You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003420_01198
You are here: Home > Sequence: MGYG000003420_01198
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA4372 sp900766785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA4372; UBA4372 sp900766785 | |||||||||||
| CAZyme ID | MGYG000003420_01198 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4161; End: 5513 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 63 | 250 | 2.2e-18 | 0.45614035087719296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 4.89e-32 | 62 | 415 | 1 | 337 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 1.06e-08 | 65 | 190 | 1 | 115 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK10206 | PRK10206 | 1.12e-04 | 107 | 221 | 35 | 145 | putative oxidoreductase; Provisional |
| PRK15057 | PRK15057 | 7.77e-04 | 70 | 183 | 3 | 118 | UDP-glucose 6-dehydrogenase; Provisional |
| pfam02737 | 3HCDH_N | 0.001 | 66 | 146 | 1 | 87 | 3-hydroxyacyl-CoA dehydrogenase, NAD binding domain. This family also includes lambda crystallin. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABR45528.1 | 5.50e-227 | 2 | 450 | 3 | 455 |
| BBK93797.1 | 5.50e-227 | 2 | 450 | 3 | 455 |
| QIX65768.1 | 5.50e-227 | 2 | 450 | 3 | 455 |
| QUT96603.1 | 5.50e-227 | 2 | 450 | 3 | 455 |
| AST52349.1 | 5.50e-227 | 2 | 450 | 3 | 455 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Z3B_B | 1.81e-07 | 136 | 339 | 61 | 250 | Lowresolution structure of RgNanOx [[Ruminococcus] gnavus] |
| 6Z3B_A | 1.84e-07 | 136 | 339 | 63 | 252 | Lowresolution structure of RgNanOx [[Ruminococcus] gnavus] |
| 6Z3C_AAA | 1.88e-07 | 136 | 339 | 69 | 258 | ChainAAA, Gfo/Idh/MocA family oxidoreductase [[Ruminococcus] gnavus],6Z3C_BBB Chain BBB, Gfo/Idh/MocA family oxidoreductase [[Ruminococcus] gnavus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9RK81 | 3.35e-09 | 6 | 225 | 7 | 214 | Glycosyl hydrolase family 109 protein OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO0529 PE=3 SV=1 |
| Q01S58 | 2.99e-08 | 65 | 251 | 43 | 229 | Glycosyl hydrolase family 109 protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6590 PE=3 SV=1 |
| A4FK61 | 2.12e-07 | 63 | 219 | 4 | 148 | Inositol 2-dehydrogenase 4 OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=iolG4 PE=3 SV=1 |
| A8H2K3 | 5.20e-07 | 6 | 234 | 3 | 217 | Glycosyl hydrolase family 109 protein OS=Shewanella pealeana (strain ATCC 700345 / ANG-SQ1) OX=398579 GN=Spea_1465 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
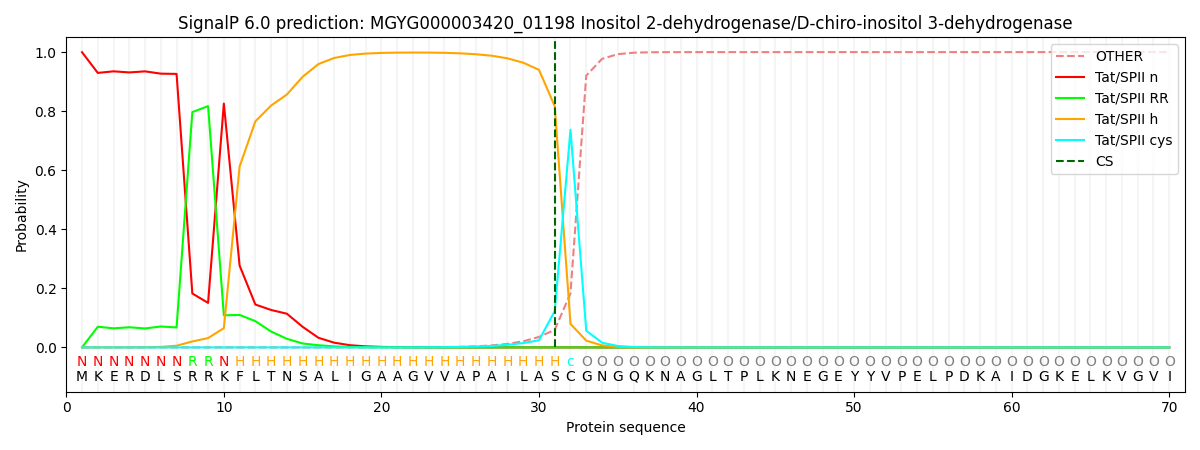
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000001 | 0.000370 | 0.999621 | 0.000000 |
