You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003583_00415
You are here: Home > Sequence: MGYG000003583_00415
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp900770195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp900770195 | |||||||||||
| CAZyme ID | MGYG000003583_00415 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17630; End: 19702 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 39 | 478 | 5.7e-100 | 0.9976076555023924 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 4.52e-94 | 42 | 476 | 1 | 373 | Glycosyl hydrolase family 9. |
| PLN00119 | PLN00119 | 1.26e-41 | 39 | 466 | 31 | 474 | endoglucanase |
| PLN02340 | PLN02340 | 1.61e-41 | 39 | 499 | 30 | 511 | endoglucanase |
| PLN02909 | PLN02909 | 2.61e-40 | 38 | 465 | 33 | 468 | Endoglucanase |
| PLN02420 | PLN02420 | 1.84e-39 | 38 | 474 | 40 | 504 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU22070.1 | 2.81e-127 | 32 | 507 | 36 | 508 |
| CAI94607.1 | 5.87e-90 | 37 | 485 | 44 | 525 |
| ACL75114.1 | 4.08e-87 | 6 | 656 | 4 | 724 |
| AEY66035.1 | 1.11e-86 | 3 | 656 | 2 | 724 |
| BAI52930.1 | 1.57e-85 | 6 | 656 | 4 | 724 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YIK_A | 1.02e-81 | 35 | 495 | 35 | 527 | ChainA, Endoglucanase [Acetivibrio thermocellus] |
| 1IA6_A | 4.16e-70 | 39 | 487 | 5 | 433 | CrystalStructure Of The Cellulase Cel9m Of C. Cellulolyticum [Ruminiclostridium cellulolyticum],1IA7_A Crystal Structure Of The Cellulase Cel9m Of C. Cellulolyticium In Complex With Cellobiose [Ruminiclostridium cellulolyticum] |
| 2XFG_A | 1.95e-54 | 39 | 485 | 25 | 464 | ChainA, ENDOGLUCANASE 1 [Acetivibrio thermocellus] |
| 1G87_A | 6.98e-51 | 39 | 511 | 5 | 464 | TheCrystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1G87_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1GA2_A The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1GA2_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1KFG_A The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum],1KFG_B The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum] |
| 1K72_A | 6.98e-51 | 39 | 511 | 5 | 464 | TheX-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum],1K72_B The X-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02934 | 1.05e-53 | 39 | 511 | 77 | 539 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
| P26224 | 3.34e-51 | 35 | 486 | 27 | 469 | Endoglucanase F OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celF PE=3 SV=1 |
| P37700 | 3.41e-50 | 1 | 511 | 1 | 499 | Endoglucanase G OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCG PE=1 SV=2 |
| P26225 | 5.09e-50 | 1 | 494 | 2 | 488 | Endoglucanase B OS=Cellulomonas fimi OX=1708 GN=cenB PE=3 SV=1 |
| P26221 | 9.98e-49 | 11 | 487 | 26 | 493 | Endoglucanase E-4 OS=Thermobifida fusca OX=2021 GN=celD PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
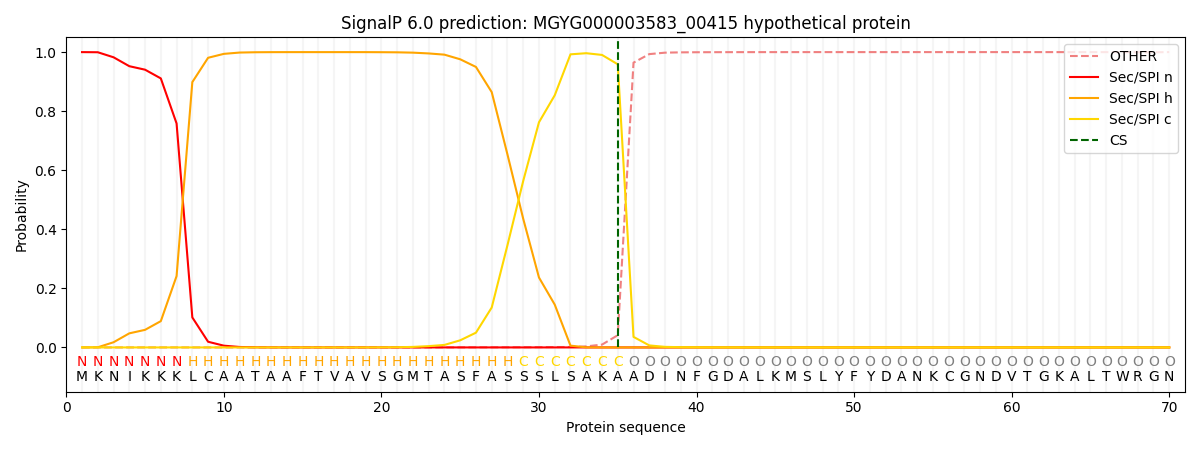
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000691 | 0.998060 | 0.000295 | 0.000382 | 0.000295 | 0.000247 |
