You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003268_00254
You are here: Home > Sequence: MGYG000003268_00254
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900762125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900762125 | |||||||||||
| CAZyme ID | MGYG000003268_00254 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5651; End: 7150 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 3 | 470 | 7.1e-59 | 0.8722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 4.10e-25 | 3 | 419 | 7 | 428 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 7.98e-09 | 1 | 346 | 4 | 340 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKH86812.1 | 1.16e-89 | 5 | 471 | 2 | 467 |
| QTO25703.1 | 3.23e-89 | 5 | 471 | 2 | 467 |
| QLK84536.1 | 3.23e-89 | 5 | 471 | 2 | 467 |
| QRP88934.1 | 5.44e-87 | 5 | 471 | 2 | 467 |
| QUU03965.1 | 5.44e-87 | 5 | 471 | 2 | 467 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2VBI7 | 1.37e-12 | 3 | 315 | 4 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=arnT PE=3 SV=1 |
| Q1C744 | 2.16e-10 | 3 | 313 | 7 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=arnT PE=3 SV=1 |
| B1JJ32 | 2.16e-10 | 3 | 313 | 7 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pseudotuberculosis serotype O:3 (strain YPIII) OX=502800 GN=arnT PE=3 SV=1 |
| B2K5L1 | 2.16e-10 | 3 | 313 | 7 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pseudotuberculosis serotype IB (strain PB1/+) OX=502801 GN=arnT PE=3 SV=1 |
| Q66A06 | 2.16e-10 | 3 | 313 | 7 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pseudotuberculosis serotype I (strain IP32953) OX=273123 GN=arnT PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.954188 | 0.045348 | 0.000176 | 0.000075 | 0.000071 | 0.000154 |
TMHMM Annotations download full data without filtering help
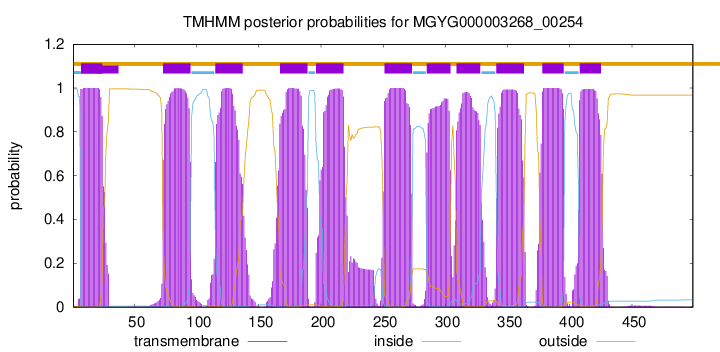
| start | end |
|---|---|
| 7 | 24 |
| 73 | 95 |
| 115 | 137 |
| 167 | 189 |
| 196 | 218 |
| 251 | 273 |
| 285 | 304 |
| 309 | 328 |
| 341 | 363 |
| 378 | 395 |
| 408 | 425 |
