gene
FY116_02345
twin-arginine translocase TatA/TatE family subunit
Accession: QEI14757
Location: 591697-591900
NCBI BlastP on this gene
Accession: QEI14757
Location: 591697-591900
NCBI BlastP on this gene
tatA
glutathione S-transferase family protein
Accession: QEI14756
Location: 590945-591628
NCBI BlastP on this gene
Accession: QEI14756
Location: 590945-591628
NCBI BlastP on this gene
FY116_02335
type II/IV secretion system protein
Accession: QEI14755
Location: 589082-590857
NCBI BlastP on this gene
Accession: QEI14755
Location: 589082-590857
NCBI BlastP on this gene
FY116_02330
455. : CP043304 Cellvibrio japonicus strain ADPT3-ISOMALTOSE chromosome. Total score: 1.0 Cumulative Blast bit score: 588
FY117_02380
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI11190
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
Accession: QEI11190
Location: 597716-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 300
Sequence coverage: 87 %
E-value: 1e-94
NCBI BlastP on this gene
FY117_02375
arabinogalactan endo-1,4-beta-galactosidase
Accession: QEI11189
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: QEI11189
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
FY117_02370
sulfite exporter TauE/SafE family protein
Accession: QEI11188
Location: 595510-596292
NCBI BlastP on this gene
Accession: QEI11188
Location: 595510-596292
NCBI BlastP on this gene
FY117_02365
FY117_02360
twin-arginine translocase subunit TatC
Accession: QEI11186
Location: 594225-595040
NCBI BlastP on this gene
Accession: QEI11186
Location: 594225-595040
NCBI BlastP on this gene
tatC
DUF839 domain-containing protein
Accession: QEI11185
Location: 592703-594100
NCBI BlastP on this gene
Accession: QEI11185
Location: 592703-594100
NCBI BlastP on this gene
FY117_02350
PEP-CTERM sorting domain-containing protein
Accession: QEI11184
Location: 592119-592676
NCBI BlastP on this gene
Accession: QEI11184
Location: 592119-592676
NCBI BlastP on this gene
FY117_02345
twin-arginine translocase TatA/TatE family subunit
Accession: QEI11183
Location: 591697-591900
NCBI BlastP on this gene
Accession: QEI11183
Location: 591697-591900
NCBI BlastP on this gene
tatA
glutathione S-transferase family protein
Accession: QEI11182
Location: 590945-591628
NCBI BlastP on this gene
Accession: QEI11182
Location: 590945-591628
NCBI BlastP on this gene
FY117_02335
type II/IV secretion system protein
Accession: QEI11181
Location: 589082-590857
NCBI BlastP on this gene
Accession: QEI11181
Location: 589082-590857
NCBI BlastP on this gene
FY117_02330
456. : CP000934 Cellvibrio japonicus Ueda107 Total score: 1.0 Cumulative Blast bit score: 587
carbohydrate binding protein, cbp35C
Accession: ACE84402
Location: 599129-606433
NCBI BlastP on this gene
Accession: ACE84402
Location: 599129-606433
NCBI BlastP on this gene
cbp35C
CJA_0493
arabinogalactan endo-1,4-beta-galactosidase, putative, gal53B
Accession: ACE86078
Location: 597653-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 299
Sequence coverage: 87 %
E-value: 3e-94
NCBI BlastP on this gene
Accession: ACE86078
Location: 597653-598894
BlastP hit with EDO09625.1
Percentage identity: 46 %
BlastP bit score: 299
Sequence coverage: 87 %
E-value: 3e-94
NCBI BlastP on this gene
gal53B
arabinogalactan endo-1,4-beta-galactosidase, putative, gal53A
Accession: ACE85511
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
Accession: ACE85511
Location: 596606-597619
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 288
Sequence coverage: 88 %
E-value: 6e-91
NCBI BlastP on this gene
gal53A-1
CJA_0490
CJA_0489
Sec-independent protein translocase TatC
Accession: ACE85846
Location: 594225-595040
NCBI BlastP on this gene
Accession: ACE85846
Location: 594225-595040
NCBI BlastP on this gene
CJA_0488
Tat (twin-arginine translocation) pathway signal sequence domain protein
Accession: ACE83245
Location: 592703-594100
NCBI BlastP on this gene
Accession: ACE83245
Location: 592703-594100
NCBI BlastP on this gene
CJA_0487
PEP-CTERM putative exosortase interaction domain protein
Accession: ACE84061
Location: 592119-592676
NCBI BlastP on this gene
Accession: ACE84061
Location: 592119-592676
NCBI BlastP on this gene
CJA_0486
twin arginine-targeting protein translocase,
Accession: ACE85402
Location: 591691-591900
NCBI BlastP on this gene
Accession: ACE85402
Location: 591691-591900
NCBI BlastP on this gene
CJA_0485
putative glutathione S-transferase
Accession: ACE85636
Location: 590945-591628
NCBI BlastP on this gene
Accession: ACE85636
Location: 590945-591628
NCBI BlastP on this gene
CJA_0484
type IV pilus biogenesis protein
Accession: ACE86015
Location: 589058-590857
NCBI BlastP on this gene
Accession: ACE86015
Location: 589058-590857
NCBI BlastP on this gene
CJA_0483
457. : CP039703 Clostridium butyricum strain 29-1 plasmid p-butyl_plas_29-1 Total score: 1.0 Cumulative Blast bit score: 572
FBD77_19750
response regulator transcription factor
Accession: QCJ04649
Location: 594955-595665
NCBI BlastP on this gene
Accession: QCJ04649
Location: 594955-595665
NCBI BlastP on this gene
FBD77_19745
FtsX-like permease family protein
Accession: FBD77_19740
Location: 592190-594783
NCBI BlastP on this gene
Accession: FBD77_19740
Location: 592190-594783
NCBI BlastP on this gene
FBD77_19740
ABC transporter ATP-binding protein
Accession: QCJ04648
Location: 591504-592190
NCBI BlastP on this gene
Accession: QCJ04648
Location: 591504-592190
NCBI BlastP on this gene
FBD77_19735
HAMP domain-containing histidine kinase
Accession: QCJ04647
Location: 590431-591420
NCBI BlastP on this gene
Accession: QCJ04647
Location: 590431-591420
NCBI BlastP on this gene
FBD77_19730
response regulator transcription factor
Accession: QCJ04646
Location: 589758-590444
NCBI BlastP on this gene
Accession: QCJ04646
Location: 589758-590444
NCBI BlastP on this gene
FBD77_19725
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ04645
Location: 588228-589253
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: QCJ04645
Location: 588228-589253
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
FBD77_19720
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ04644
Location: 586817-587815
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: QCJ04644
Location: 586817-587815
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
FBD77_19715
FBD77_19710
FBD77_19705
DUF3502 domain-containing protein
Accession: QCJ04641
Location: 582723-584267
NCBI BlastP on this gene
Accession: QCJ04641
Location: 582723-584267
NCBI BlastP on this gene
FBD77_19700
DUF3502 domain-containing protein
