You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002584_00419
You are here: Home > Sequence: MGYG000002584_00419
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp900545155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp900545155 | |||||||||||
| CAZyme ID | MGYG000002584_00419 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13748; End: 16549 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 36 | 529 | 1e-73 | 0.5132978723404256 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 5.43e-20 | 32 | 444 | 5 | 413 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.41e-17 | 31 | 334 | 2 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 4.13e-12 | 107 | 460 | 113 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 7.03e-11 | 209 | 314 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 4.97e-10 | 42 | 183 | 4 | 155 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUI45420.1 | 0.0 | 19 | 933 | 10 | 923 |
| QTO27419.1 | 0.0 | 19 | 933 | 10 | 923 |
| QCQ31726.1 | 0.0 | 19 | 933 | 10 | 923 |
| QRM69893.1 | 0.0 | 19 | 933 | 10 | 923 |
| QCQ49396.1 | 0.0 | 19 | 933 | 10 | 923 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 2.42e-15 | 41 | 469 | 41 | 463 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 4CU6_A | 2.53e-10 | 103 | 437 | 74 | 428 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU7_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU8_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4] |
| 4CUC_A | 2.53e-10 | 103 | 437 | 74 | 428 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA. [Streptococcus pneumoniae TIGR4] |
| 1BHG_A | 5.08e-08 | 36 | 437 | 15 | 431 | HumanBeta-Glucuronidase At 2.6 A Resolution [Homo sapiens],1BHG_B Human Beta-Glucuronidase At 2.6 A Resolution [Homo sapiens],3HN3_A Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_B Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_D Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_E Human beta-glucuronidase at 1.7 A resolution [Homo sapiens] |
| 3DEC_A | 3.20e-07 | 41 | 443 | 46 | 466 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26257 | 2.65e-16 | 37 | 459 | 2 | 411 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| P77989 | 1.10e-10 | 105 | 459 | 58 | 410 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P08236 | 2.88e-07 | 36 | 437 | 35 | 451 | Beta-glucuronidase OS=Homo sapiens OX=9606 GN=GUSB PE=1 SV=2 |
| Q5R5N6 | 8.62e-07 | 36 | 437 | 35 | 451 | Beta-glucuronidase OS=Pongo abelii OX=9601 GN=GUSB PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
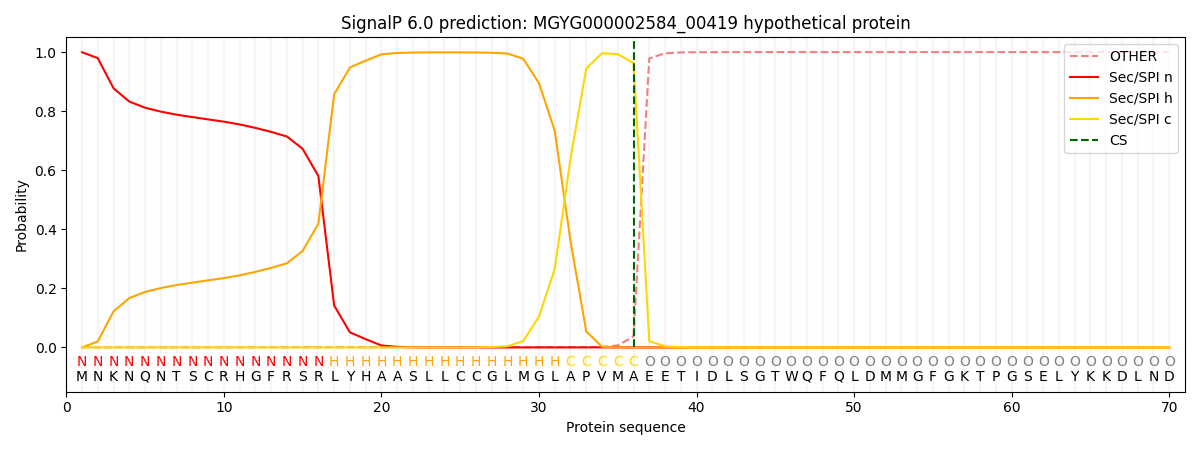
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001316 | 0.997306 | 0.000654 | 0.000265 | 0.000228 | 0.000214 |

