You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002533_01515
You are here: Home > Sequence: MGYG000002533_01515
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio vulnificus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio vulnificus | |||||||||||
| CAZyme ID | MGYG000002533_01515 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 148909; End: 152526 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd12204 | CBD_like | 1.60e-16 | 457 | 503 | 1 | 47 | Cellulose-binding domain, chitinase and related proteins. This group contains proteins related to the cellulose-binding domain of Erwinia chrysanthemi endoglucanase Z (EGZ) and Serratia marcescens chitinase B (ChiB). Gram negative plant parasite Erwinia chrysanthemi produces a variety of depolymerizing enzymes to metabolize pectin and cellulose on the host plant. Cellulase EGZ has a modular structure, with N-terminal catalytic domain linked to a C-terminal cellulose-binding domain (CBD). CBD mediates the secretion activity of EGZ. Chitinases allow certain bacteria to utilize chitin as a energy source. Typically, non-plant chitinases are of the glycosidase family 18. |
| pfam14600 | CBM_5_12_2 | 3.10e-15 | 453 | 507 | 1 | 62 | Cellulose-binding domain. This C-terminal domain belongs to the CAZy family of carbohydrate-binding domains that are associated with glycosyl-hydrolases. It is suggested to bind cellulose. |
| pfam17957 | Big_7 | 1.70e-06 | 158 | 222 | 3 | 67 | Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
| pfam17957 | Big_7 | 8.92e-06 | 354 | 423 | 1 | 67 | Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
| pfam17957 | Big_7 | 4.33e-05 | 257 | 324 | 2 | 67 | Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIL73261.1 | 0.0 | 1 | 1205 | 1 | 1205 |
| QBN15830.1 | 0.0 | 1 | 1205 | 1 | 1205 |
| AAO07745.2 | 0.0 | 1 | 1205 | 1 | 1205 |
| AMG10435.1 | 0.0 | 1 | 1205 | 1 | 1205 |
| AUJ36769.1 | 0.0 | 1 | 1205 | 1 | 1205 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1AIW_A | 1.46e-08 | 454 | 507 | 2 | 62 | NMRSTRUCTURES OF THE CELLULOSE-BINDING DOMAIN OF THE ENDOGLUCANASE Z FROM ERWINIA CHRYSANTHEMI, 23 STRUCTURES [Dickeya dadantii 3937] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
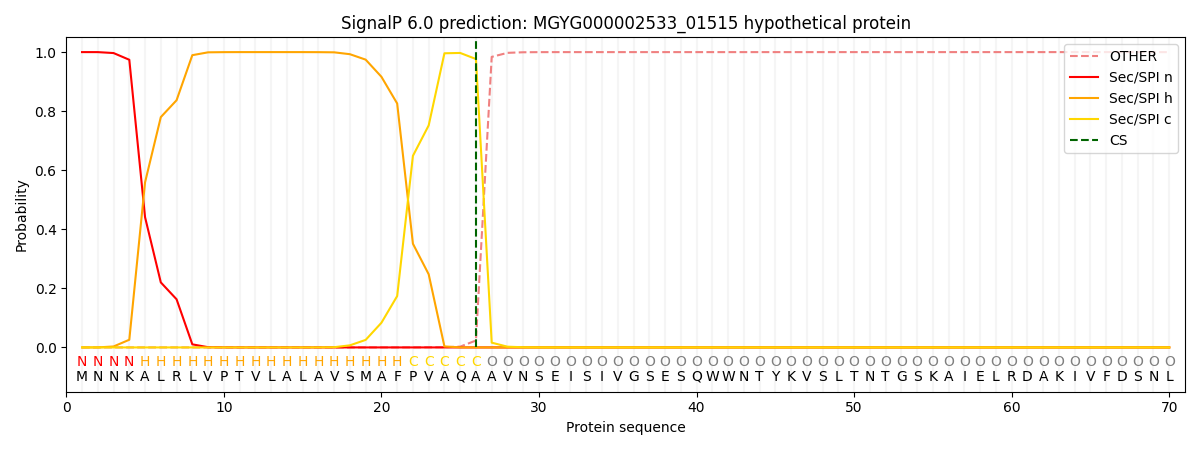
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000351 | 0.998729 | 0.000226 | 0.000230 | 0.000222 | 0.000203 |