Accession: QCJ04640
Location: 580948-582486
NCBI BlastP on this gene
Accession: QCJ04640
Location: 580948-582486
NCBI BlastP on this gene
FBD77_19695
carbohydrate ABC transporter permease
Accession: QCJ04639
Location: 579860-580750
NCBI BlastP on this gene
Accession: QCJ04639
Location: 579860-580750
NCBI BlastP on this gene
FBD77_19690
458. : CP016333 Clostridium butyricum strain TK520 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
BBB49_18970
two-component system response regulator
Accession: AOR96152
Location: 601479-602189
NCBI BlastP on this gene
Accession: AOR96152
Location: 601479-602189
NCBI BlastP on this gene
BBB49_18975
BBB49_18980
peptide ABC transporter ATP-binding protein
Accession: AOR96154
Location: 604955-605641
NCBI BlastP on this gene
Accession: AOR96154
Location: 604955-605641
NCBI BlastP on this gene
BBB49_18985
two-component sensor histidine kinase
Accession: AOR96155
Location: 605725-606714
NCBI BlastP on this gene
Accession: AOR96155
Location: 605725-606714
NCBI BlastP on this gene
BBB49_18990
BBB49_18995
arabinogalactan endo-1,4-beta-galactosidase
Accession: AOR96157
Location: 607894-608919
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: AOR96157
Location: 607894-608919
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
BBB49_19000
arabinogalactan endo-1,4-beta-galactosidase
Accession: AOR96340
Location: 609332-610330
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: AOR96340
Location: 609332-610330
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
BBB49_19005
BBB49_19010
BBB49_19015
BBB49_19020
BBB49_19025
459. : CP014705 Clostridium butyricum strain TOA chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
AZ909_19500
two-component system response regulator
Accession: ANF16241
Location: 642248-642958
NCBI BlastP on this gene
Accession: ANF16241
Location: 642248-642958
NCBI BlastP on this gene
AZ909_19505
AZ909_19510
peptide ABC transporter ATP-binding protein
Accession: ANF16243
Location: 645724-646410
NCBI BlastP on this gene
Accession: ANF16243
Location: 645724-646410
NCBI BlastP on this gene
AZ909_19515
two-component sensor histidine kinase
Accession: ANF16244
Location: 646494-647483
NCBI BlastP on this gene
Accession: ANF16244
Location: 646494-647483
NCBI BlastP on this gene
AZ909_19520
AZ909_19525
arabinogalactan endo-1,4-beta-galactosidase
Accession: ANF16246
Location: 648663-649688
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: ANF16246
Location: 648663-649688
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
AZ909_19530
arabinogalactan endo-1,4-beta-galactosidase
Accession: ANF16398
Location: 650101-651099
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: ANF16398
Location: 650101-651099
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
AZ909_19535
AZ909_19540
AZ909_19545
AZ909_19550
AZ909_19555
460. : CP013489 Clostridium butyricum strain KNU-L09 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
ATN24_17010
two-component system response regulator
Accession: ALR90169
Location: 6115-6825
NCBI BlastP on this gene
Accession: ALR90169
Location: 6115-6825
NCBI BlastP on this gene
ATN24_17015
ATN24_17020
peptide ABC transporter ATP-binding protein
Accession: ALR90171
Location: 9591-10277
NCBI BlastP on this gene
Accession: ALR90171
Location: 9591-10277
NCBI BlastP on this gene
ATN24_17025
ATN24_17030
two-component system response regulator
Accession: ALR90173
Location: 11337-12023
NCBI BlastP on this gene
Accession: ALR90173
Location: 11337-12023
NCBI BlastP on this gene
ATN24_17035
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALR90174
Location: 12530-13555
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: ALR90174
Location: 12530-13555
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
ATN24_17040
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALR90819
Location: 13968-14966
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: ALR90819
Location: 13968-14966
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
ATN24_17045
ATN24_17050
ATN24_17055
ATN24_17060
461. : CP013353 Clostridium butyricum strain JKY6D1 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 572
ATD26_19465
two-component system response regulator
Accession: ALS19054
Location: 573180-573890
NCBI BlastP on this gene
Accession: ALS19054
Location: 573180-573890
NCBI BlastP on this gene
ATD26_19470
ATD26_19475
peptide ABC transporter ATP-binding protein
Accession: ALS19056
Location: 576656-577342
NCBI BlastP on this gene
Accession: ALS19056
Location: 576656-577342
NCBI BlastP on this gene
ATD26_19480
ATD26_19485
two-component system response regulator
Accession: ALS19058
Location: 578402-579088
NCBI BlastP on this gene
Accession: ALS19058
Location: 578402-579088
NCBI BlastP on this gene
ATD26_19490
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALS19059
Location: 579595-580620
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
Accession: ALS19059
Location: 579595-580620
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 305
Sequence coverage: 98 %
E-value: 2e-97
NCBI BlastP on this gene
ATD26_19495
arabinogalactan endo-1,4-beta-galactosidase
Accession: ALS19254
Location: 581033-582031
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: ALS19254
Location: 581033-582031
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
ATD26_19500
ATD26_19505
ATD26_19510
ATD26_19515
ATD26_19520
462. : CP039706 Clostridium butyricum strain 4-1 plasmid p-butyl_plas_4-1 Total score: 1.0 Cumulative Blast bit score: 571
FBD76_18270
response regulator transcription factor
Accession: QCJ08290
Location: 130688-131398
NCBI BlastP on this gene
Accession: QCJ08290
Location: 130688-131398
NCBI BlastP on this gene
FBD76_18275
FBD76_18280
ABC transporter ATP-binding protein
Accession: QCJ08292
Location: 134164-134850
NCBI BlastP on this gene
Accession: QCJ08292
Location: 134164-134850
NCBI BlastP on this gene
FBD76_18285
HAMP domain-containing histidine kinase
Accession: QCJ08293
Location: 134934-135923
NCBI BlastP on this gene
Accession: QCJ08293
Location: 134934-135923
NCBI BlastP on this gene
FBD76_18290
response regulator transcription factor
Accession: QCJ08294
Location: 135910-136596
NCBI BlastP on this gene
Accession: QCJ08294
Location: 135910-136596
NCBI BlastP on this gene
FBD76_18295
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ08295
Location: 137102-138127
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
Accession: QCJ08295
Location: 137102-138127
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
FBD76_18300
arabinogalactan endo-1,4-beta-galactosidase
