You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001981_01250
You are here: Home > Sequence: MGYG000001981_01250
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-617 sp000431275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-617; CAG-617 sp000431275 | |||||||||||
| CAZyme ID | MGYG000001981_01250 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7616; End: 10219 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 544 | 754 | 1.1e-16 | 0.7466216216216216 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 1.34e-11 | 30 | 168 | 1 | 148 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam00704 | Glyco_hydro_18 | 2.66e-09 | 544 | 747 | 73 | 307 | Glycosyl hydrolases family 18. |
| smart00560 | LamGL | 3.15e-08 | 50 | 165 | 4 | 130 | LamG-like jellyroll fold domain. |
| TIGR04183 | Por_Secre_tail | 3.77e-08 | 798 | 865 | 3 | 70 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| smart00636 | Glyco_18 | 3.36e-06 | 550 | 747 | 82 | 334 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61689.1 | 0.0 | 10 | 866 | 11 | 866 |
| QUT78021.1 | 3.41e-265 | 19 | 864 | 15 | 848 |
| BCI61676.1 | 3.34e-135 | 19 | 779 | 21 | 764 |
| AHF11971.1 | 5.28e-132 | 12 | 863 | 14 | 846 |
| QUT75436.1 | 9.10e-108 | 16 | 851 | 18 | 837 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
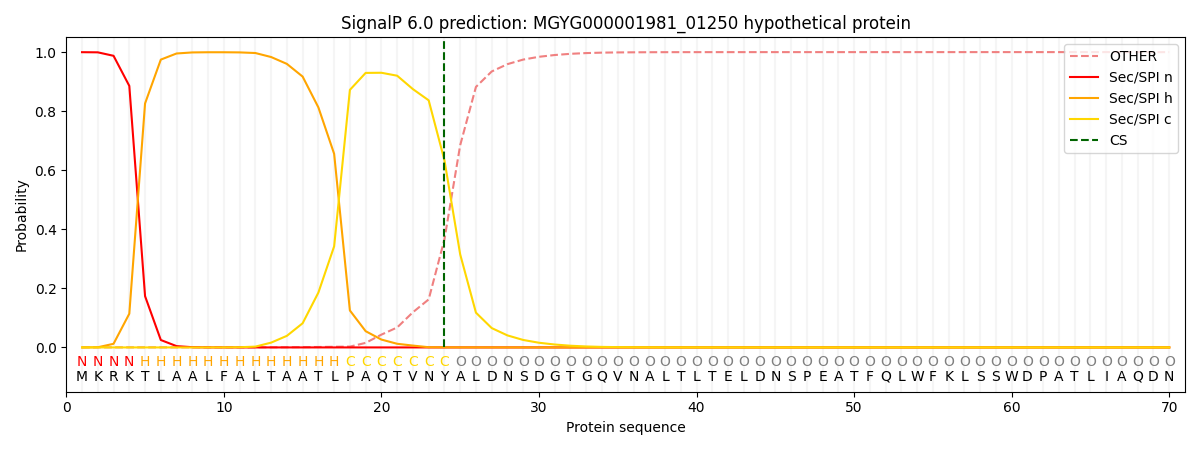
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001296 | 0.997621 | 0.000298 | 0.000284 | 0.000240 | 0.000225 |
