You are browsing environment: HUMAN GUT
MGYG000002414_00590
Basic Information
help
Species
Paenibacillus odorifer
Lineage
Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer
CAZyme ID
MGYG000002414_00590
CAZy Family
GT83
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
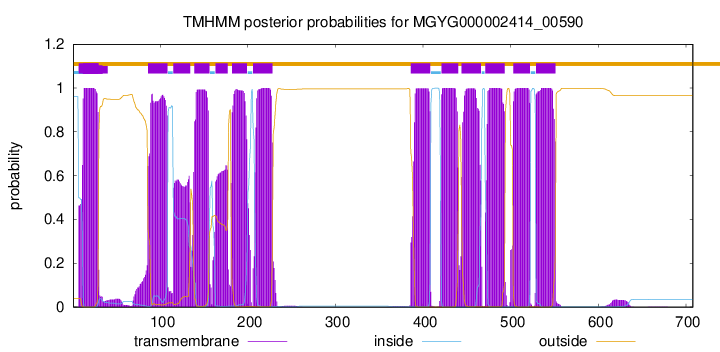
CGC
Molecular Weight
Isoelectric Point
708
74816.63
9.1796
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002414
6802552
Isolate
South Korea
Asia
Gene Location
Start: 658065;
End: 660191
Strand: +
No EC number prediction in MGYG000002414_00590.
Family
Start
End
Evalue
family coverage
GT83
11
261
4.8e-32
0.43703703703703706
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
2.04e-42
20
662
17
512
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
pfam13231
PMT_2
1.79e-29
66
224
1
160
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366.
more
pfam11298
DUF3099
0.001
444
484
20
55
Protein of unknown function (DUF3099). Some members in this family of proteins are annotated as membrane proteins however this cannot be confirmed. Currently no function is known.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O34575
4.08e-217
5
691
4
669
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2
more
P37483
1.75e-194
10
696
7
661
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2
more
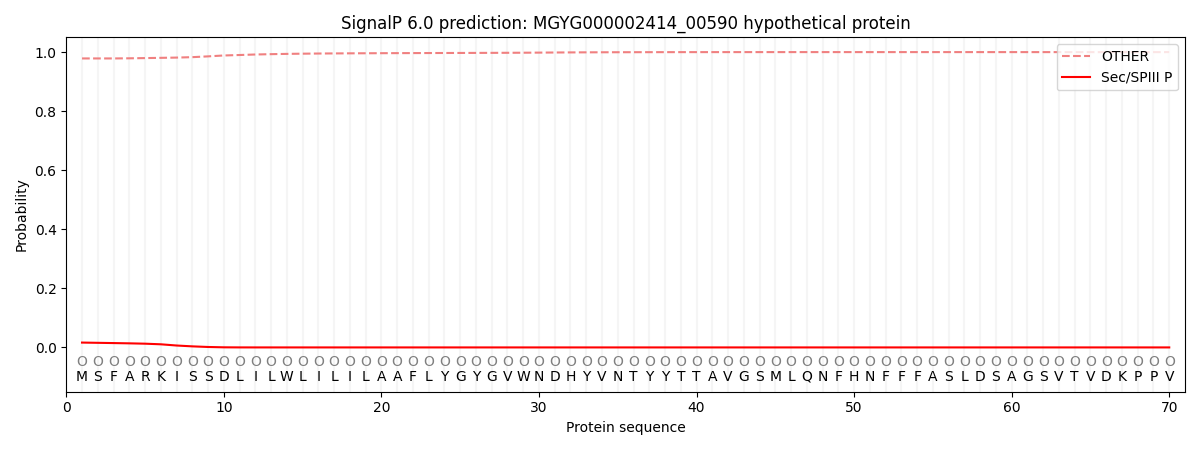
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.978704
0.003852
0.000946
0.000036
0.000019
0.016472
start
end
7
29
86
108
115
134
139
156
163
177
182
199
206
228
386
408
421
440
444
466
471
493
503
522
529
551