Accession: QCJ08778
Location: 138540-139538
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: QCJ08778
Location: 138540-139538
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
FBD76_18305
FBD76_18310
extracellular solute-binding protein
Accession: QCJ08297
Location: 142088-143632
NCBI BlastP on this gene
Accession: QCJ08297
Location: 142088-143632
NCBI BlastP on this gene
FBD76_18315
DUF3502 domain-containing protein
Accession: QCJ08298
Location: 143869-145407
NCBI BlastP on this gene
Accession: QCJ08298
Location: 143869-145407
NCBI BlastP on this gene
FBD76_18320
carbohydrate ABC transporter permease
Accession: QCJ08299
Location: 145605-146495
NCBI BlastP on this gene
Accession: QCJ08299
Location: 145605-146495
NCBI BlastP on this gene
FBD76_18325
463. : CP030776 Clostridium butyricum strain S-45-5 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 571
DRB99_19095
DRB99_19100
DRB99_19105
ABC transporter ATP-binding protein
Accession: AXB87024
Location: 402998-403684
NCBI BlastP on this gene
Accession: AXB87024
Location: 402998-403684
NCBI BlastP on this gene
DRB99_19110
DRB99_19115
DRB99_19120
arabinogalactan endo-1,4-beta-galactosidase
Accession: AXB87027
Location: 405936-406961
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
Accession: AXB87027
Location: 405936-406961
BlastP hit with EDO09625.1
Percentage identity: 44 %
BlastP bit score: 304
Sequence coverage: 98 %
E-value: 6e-97
NCBI BlastP on this gene
DRB99_19125
arabinogalactan endo-1,4-beta-galactosidase
Accession: AXB87306
Location: 407374-408372
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
Accession: AXB87306
Location: 407374-408372
BlastP hit with EDO09625.1
Percentage identity: 43 %
BlastP bit score: 267
Sequence coverage: 88 %
E-value: 1e-82
NCBI BlastP on this gene
DRB99_19130
DRB99_19135
extracellular solute-binding protein
Accession: AXB87029
Location: 410922-412466
NCBI BlastP on this gene
Accession: AXB87029
Location: 410922-412466
NCBI BlastP on this gene
DRB99_19140
DUF3502 domain-containing protein
Accession: AXB87030
Location: 412703-414241
NCBI BlastP on this gene
Accession: AXB87030
Location: 412703-414241
NCBI BlastP on this gene
DRB99_19145
carbohydrate ABC transporter permease
Accession: AXB87031
Location: 414439-415329
NCBI BlastP on this gene
Accession: AXB87031
Location: 414439-415329
NCBI BlastP on this gene
DRB99_19150
464. : CP012643 Rufibacter tibetensis strain 1351 Total score: 1.0 Cumulative Blast bit score: 521
DC20_01105
DC20_01110
DC20_01115
DC20_01120
SusC/RagA family TonB-linked outer membrane protein
Accession: ALI97834
Location: 297461-300445
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 521
Sequence coverage: 105 %
E-value: 2e-164
NCBI BlastP on this gene
Accession: ALI97834
Location: 297461-300445
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 521
Sequence coverage: 105 %
E-value: 2e-164
NCBI BlastP on this gene
DC20_01125
DC20_01130
DC20_01135
DC20_01140
DC20_01145
465. : AP017422 Filimonas lacunae DNA Total score: 1.0 Cumulative Blast bit score: 520
FLA_3189
FLA_3190
FLA_3191
rhamnogalacturonan acetylesterase
Accession: BAV07169
Location: 3897284-3899215
NCBI BlastP on this gene
Accession: BAV07169
Location: 3897284-3899215
NCBI BlastP on this gene
FLA_3192
FLA_3193
outer membrane protein SusC, starch binding
Accession: BAV07171
Location: 3900777-3904079
BlastP hit with EDO09628.1
Percentage identity: 36 %
BlastP bit score: 520
Sequence coverage: 102 %
E-value: 5e-163
NCBI BlastP on this gene
Accession: BAV07171
Location: 3900777-3904079
BlastP hit with EDO09628.1
Percentage identity: 36 %
BlastP bit score: 520
Sequence coverage: 102 %
E-value: 5e-163
NCBI BlastP on this gene
FLA_3194
FLA_3195
RNA polymerase ECF-type sigma factor
Accession: BAV07173
Location: 3905487-3906071
NCBI BlastP on this gene
Accession: BAV07173
Location: 3905487-3906071
NCBI BlastP on this gene
FLA_3196
FLA_3197
FLA_3198
transcriptional regulator, AraC family
Accession: BAV07176
Location: 3908412-3909401
NCBI BlastP on this gene
Accession: BAV07176
Location: 3908412-3909401
NCBI BlastP on this gene
FLA_3199
FLA_3200
FLA_3201
466. : CP017774 Flavobacterium commune strain PK15 chromosome Total score: 1.0 Cumulative Blast bit score: 506
SusC/RagA family TonB-linked outer membrane protein
Accession: AOZ99793
Location: 2446153-2449416
BlastP hit with EDO09628.1
Percentage identity: 35 %
BlastP bit score: 506
Sequence coverage: 103 %
E-value: 6e-158
NCBI BlastP on this gene
Accession: AOZ99793
Location: 2446153-2449416
BlastP hit with EDO09628.1
Percentage identity: 35 %
BlastP bit score: 506
Sequence coverage: 103 %
E-value: 6e-158
NCBI BlastP on this gene
BIW12_10290
BIW12_10285
BIW12_10280
BIW12_10275
BIW12_10270
467. : AP018694 Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 496
SusC, outer membrane protein
Accession: BBE18918
Location: 3446242-3449412
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 497
Sequence coverage: 101 %
E-value: 1e-154
NCBI BlastP on this gene
Accession: BBE18918
Location: 3446242-3449412
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 497
Sequence coverage: 101 %
E-value: 1e-154
NCBI BlastP on this gene
AQPE_3088
AQPE_3087
RNA polymerase ECF-type sigma factor
Accession: BBE18916
Location: 3444182-3444805
NCBI BlastP on this gene
Accession: BBE18916
Location: 3444182-3444805
NCBI BlastP on this gene
AQPE_3086
AQPE_3085
AQPE_3084
two-component system sensor histidine kinase
Accession: BBE18913
Location: 3440824-3442479
NCBI BlastP on this gene
Accession: BBE18913
Location: 3440824-3442479
NCBI BlastP on this gene
AQPE_3083
AQPE_3082
468. : CP049857 Dysgonomonas sp. HDW5A chromosome Total score: 1.0 Cumulative Blast bit score: 491
helix-turn-helix transcriptional regulator
Accession: QIK59404
Location: 1427691-1428116
NCBI BlastP on this gene
Accession: QIK59404
Location: 1427691-1428116
NCBI BlastP on this gene
G7050_05960
glycoside hydrolase family 97 protein
Accession: QIK59403
Location: 1425506-1427500
NCBI BlastP on this gene
Accession: QIK59403
Location: 1425506-1427500
NCBI BlastP on this gene
G7050_05955
G7050_05950
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIK59401
Location: 1422093-1423607
NCBI BlastP on this gene
Accession: QIK59401
Location: 1422093-1423607
NCBI BlastP on this gene
G7050_05945
TonB-dependent receptor
Accession: QIK59400
Location: 1419121-1422057
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 491
Sequence coverage: 102 %
E-value: 2e-153
NCBI BlastP on this gene
Accession: QIK59400
