You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003242_00746
You are here: Home > Sequence: MGYG000003242_00746
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia sp000437675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia sp000437675 | |||||||||||
| CAZyme ID | MGYG000003242_00746 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2785; End: 5466 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 39 | 605 | 5e-94 | 0.6090425531914894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 5.02e-54 | 95 | 608 | 82 | 563 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 1.07e-44 | 27 | 461 | 56 | 463 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 2.34e-43 | 42 | 466 | 10 | 428 | beta-D-glucuronidase; Provisional |
| PRK09525 | lacZ | 5.07e-27 | 137 | 441 | 158 | 457 | beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 4.62e-20 | 332 | 601 | 8 | 289 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT85817.1 | 0.0 | 6 | 893 | 5 | 892 |
| QEW35299.1 | 0.0 | 13 | 893 | 21 | 901 |
| QUT56571.1 | 0.0 | 13 | 893 | 21 | 901 |
| QQY42842.1 | 0.0 | 13 | 893 | 12 | 892 |
| ABR38111.1 | 0.0 | 13 | 893 | 21 | 901 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T98_A | 6.19e-62 | 46 | 811 | 29 | 807 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 5UJ6_A | 2.15e-61 | 28 | 460 | 11 | 452 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 6D50_A | 5.82e-61 | 42 | 460 | 33 | 460 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
| 3FN9_A | 3.49e-60 | 42 | 652 | 5 | 616 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6D8G_A | 1.63e-59 | 42 | 460 | 33 | 460 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P77989 | 2.00e-104 | 41 | 822 | 2 | 735 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P26257 | 1.30e-63 | 42 | 781 | 2 | 710 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| A7LXS9 | 9.59e-58 | 40 | 811 | 40 | 835 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| T2KN75 | 7.57e-55 | 37 | 810 | 21 | 781 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| T2KPJ7 | 2.98e-52 | 30 | 652 | 39 | 668 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
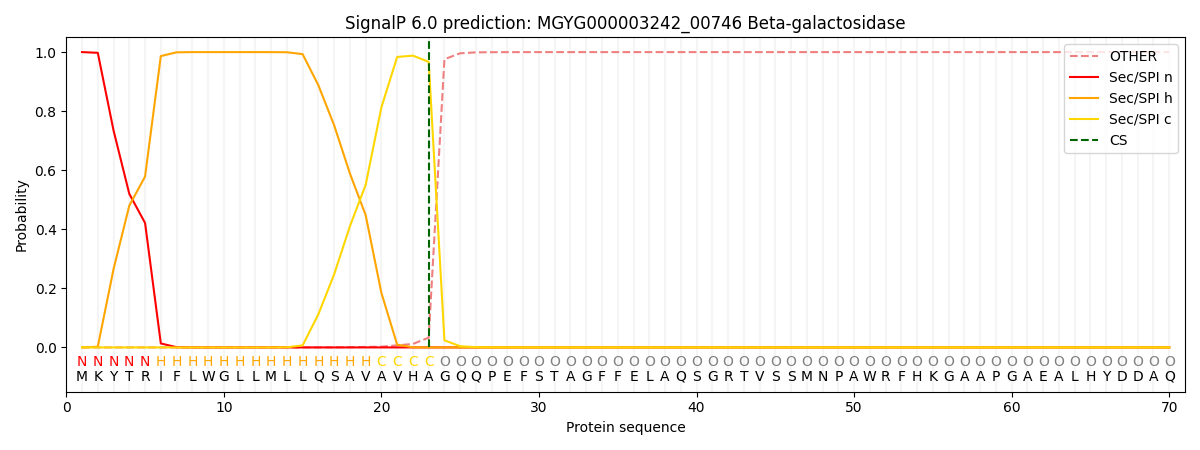
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000295 | 0.998983 | 0.000203 | 0.000173 | 0.000166 | 0.000162 |
