You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003375_00656
You are here: Home > Sequence: MGYG000003375_00656
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium striatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium striatum | |||||||||||
| CAZyme ID | MGYG000003375_00656 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37317; End: 38261 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 74 | 297 | 2e-19 | 0.9030837004405287 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 6.64e-19 | 64 | 314 | 35 | 307 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| TIGR01840 | esterase_phb | 5.02e-06 | 114 | 206 | 34 | 132 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| COG1506 | DAP2 | 5.22e-05 | 78 | 234 | 379 | 570 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG4099 | COG4099 | 1.06e-04 | 74 | 230 | 170 | 330 | Predicted peptidase [General function prediction only]. |
| pfam10503 | Esterase_phd | 0.001 | 78 | 216 | 2 | 139 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSB17085.1 | 5.69e-17 | 63 | 312 | 46 | 286 |
| QLK00730.1 | 6.34e-17 | 63 | 312 | 55 | 297 |
| CAN90905.1 | 3.22e-16 | 63 | 311 | 243 | 468 |
| BAJ10857.1 | 8.92e-16 | 75 | 310 | 39 | 259 |
| AGP34778.1 | 1.08e-15 | 64 | 312 | 246 | 471 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0CDX2 | 1.06e-15 | 77 | 311 | 43 | 267 | Probable feruloyl esterase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=faeC PE=3 SV=1 |
| Q5B2G3 | 6.94e-15 | 77 | 311 | 43 | 267 | Feruloyl esterase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=faeC PE=1 SV=1 |
| A1C9D4 | 3.49e-12 | 77 | 311 | 45 | 269 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
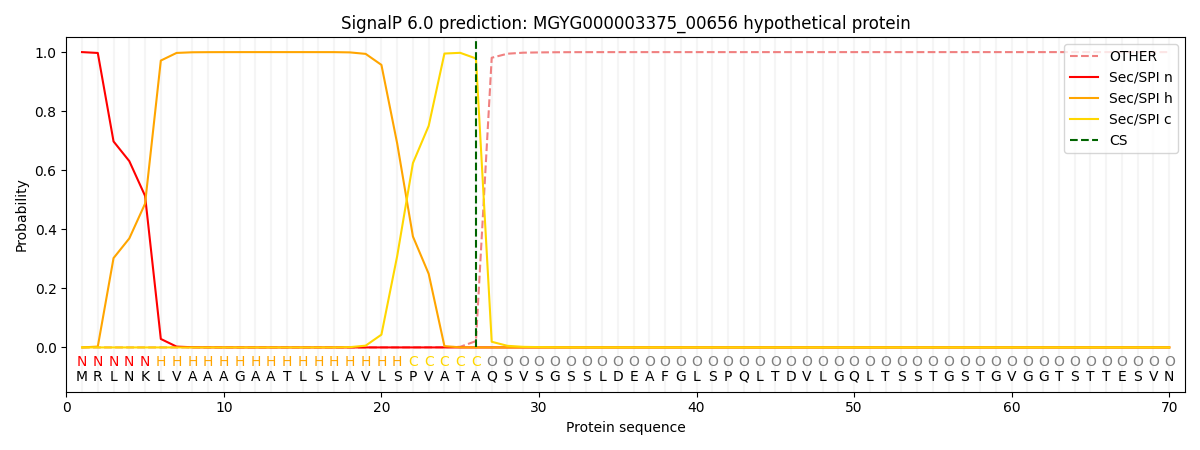
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000221 | 0.999101 | 0.000179 | 0.000170 | 0.000158 | 0.000144 |