Location: 1419121-1422057
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 491
Sequence coverage: 102 %
E-value: 2e-153
NCBI BlastP on this gene
G7050_05940
G7050_05935
DUF5004 domain-containing protein
Accession: QIK59398
Location: 1417190-1417720
NCBI BlastP on this gene
Accession: QIK59398
Location: 1417190-1417720
NCBI BlastP on this gene
G7050_05930
G7050_05925
glycoside hydrolase family 97 protein
Accession: QIK59396
Location: 1412878-1414860
NCBI BlastP on this gene
Accession: QIK59396
Location: 1412878-1414860
NCBI BlastP on this gene
G7050_05920
469. : CP049858 Dysgonomonas sp. HDW5B chromosome Total score: 1.0 Cumulative Blast bit score: 488
helix-turn-helix transcriptional regulator
Accession: QIK53955
Location: 1519538-1519963
NCBI BlastP on this gene
Accession: QIK53955
Location: 1519538-1519963
NCBI BlastP on this gene
G7051_06225
glycoside hydrolase family 97 protein
Accession: QIK53954
Location: 1517444-1519438
NCBI BlastP on this gene
Accession: QIK53954
Location: 1517444-1519438
NCBI BlastP on this gene
G7051_06220
G7051_06215
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIK53952
Location: 1514152-1515666
NCBI BlastP on this gene
Accession: QIK53952
Location: 1514152-1515666
NCBI BlastP on this gene
G7051_06210
TonB-dependent receptor
Accession: QIK53951
Location: 1511180-1514116
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 488
Sequence coverage: 102 %
E-value: 5e-152
NCBI BlastP on this gene
Accession: QIK53951
Location: 1511180-1514116
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 488
Sequence coverage: 102 %
E-value: 5e-152
NCBI BlastP on this gene
G7051_06205
G7051_06200
DUF5004 domain-containing protein
Accession: QIK53949
Location: 1509278-1509808
NCBI BlastP on this gene
Accession: QIK53949
Location: 1509278-1509808
NCBI BlastP on this gene
G7051_06195
G7051_06190
glycoside hydrolase family 97 protein
Accession: QIK53947
Location: 1504966-1506948
NCBI BlastP on this gene
Accession: QIK53947
Location: 1504966-1506948
NCBI BlastP on this gene
G7051_06185
470. : AP018694 Prolixibacteraceae bacterium MeG22 DNA Total score: 1.0 Cumulative Blast bit score: 488
AQPE_0220
AQPE_0219
AQPE_0218
AQPE_0217
SusC, outer membrane protein
Accession: BBE16079
Location: 257808-261026
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 488
Sequence coverage: 103 %
E-value: 3e-151
NCBI BlastP on this gene
Accession: BBE16079
Location: 257808-261026
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 488
Sequence coverage: 103 %
E-value: 3e-151
NCBI BlastP on this gene
AQPE_0216
AQPE_0215
RNA polymerase ECF-type sigma factor
Accession: BBE16077
Location: 255774-256430
NCBI BlastP on this gene
Accession: BBE16077
Location: 255774-256430
NCBI BlastP on this gene
AQPE_0214
AQPE_0213
AQPE_0212
RNA polymerase ECF-type sigma factor
Accession: BBE16074
Location: 253457-254053
NCBI BlastP on this gene
Accession: BBE16074
Location: 253457-254053
NCBI BlastP on this gene
AQPE_0211
AQPE_0210
AQPE_0209
471. : CP003557 Melioribacter roseus P3M Total score: 1.0 Cumulative Blast bit score: 485
nicotinate-nucleotide pyrophosphorylase
Accession: AFN74194
Location: 1088098-1088952
NCBI BlastP on this gene
Accession: AFN74194
Location: 1088098-1088952
NCBI BlastP on this gene
MROS_0953
MROS_0954
MROS_0955
MROS_0956
phosphoribulokinase/uridine kinase
Accession: AFN74198
Location: 1091578-1092921
NCBI BlastP on this gene
Accession: AFN74198
Location: 1091578-1092921
NCBI BlastP on this gene
MROS_0957
MROS_0958
MROS_0959
Glycosyl hydrolase family 5
Accession: AFN74201
Location: 1094686-1097229
BlastP hit with EDO09630.1
Percentage identity: 34 %
BlastP bit score: 485
Sequence coverage: 100 %
E-value: 7e-154
NCBI BlastP on this gene
Accession: AFN74201
Location: 1094686-1097229
BlastP hit with EDO09630.1
Percentage identity: 34 %
BlastP bit score: 485
Sequence coverage: 100 %
E-value: 7e-154
NCBI BlastP on this gene
MROS_0960
arabinogalactan endo-1,4-beta-galactosidase
Accession: AFN74202
Location: 1097226-1099451
NCBI BlastP on this gene
Accession: AFN74202
Location: 1097226-1099451
NCBI BlastP on this gene
MROS_0961
472. : CP050956 Parabacteroides distasonis strain FDAARGOS_615 chromosome. Total score: 1.0 Cumulative Blast bit score: 483
FOB23_06895
FOB23_06890
galactosamine-6-phosphate isomerase
Accession: QIX64863
Location: 1618136-1618858
NCBI BlastP on this gene
Accession: QIX64863
Location: 1618136-1618858
NCBI BlastP on this gene
FOB23_06885
DUF1080 domain-containing protein
Accession: QIX64862
Location: 1617323-1618129
NCBI BlastP on this gene
Accession: QIX64862
Location: 1617323-1618129
NCBI BlastP on this gene
FOB23_06880
Gfo/Idh/MocA family oxidoreductase
Accession: QIX64861
Location: 1615680-1617224
NCBI BlastP on this gene
Accession: QIX64861
Location: 1615680-1617224
NCBI BlastP on this gene
FOB23_06875
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIX64860
Location: 1614039-1615607
NCBI BlastP on this gene
Accession: QIX64860
Location: 1614039-1615607
NCBI BlastP on this gene
FOB23_06870
TonB-dependent receptor
Accession: QIX64859
Location: 1610876-1614019
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 7e-150
NCBI BlastP on this gene
Accession: QIX64859
Location: 1610876-1614019
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 7e-150
NCBI BlastP on this gene
FOB23_06865
DUF4974 domain-containing protein
Accession: QIX64858
Location: 1609639-1610613
NCBI BlastP on this gene
Accession: QIX64858
Location: 1609639-1610613
NCBI BlastP on this gene
FOB23_06860
N-acetylmuramoyl-L-alanine amidase
Accession: QIX64857
Location: 1609098-1609568
NCBI BlastP on this gene
Accession: QIX64857
Location: 1609098-1609568
NCBI BlastP on this gene
FOB23_06855
FOB23_06850
FOB23_06845
FOB23_06840
FOB23_06835
473. : CP034563 Flammeovirga pectinis strain L12M1 chromosome 2 Total score: 1.0 Cumulative Blast bit score: 483
TonB-dependent receptor
Accession: AZQ65029
Location: 604756-607773
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 484
Sequence coverage: 107 %
E-value: 2e-150
NCBI BlastP on this gene
Accession: AZQ65029
Location: 604756-607773
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 484
Sequence coverage: 107 %
E-value: 2e-150
NCBI BlastP on this gene
EI427_22680
EI427_22675
hybrid sensor histidine kinase/response regulator
Accession: AZQ65027
Location: 598510-602772
NCBI BlastP on this gene
Accession: AZQ65027
Location: 598510-602772
NCBI BlastP on this gene
EI427_22670
474. : AP019729 Parabacteroides distasonis NBRC 113806 DNA Total score: 1.0 Cumulative Blast bit score: 483
DN0286_31590
DN0286_31580
glucosamine-6-phosphate deaminase
Accession: BBK92871
Location: 3824100-3824822
NCBI BlastP on this gene
Accession: BBK92871
Location: 3824100-3824822
NCBI BlastP on this gene
agaI
DN0286_31560
DN0286_31550
DN0286_31540
SusC/RagA family TonB-linked outer membrane protein
Accession: BBK92867
Location: 3816822-3819983
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 6e-150
NCBI BlastP on this gene
Accession: BBK92867
Location: 3816822-3819983
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 484
Sequence coverage: 100 %
E-value: 6e-150
NCBI BlastP on this gene
DN0286_31530
iron dicitrate transporter FecR
Accession: BBK92866
Location: 3815603-3816577
NCBI BlastP on this gene
Accession: BBK92866
Location: 3815603-3816577
NCBI BlastP on this gene
DN0286_31520
N-acetylmuramoyl-L-alanine amidase
Accession: BBK92865
Location: 3815062-3815532
NCBI BlastP on this gene
Accession: BBK92865
Location: 3815062-3815532
NCBI BlastP on this gene
DN0286_31510
DN0286_31500
DN0286_31490
DN0286_31480
DN0286_31470
DN0286_31460
475. : CP040468 Parabacteroides distasonis strain CavFT-hAR46 chromosome Total score: 1.0 Cumulative Blast bit score: 481
TonB-dependent receptor
Accession: QCY57978
Location: 4269079-4272222
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 481
Sequence coverage: 100 %
E-value: 1e-148
NCBI BlastP on this gene
Accession: QCY57978
Location: 4269079-4272222
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 481
Sequence coverage: 100 %
E-value: 1e-148
NCBI BlastP on this gene
FE931_18250
DUF4974 domain-containing protein
Accession: QCY57977
Location: 4267843-4268817
NCBI BlastP on this gene
Accession: QCY57977
Location: 4267843-4268817
NCBI BlastP on this gene
FE931_18245
N-acetylmuramoyl-L-alanine amidase
Accession: QCY57976
Location: 4267302-4267772
NCBI BlastP on this gene
Accession: QCY57976
Location: 4267302-4267772
NCBI BlastP on this gene
FE931_18240
FE931_18235
FE931_18230
FE931_18225
FE931_18220
FE931_18215
476. : CP022754 Parabacteroides sp. CT06 chromosome Total score: 1.0 Cumulative Blast bit score: 479
SusC/RagA family TonB-linked outer membrane protein
Accession: AST55569
Location: 4699591-4702734
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 2e-148
NCBI BlastP on this gene
Accession: AST55569
Location: 4699591-4702734
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 2e-148
NCBI BlastP on this gene
CI960_20600
CI960_20595
N-acetylmuramoyl-L-alanine amidase
Accession: AST55567
Location: 4697814-4698284
NCBI BlastP on this gene
Accession: AST55567
Location: 4697814-4698284
NCBI BlastP on this gene
CI960_20590
CI960_20585
CI960_20580
CI960_20575
CI960_20570
CI960_20565
477. : CP000140 Parabacteroides distasonis ATCC 8503 Total score: 1.0 Cumulative Blast bit score: 479
putative outer membrane protein, probably involved in nutrient binding
Accession: ABR44581
Location: 3420940-3424152
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 4e-148
NCBI BlastP on this gene
Accession: ABR44581
Location: 3420940-3424152
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 480
Sequence coverage: 100 %
E-value: 4e-148
NCBI BlastP on this gene
BDI_2873
BDI_2872
N-acetylmuramoyl-L-alanine amidase
Accession: ABR44579
Location: 3419232-3419702
NCBI BlastP on this gene
Accession: ABR44579
Location: 3419232-3419702
NCBI BlastP on this gene
BDI_2871
conserved hypothetical protein
Accession: ABR44578
Location: 3418801-3419235
NCBI BlastP on this gene
Accession: ABR44578
Location: 3418801-3419235
NCBI BlastP on this gene
BDI_2870
conserved hypothetical protein
Accession: ABR44577
Location: 3417769-3418398
NCBI BlastP on this gene
Accession: ABR44577
Location: 3417769-3418398
NCBI BlastP on this gene
BDI_2869
conserved hypothetical protein
Accession: ABR44576
Location: 3416466-3417704
NCBI BlastP on this gene
Accession: ABR44576
Location: 3416466-3417704
NCBI BlastP on this gene
BDI_2868
conserved hypothetical protein
Accession: ABR44575
Location: 3416064-3416285
NCBI BlastP on this gene
Accession: ABR44575
Location: 3416064-3416285
NCBI BlastP on this gene
BDI_2867
BDI_2866
conserved hypothetical protein
Accession: ABR44573
Location: 3415130-3415357
NCBI BlastP on this gene
Accession: ABR44573
Location: 3415130-3415357
NCBI BlastP on this gene
BDI_2865
BDI_2864
478. : CP012801 Bacteroides cellulosilyticus strain WH2 Total score: 1.0 Cumulative Blast bit score: 476
BcellWH2_03128
BcellWH2_03129
BcellWH2_03130
TonB dependent receptor
Accession: ALJ60368
Location: 4010059-4013079
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 476
Sequence coverage: 104 %
E-value: 2e-147
NCBI BlastP on this gene
Accession: ALJ60368
Location: 4010059-4013079
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 476
Sequence coverage: 104 %
E-value: 2e-147
NCBI BlastP on this gene
BcellWH2_03131
479. : CP003560 Flammeovirga sp. MY04 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 474
Ribonucleotide reductase of class Ia, beta subunit
Accession: ANQ48086
Location: 898308-899285
NCBI BlastP on this gene
Accession: ANQ48086
Location: 898308-899285
NCBI BlastP on this gene
MY04_0704
Ribonucleotide reductase of class Ia, alpha subunit
Accession: ANQ48087
Location: 899328-901016
NCBI BlastP on this gene
Accession: ANQ48087
Location: 899328-901016
NCBI BlastP on this gene
MY04_0705
Endonuclease/exonuclease/phosphatase family protein
Accession: ANQ48088
Location: 901858-902895
NCBI BlastP on this gene
Accession: ANQ48088
Location: 901858-902895
NCBI BlastP on this gene
MY04_0706
MY04_0707
SusC-like TonB-dependent receptor
Accession: ANQ48090
Location: 904675-907602
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 474
Sequence coverage: 104 %
E-value: 1e-146
NCBI BlastP on this gene
Accession: ANQ48090
Location: 904675-907602
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 474
Sequence coverage: 104 %
E-value: 1e-146
NCBI BlastP on this gene
MY04_0708
MY04_0709
Phosphoserine phosphatase/homoserine phosphotransferase bifunctional protein
Accession: ANQ48092
Location: 912287-913288
NCBI BlastP on this gene
Accession: ANQ48092
Location: 912287-913288
NCBI BlastP on this gene
MY04_0710
Aldose epimerase family protein
Accession: ANQ48093
Location: 913433-914329
NCBI BlastP on this gene
Accession: ANQ48093
Location: 913433-914329
NCBI BlastP on this gene
MY04_0711
480. : CP031965 Aquimarina sp. AD10 chromosome Total score: 1.0 Cumulative Blast bit score: 472
D1816_20035
D1816_20030
RagB/SusD family nutrient uptake outer membrane protein
Accession: AXT62551
Location: 4829941-4831428
NCBI BlastP on this gene
Accession: AXT62551
Location: 4829941-4831428
NCBI BlastP on this gene
D1816_20025
TonB-dependent receptor
Accession: AXT62550
Location: 4826939-4829917
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 472
Sequence coverage: 102 %
E-value: 6e-146
NCBI BlastP on this gene
Accession: AXT62550
Location: 4826939-4829917
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 472
Sequence coverage: 102 %
E-value: 6e-146
NCBI BlastP on this gene
D1816_20020
D1816_20015
LacI family transcriptional regulator
Accession: AXT62548
Location: 4823257-4824288
NCBI BlastP on this gene
Accession: AXT62548
Location: 4823257-4824288
NCBI BlastP on this gene
D1816_20010
LacI family transcriptional regulator
Accession: AXT62547
Location: 4822205-4823218
NCBI BlastP on this gene
Accession: AXT62547
Location: 4822205-4823218
NCBI BlastP on this gene
D1816_20005
D1816_20000
481. : CP011531 Bacteroides dorei CL03T12C01 Total score: 1.0 Cumulative Blast bit score: 472
ABI39_03940
ABI39_03935
glycerophosphodiester phosphodiesterase
Accession: AND18685
Location: 1098725-1099537
NCBI BlastP on this gene
Accession: AND18685
Location: 1098725-1099537
NCBI BlastP on this gene
ABI39_03930
glycan metabolism protein RagB
Accession: AND18684
Location: 1097011-1098651
NCBI BlastP on this gene
Accession: AND18684
Location: 1097011-1098651
NCBI BlastP on this gene
ABI39_03925
TonB-dependent receptor
Accession: AND18683
Location: 1093967-1096990
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 472
Sequence coverage: 105 %
E-value: 1e-145
NCBI BlastP on this gene
Accession: AND18683
Location: 1093967-1096990
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 472
Sequence coverage: 105 %
E-value: 1e-145
NCBI BlastP on this gene
ABI39_03920
ABI39_03915
alpha-N-acetylglucosaminidase
Accession: AND18681
Location: 1090501-1092765
NCBI BlastP on this gene
Accession: AND18681
Location: 1090501-1092765
NCBI BlastP on this gene
ABI39_03910
ABI39_03905
482. : CP043529 Bacteroides vulgatus strain VIC01 chromosome Total score: 1.0 Cumulative Blast bit score: 471
Exo-poly-alpha-D-galacturonosidase
Accession: QEW34965
Location: 521778-523190
NCBI BlastP on this gene
Accession: QEW34965
Location: 521778-523190
NCBI BlastP on this gene
pehX_1
VIC01_00415
Glycerophosphodiester phosphodiesterase
Accession: QEW34967
Location: 525569-526381
NCBI BlastP on this gene
Accession: QEW34967
Location: 525569-526381
NCBI BlastP on this gene
glpQ
susD_2
TonB-dependent receptor SusC
Accession: QEW34969
Location: 528116-531139
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 3e-145
NCBI BlastP on this gene
Accession: QEW34969
Location: 528116-531139
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 3e-145
NCBI BlastP on this gene
susC_12
VIC01_00419
VIC01_00420
todS_4
483. : CP013020 Bacteroides vulgatus strain mpk genome. Total score: 1.0 Cumulative Blast bit score: 471
BvMPK_0613
BvMPK_0612
BvMPK_0611
Glycerophosphoryl diester phosphodiesterase
Accession: ALK83231
Location: 717256-718065
NCBI BlastP on this gene
Accession: ALK83231
Location: 717256-718065
NCBI BlastP on this gene
BvMPK_0610
BvMPK_0609
SusC, outer membrane protein involved in starch binding
Accession: ALK83229
Location: 712498-715521
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 2e-145
NCBI BlastP on this gene
Accession: ALK83229
Location: 712498-715521
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 471
Sequence coverage: 105 %
E-value: 2e-145
NCBI BlastP on this gene
BvMPK_0608
N-acetylglucosamine related transporter, NagX
Accession: ALK83228
Location: 711304-712422
NCBI BlastP on this gene
Accession: ALK83228
Location: 711304-712422
NCBI BlastP on this gene
BvMPK_0607
BvMPK_0606
BvMPK_0605
BvMPK_0604
two-component system sensor histidine
Accession: ALK83224
Location: 703690-707910
NCBI BlastP on this gene
Accession: ALK83224
Location: 703690-707910
NCBI BlastP on this gene
BvMPK_0603
484. : CP048115 Mucilaginibacter sp. 14171R-50 chromosome Total score: 1.0 Cumulative Blast bit score: 469
VCBS repeat-containing protein
Accession: QHS57762
Location: 4475985-4479320
NCBI BlastP on this gene
Accession: QHS57762
Location: 4475985-4479320
NCBI BlastP on this gene
GWR56_20245
VCBS repeat-containing protein
Accession: QHS57763
Location: 4479327-4482623
NCBI BlastP on this gene
Accession: QHS57763
Location: 4479327-4482623
NCBI BlastP on this gene
GWR56_20250
RagB/SusD family nutrient uptake outer membrane protein
Accession: QHS57764
Location: 4482710-4484248
NCBI BlastP on this gene
Accession: QHS57764
Location: 4482710-4484248
NCBI BlastP on this gene
GWR56_20255
SusC/RagA family TonB-linked outer membrane protein
Accession: QHS57765
Location: 4484262-4487297
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 469
Sequence coverage: 107 %
E-value: 2e-144
NCBI BlastP on this gene
Accession: QHS57765
Location: 4484262-4487297
BlastP hit with EDO09628.1
Percentage identity: 34 %
BlastP bit score: 469
Sequence coverage: 107 %
E-value: 2e-144
NCBI BlastP on this gene
GWR56_20260
485. : CP032869 Mucilaginibacter sp. HYN0043 chromosome Total score: 1.0 Cumulative Blast bit score: 468
HYN43_010400
HYN43_010405
RagB/SusD family nutrient uptake outer membrane protein
Accession: AYL95674
Location: 2639607-2641169
NCBI BlastP on this gene
Accession: AYL95674
Location: 2639607-2641169
NCBI BlastP on this gene
HYN43_010410
SusC/RagA family TonB-linked outer membrane protein
Accession: AYL95675
Location: 2641156-2644197
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 468
Sequence coverage: 107 %
E-value: 4e-144
NCBI BlastP on this gene
Accession: AYL95675
Location: 2641156-2644197
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 468
Sequence coverage: 107 %
E-value: 4e-144
NCBI BlastP on this gene
HYN43_010415
486. : CP029255 Flavobacterium crocinum strain HYN0056 chromosome Total score: 1.0 Cumulative Blast bit score: 464
HYN56_21310
HYN56_21315
HYN56_21320
RagB/SusD family nutrient uptake outer membrane protein
Accession: AWK06633
Location: 5005723-5007234
NCBI BlastP on this gene
Accession: AWK06633
Location: 5005723-5007234
NCBI BlastP on this gene
HYN56_21325
SusC/RagA family TonB-linked outer membrane protein
Accession: AWK07537
Location: 5007262-5010243
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 465
Sequence coverage: 105 %
E-value: 4e-143
NCBI BlastP on this gene
Accession: AWK07537
Location: 5007262-5010243
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 465
Sequence coverage: 105 %
E-value: 4e-143
NCBI BlastP on this gene
HYN56_21330
487. : CP030261 Flavobacterium sp. HYN0086 chromosome Total score: 1.0 Cumulative Blast bit score: 464
HYN86_17735
HYN86_17740
glycoside hydrolase family 97 protein
Accession: AXB58339
Location: 4112970-4114988
NCBI BlastP on this gene
Accession: AXB58339
Location: 4112970-4114988
NCBI BlastP on this gene
HYN86_17745
HYN86_17750
RagB/SusD family nutrient uptake outer membrane protein
Accession: AXB58341
Location: 4116458-4117969
NCBI BlastP on this gene
Accession: AXB58341
Location: 4116458-4117969
NCBI BlastP on this gene
HYN86_17755
SusC/RagA family TonB-linked outer membrane protein
Accession: AXB58342
Location: 4117998-4120979
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 105 %
E-value: 6e-143
NCBI BlastP on this gene
Accession: AXB58342
Location: 4117998-4120979
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 105 %
E-value: 6e-143
NCBI BlastP on this gene
HYN86_17760
488. : CP028136 Gramella fulva strain SH35 Total score: 1.0 Cumulative Blast bit score: 464
DUF4861 domain-containing protein
Accession: AVR47419
Location: 2298992-2300293
NCBI BlastP on this gene
Accession: AVR47419
Location: 2298992-2300293
NCBI BlastP on this gene
C7S20_10250
C7S20_10245
C7S20_10240
RagB/SusD family nutrient uptake outer membrane protein
Accession: AVR45611
Location: 2293252-2294820
NCBI BlastP on this gene
Accession: AVR45611
Location: 2293252-2294820
NCBI BlastP on this gene
C7S20_10235
TonB-dependent receptor
Accession: AVR45610
Location: 2290258-2293239
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 104 %
E-value: 9e-143
NCBI BlastP on this gene
Accession: AVR45610
Location: 2290258-2293239
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 464
Sequence coverage: 104 %
E-value: 9e-143
NCBI BlastP on this gene
C7S20_10230
C7S20_10225
SusC/RagA family TonB-linked outer membrane protein
Accession: AVR45608
Location: 2284759-2287923
NCBI BlastP on this gene
Accession: AVR45608
Location: 2284759-2287923
NCBI BlastP on this gene
C7S20_10220
C7S20_10215
489. : CP031965 Aquimarina sp. AD10 chromosome Total score: 1.0 Cumulative Blast bit score: 462
SusC/RagA family TonB-linked outer membrane protein
Accession: AXT62743
Location: 5099255-5102542
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 463
Sequence coverage: 103 %
E-value: 2e-141
NCBI BlastP on this gene
Accession: AXT62743
Location: 5099255-5102542
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 463
Sequence coverage: 103 %
E-value: 2e-141
NCBI BlastP on this gene
D1816_21150
DUF4974 domain-containing protein
Accession: AXT62742
Location: 5097952-5099118
NCBI BlastP on this gene
Accession: AXT62742
Location: 5097952-5099118
NCBI BlastP on this gene
D1816_21145
RNA polymerase sigma-70 factor
Accession: AXT62741
Location: 5097256-5097852
NCBI BlastP on this gene
Accession: AXT62741
Location: 5097256-5097852
NCBI BlastP on this gene
D1816_21140
D1816_21135
DUF2452 domain-containing protein
Accession: AXT62739
Location: 5095059-5095457
NCBI BlastP on this gene
Accession: AXT62739
Location: 5095059-5095457
NCBI BlastP on this gene
D1816_21130
D1816_21125
deoxyribodipyrimidine photolyase
Accession: AXT62737
Location: 5093292-5094782
NCBI BlastP on this gene
Accession: AXT62737
Location: 5093292-5094782
NCBI BlastP on this gene
D1816_21120
cryptochrome/photolyase family protein
Accession: AXT62736
Location: 5091770-5093302
NCBI BlastP on this gene
Accession: AXT62736
Location: 5091770-5093302
NCBI BlastP on this gene
D1816_21115
490. : CP003346 Echinicola vietnamensis DSM 17526 Total score: 1.0 Cumulative Blast bit score: 462
Protein of unknown function (DUF2726)
Accession: AGA76749
Location: 508718-509575
NCBI BlastP on this gene
Accession: AGA76749
Location: 508718-509575
NCBI BlastP on this gene
Echvi_0464
Echvi_0465
Echvi_0466
Echvi_0467
Echvi_0468
TonB-linked outer membrane protein, SusC/RagA family
Accession: AGA76754
Location: 514799-517885
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 463
Sequence coverage: 102 %
E-value: 5e-142
NCBI BlastP on this gene
Accession: AGA76754
Location: 514799-517885
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 463
Sequence coverage: 102 %
E-value: 5e-142
NCBI BlastP on this gene
Echvi_0469
beta-ketoacyl-acyl-carrier-protein synthase II
Accession: AGA76755
Location: 518337-519581
NCBI BlastP on this gene
Accession: AGA76755
Location: 518337-519581
NCBI BlastP on this gene
Echvi_0470
transcriptional regulator, tetR family
Accession: AGA76756
Location: 519578-520138
NCBI BlastP on this gene
Accession: AGA76756
Location: 519578-520138
NCBI BlastP on this gene
Echvi_0471
Echvi_0472
putative oxidoreductase, aryl-alcohol dehydrogenase like protein
Accession: AGA76758
Location: 520723-521754
NCBI BlastP on this gene
Accession: AGA76758
Location: 520723-521754
NCBI BlastP on this gene
Echvi_0473
Echvi_0474
Echvi_0475
Calcineurin-like phosphoesterase
Accession: AGA76761
Location: 522675-523334
NCBI BlastP on this gene
Accession: AGA76761
Location: 522675-523334
NCBI BlastP on this gene
Echvi_0476
Echvi_0477
491. : CP003349 Solitalea canadensis DSM 3403 Total score: 1.0 Cumulative Blast bit score: 459
TonB-linked outer membrane protein, SusC/RagA family
Accession: AFD07962
Location: 3474885-3477860
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 459
Sequence coverage: 100 %
E-value: 8e-141
NCBI BlastP on this gene
Accession: AFD07962
Location: 3474885-3477860
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 459
Sequence coverage: 100 %
E-value: 8e-141
NCBI BlastP on this gene
Solca_2943
Solca_2942
Solca_2941
Solca_2940
Solca_2939
dehydrogenase of unknown specificity, short-chain alcohol dehydrogenase like protein
Accession: AFD07957
Location: 3469445-3470197
NCBI BlastP on this gene
Accession: AFD07957
Location: 3469445-3470197
NCBI BlastP on this gene
Solca_2938
dehydrogenase of unknown specificity, short-chain alcohol dehydrogenase like protein
Accession: AFD07956
Location: 3468652-3469407
NCBI BlastP on this gene
Accession: AFD07956
Location: 3468652-3469407
NCBI BlastP on this gene
Solca_2937
492. : CP014773 Mucilaginibacter sp. PAMC 26640 chromosome Total score: 1.0 Cumulative Blast bit score: 457
A0256_11575
A0256_11570
A0256_11565
hypothetical protein
Accession: AMR32010
Location: 2645392-2648421
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 4e-140
NCBI BlastP on this gene
Accession: AMR32010
Location: 2645392-2648421
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 4e-140
NCBI BlastP on this gene
A0256_11560
A0256_11555
adenine phosphoribosyltransferase
Accession: AMR32008
Location: 2642424-2642951
NCBI BlastP on this gene
Accession: AMR32008
Location: 2642424-2642951
NCBI BlastP on this gene
A0256_11550
A0256_11545
A0256_11540
A0256_11535
493. : CP001807 Rhodothermus marinus DSM 4252 Total score: 1.0 Cumulative Blast bit score: 457
Rmar_1759
Rmar_1760
Rmar_1761
TonB-dependent receptor plug
Accession: ACY48646
Location: 2061694-2064765
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 103 %
E-value: 4e-140
NCBI BlastP on this gene
Accession: ACY48646
Location: 2061694-2064765
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 103 %
E-value: 4e-140
NCBI BlastP on this gene
Rmar_1762
494. : AP019797 Rhodothermus marinus AA3-38 DNA Total score: 1.0 Cumulative Blast bit score: 457
RmaAA338_19320
RmaAA338_19330
RmaAA338_19340
SusC/RagA family TonB-linked outer membrane protein
Accession: BBM73070
Location: 2255167-2258229
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: BBM73070
Location: 2255167-2258229
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
RmaAA338_19350
495. : AP019796 Rhodothermus marinus AA2-13 DNA Total score: 1.0 Cumulative Blast bit score: 457
RmaAA213_19270
RmaAA213_19280
RmaAA213_19290
SusC/RagA family TonB-linked outer membrane protein
Accession: BBM70084
Location: 2257950-2261012
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: BBM70084
Location: 2257950-2261012
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 457
Sequence coverage: 106 %
E-value: 5e-140
NCBI BlastP on this gene
RmaAA213_19300
496. : CP048409 Draconibacterium sp. M1 chromosome Total score: 1.0 Cumulative Blast bit score: 456
TonB-dependent receptor
Accession: QIA07694
Location: 2023131-2026043
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 104 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: QIA07694
Location: 2023131-2026043
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 104 %
E-value: 5e-140
NCBI BlastP on this gene
G0Q07_08125
substrate-binding domain-containing protein
Accession: QIA07693
Location: 2020007-2022661
NCBI BlastP on this gene
Accession: QIA07693
Location: 2020007-2022661
NCBI BlastP on this gene
G0Q07_08120
G0Q07_08115
G0Q07_08110
497. : CP034562 Flammeovirga pectinis strain L12M1 chromosome 1 Total score: 1.0 Cumulative Blast bit score: 456
TonB-dependent receptor
Accession: AZQ62316
Location: 2184855-2187782
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 105 %
E-value: 5e-140
NCBI BlastP on this gene
Accession: AZQ62316
Location: 2184855-2187782
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 105 %
E-value: 5e-140
NCBI BlastP on this gene
EI427_08710
EI427_08705
DUF1349 domain-containing protein
Accession: AZQ62314
Location: 2179522-2180223
NCBI BlastP on this gene
Accession: AZQ62314
Location: 2179522-2180223
NCBI BlastP on this gene
EI427_08700
aldose 1-epimerase family protein
Accession: AZQ62313
Location: 2178480-2179400
NCBI BlastP on this gene
Accession: AZQ62313
Location: 2178480-2179400
NCBI BlastP on this gene
EI427_08695
498. : CP025791 Flavivirga eckloniae strain ECD14 chromosome Total score: 1.0 Cumulative Blast bit score: 456
C1H87_07285
C1H87_07290
C1H87_07295
C1H87_07300
C1H87_07305
C1H87_07310
TonB-dependent receptor
Accession: AUP78528
Location: 1753729-1756725
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 456
Sequence coverage: 101 %
E-value: 1e-139
NCBI BlastP on this gene
Accession: AUP78528
Location: 1753729-1756725
BlastP hit with EDO09628.1
Percentage identity: 33 %
BlastP bit score: 456
Sequence coverage: 101 %
E-value: 1e-139
NCBI BlastP on this gene
C1H87_07315
C1H87_07320
C1H87_07325
C1H87_07330
499. : CP023254 Chitinophaga sp. MD30 chromosome. Total score: 1.0 Cumulative Blast bit score: 456
metal-independent alpha-mannosidase
Accession: ASZ10680
Location: 1676942-1678372
NCBI BlastP on this gene
Accession: ASZ10680
Location: 1676942-1678372
NCBI BlastP on this gene
CK934_06640
CK934_06635
RagB/SusD family nutrient uptake outer membrane protein
Accession: ASZ10678
Location: 1671695-1673245
NCBI BlastP on this gene
Accession: ASZ10678
Location: 1671695-1673245
NCBI BlastP on this gene
CK934_06630
hypothetical protein
Accession: ASZ10677
Location: 1668704-1671673
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 102 %
E-value: 1e-139
NCBI BlastP on this gene
Accession: ASZ10677
Location: 1668704-1671673
BlastP hit with EDO09628.1
Percentage identity: 32 %
BlastP bit score: 456
Sequence coverage: 102 %
E-value: 1e-139
NCBI BlastP on this gene
CK934_06625
CK934_06620
CK934_06615
CK934_06610
tRNA-specific adenosine deaminase
Accession: ASZ10673
Location: 1662805-1663245
NCBI BlastP on this gene
Accession: ASZ10673
Location: 1662805-1663245
NCBI BlastP on this gene
CK934_06605
TIGR00730 family Rossman fold protein
Accession: ASZ10672
Location: 1662196-1662780
NCBI BlastP on this gene
Accession: ASZ10672
Location: 1662196-1662780
NCBI BlastP on this gene
CK934_06600
500. : CP050956 Parabacteroides distasonis strain FDAARGOS_615 chromosome. Total score: 1.0 Cumulative Blast bit score: 454
multidrug DMT transporter permease
Accession: QIX66393
Location: 3696407-3697408
NCBI BlastP on this gene
Accession: QIX66393
Location: 3696407-3697408
NCBI BlastP on this gene
FOB23_15395
FOB23_15390
ADP-ribosylglycohydrolase family protein
Accession: QIX66391
Location: 3693815-3695371
NCBI BlastP on this gene
Accession: QIX66391
Location: 3693815-3695371
NCBI BlastP on this gene
FOB23_15385
rbsK
RagB/SusD family nutrient uptake outer membrane protein
Accession: QIX66389
Location: 3691335-3692852
NCBI BlastP on this gene
Accession: QIX66389
Location: 3691335-3692852
NCBI BlastP on this gene
FOB23_15375
TonB-dependent receptor
Accession: QIX66388
Location: 3688350-3691322
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 454
Sequence coverage: 105 %
E-value: 3e-139
NCBI BlastP on this gene
Accession: QIX66388
Location: 3688350-3691322
BlastP hit with EDO09628.1
Percentage identity: 31 %
BlastP bit score: 454
Sequence coverage: 105 %
E-value: 3e-139
NCBI BlastP on this gene
FOB23_15370
FOB23_15365
helix-turn-helix transcriptional regulator
Accession: QIX66386
Location: 3686168-3686521
NCBI BlastP on this gene
Accession: QIX66386
Location: 3686168-3686521
NCBI BlastP on this gene
FOB23_15360
FOB23_15355
FOB23_15350
FOB23_15345
FOB23_15340

Detecting sequence homology at the gene cluster level with MultiGeneBlast.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.
Marnix H. Medema, Rainer Breitling & Eriko Takano (2013)
Molecular Biology and Evolution , 30: 1218-1223.